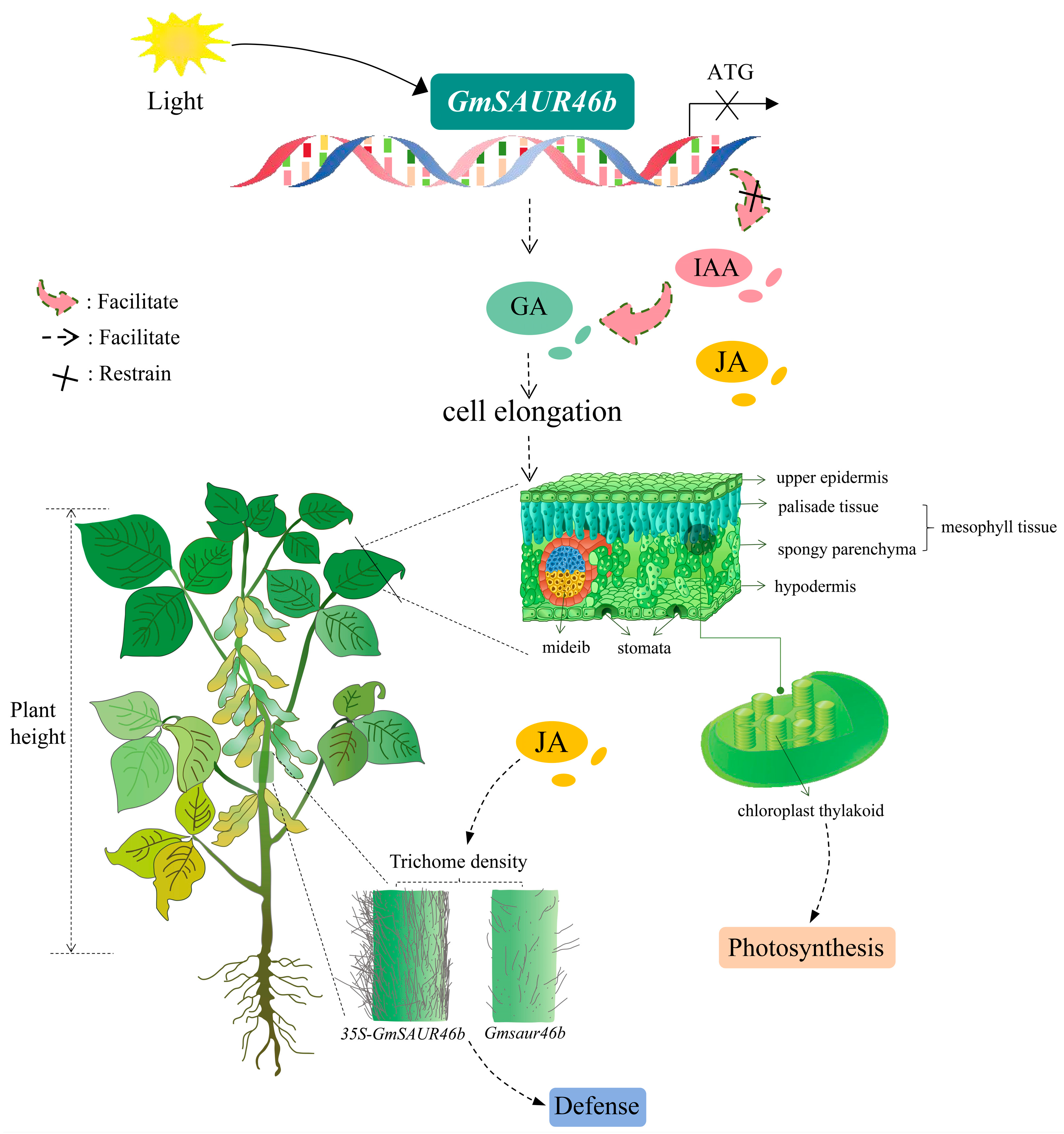
GmSAUR46b Integrates Light Signals to Regulate Leaf Midrib Thickness and Stem Trichome Density in Soybean
Abstract
1. Introduction
2. Results
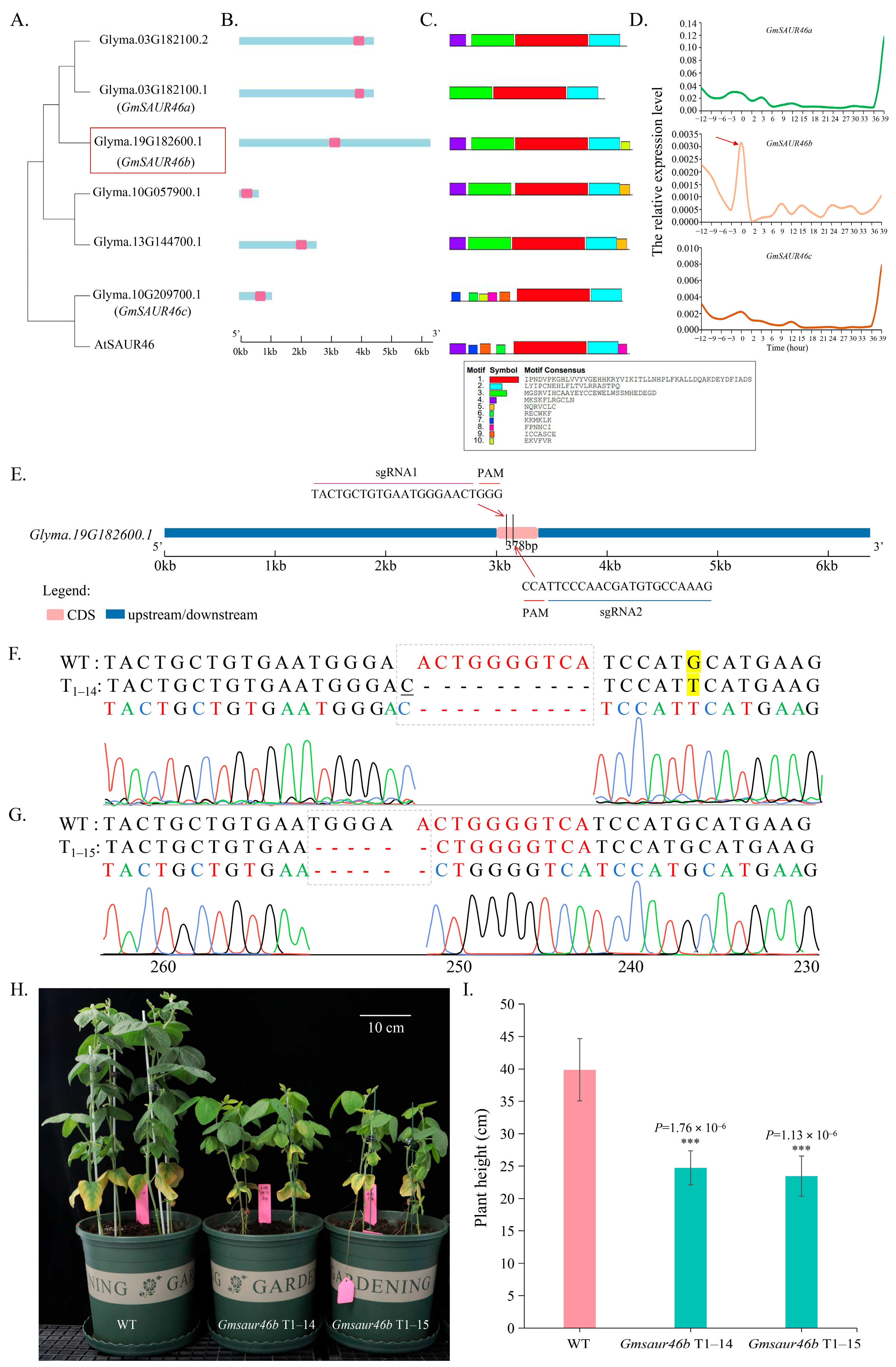
2.1. Identification and Functional Screening of GmSAUR46b as a Candidate Gene Responsive to Light Signaling
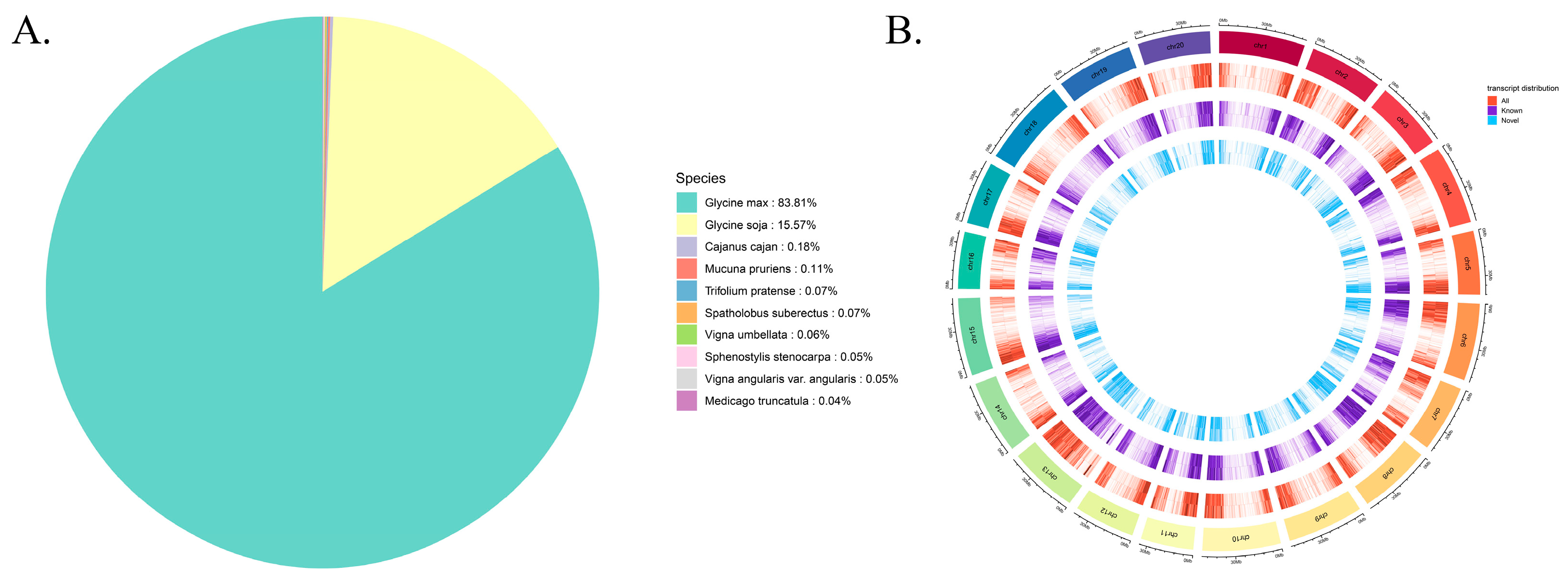
2.2. Identification and Characterization of Novel Transcripts
2.3. Functional Annotation of New Transcripts via GO, KEGG, and KOG Analyses
2.4. Evolutionary and Genomic Distribution Analyses of Transcripts
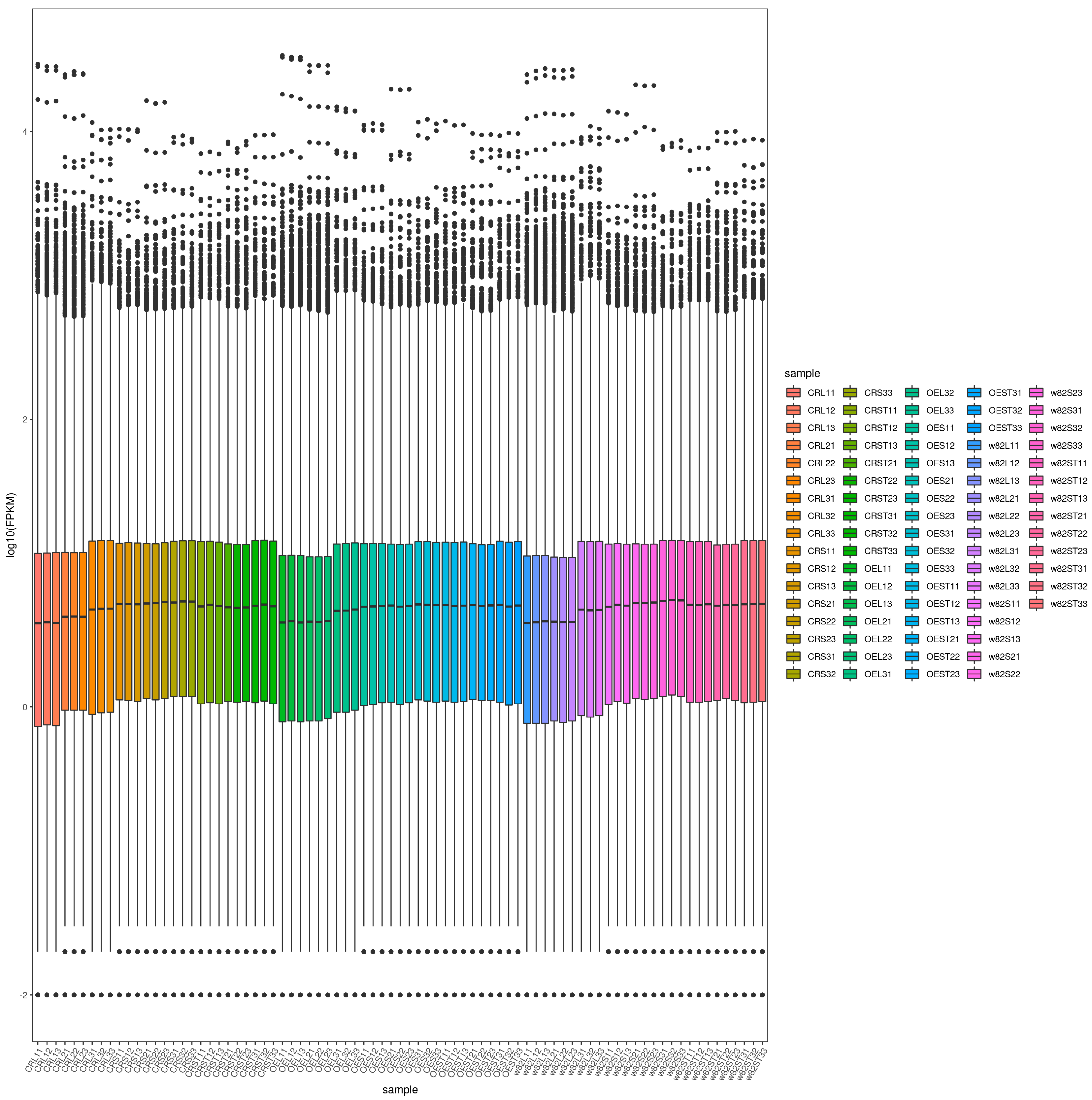
2.5. Distribution of Gene Expression Levels Across Samples
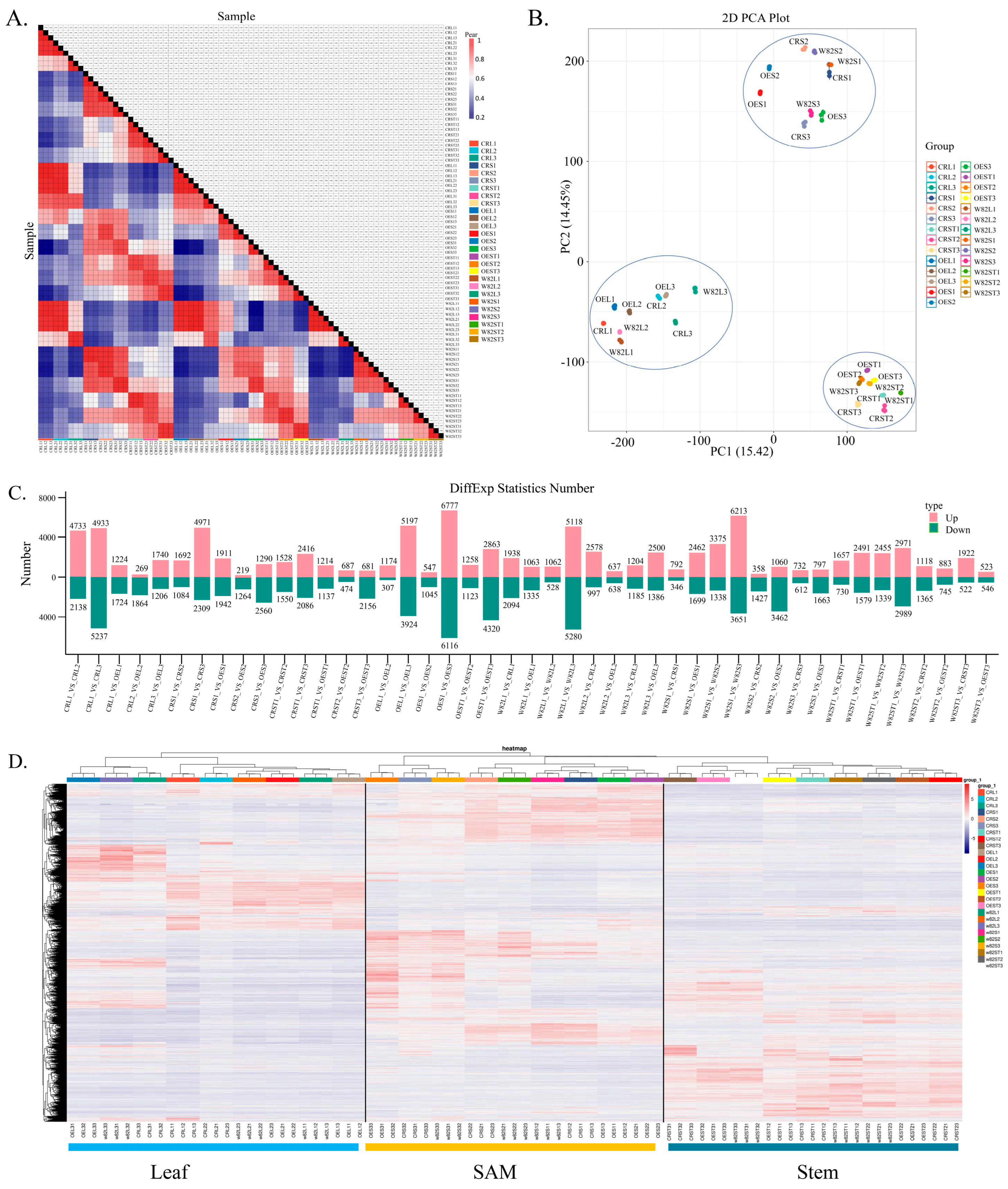
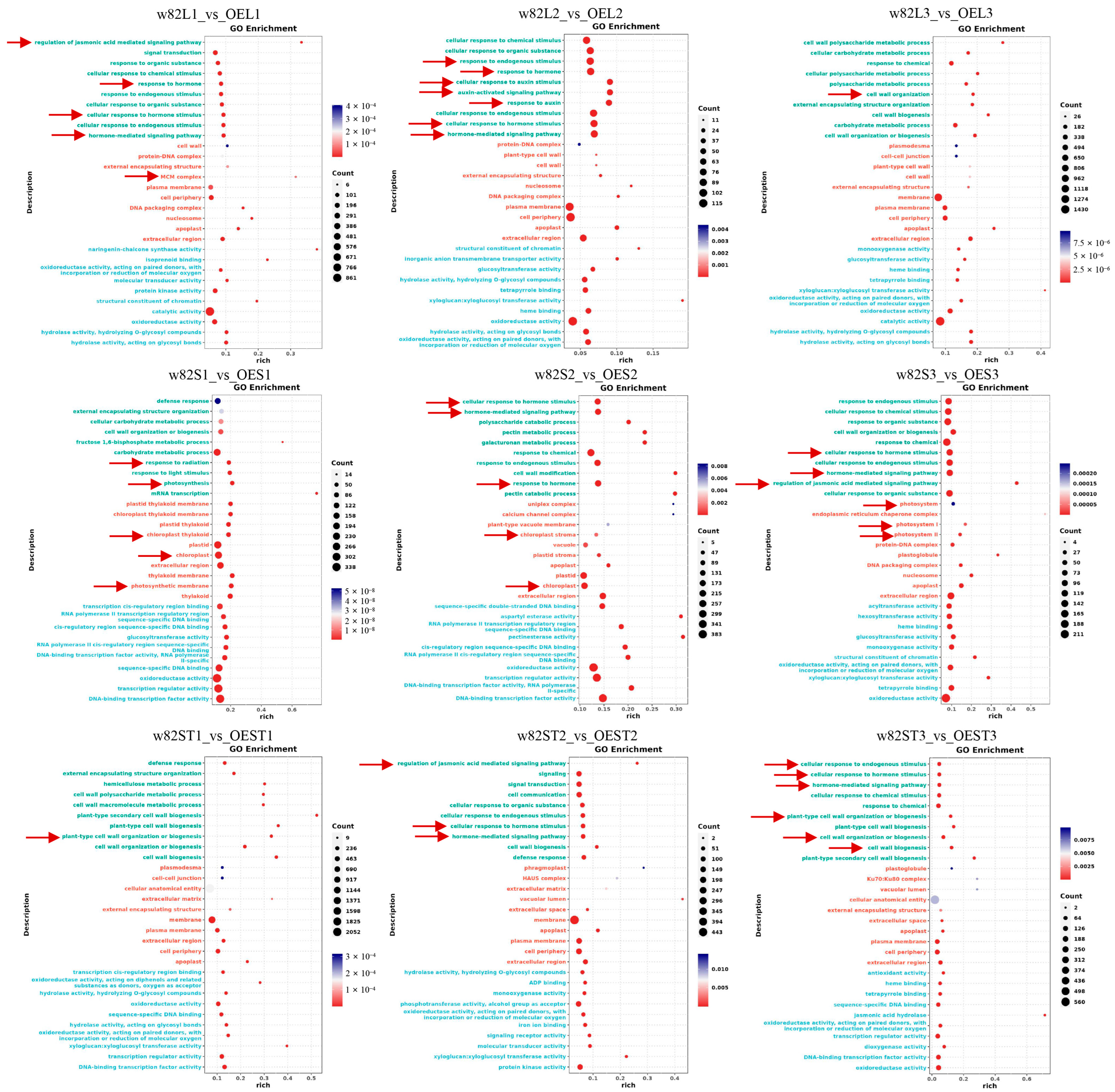
2.6. Transcriptomic Analysis Reveals Enrichment of DEGs in Hormone and Photosynthesis Pathways
2.7. Functional Enrichment Analysis of DEGs
2.8. Analysis of Alternative Splicing Events and Transcription Factor Families
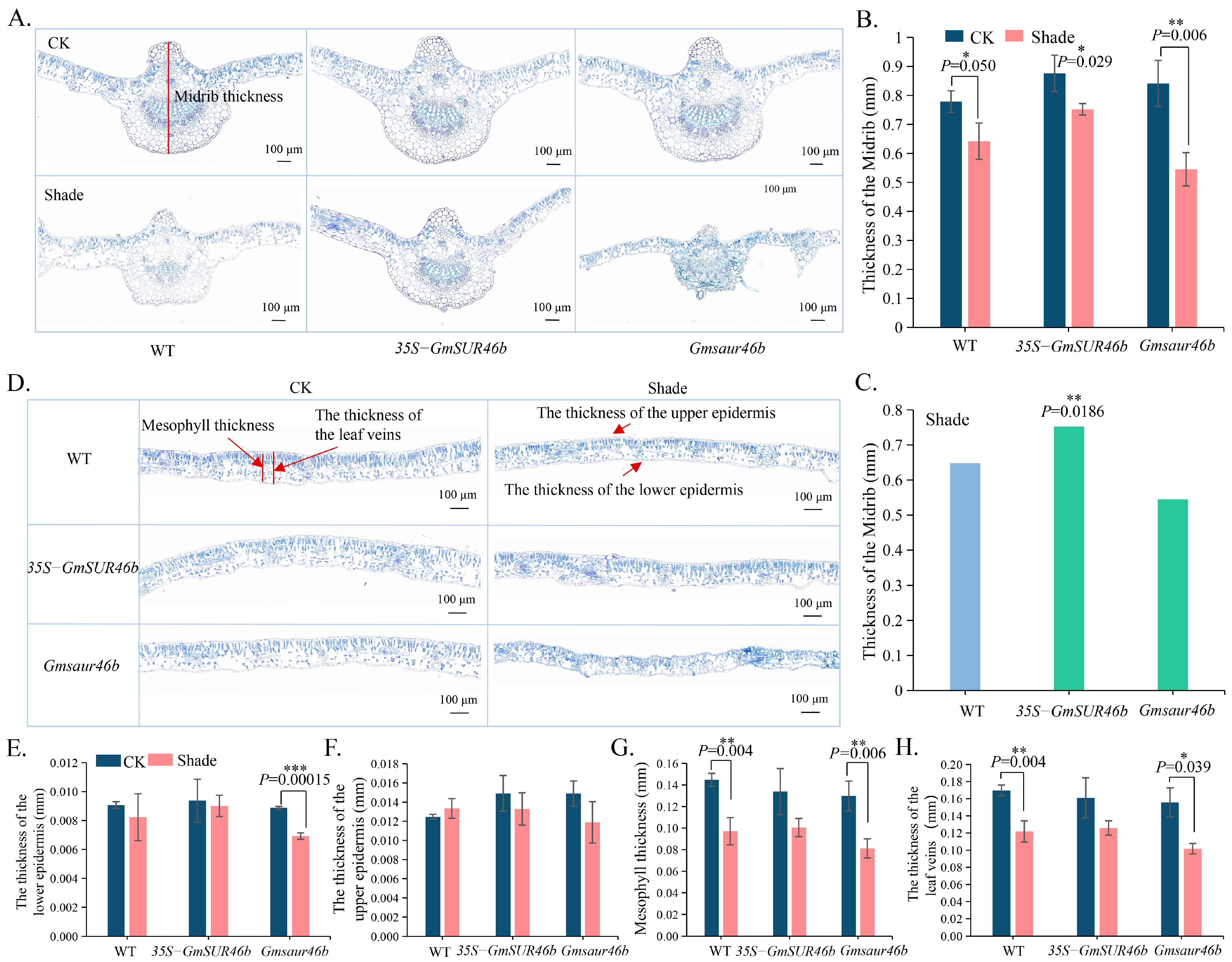
2.9. GmSAUR46b Specifically Regulates the Leaf Midrib Thickness in a Light-Dependent Manner and Modulates the Epidermal/Mesophyll Responses to Light Signals
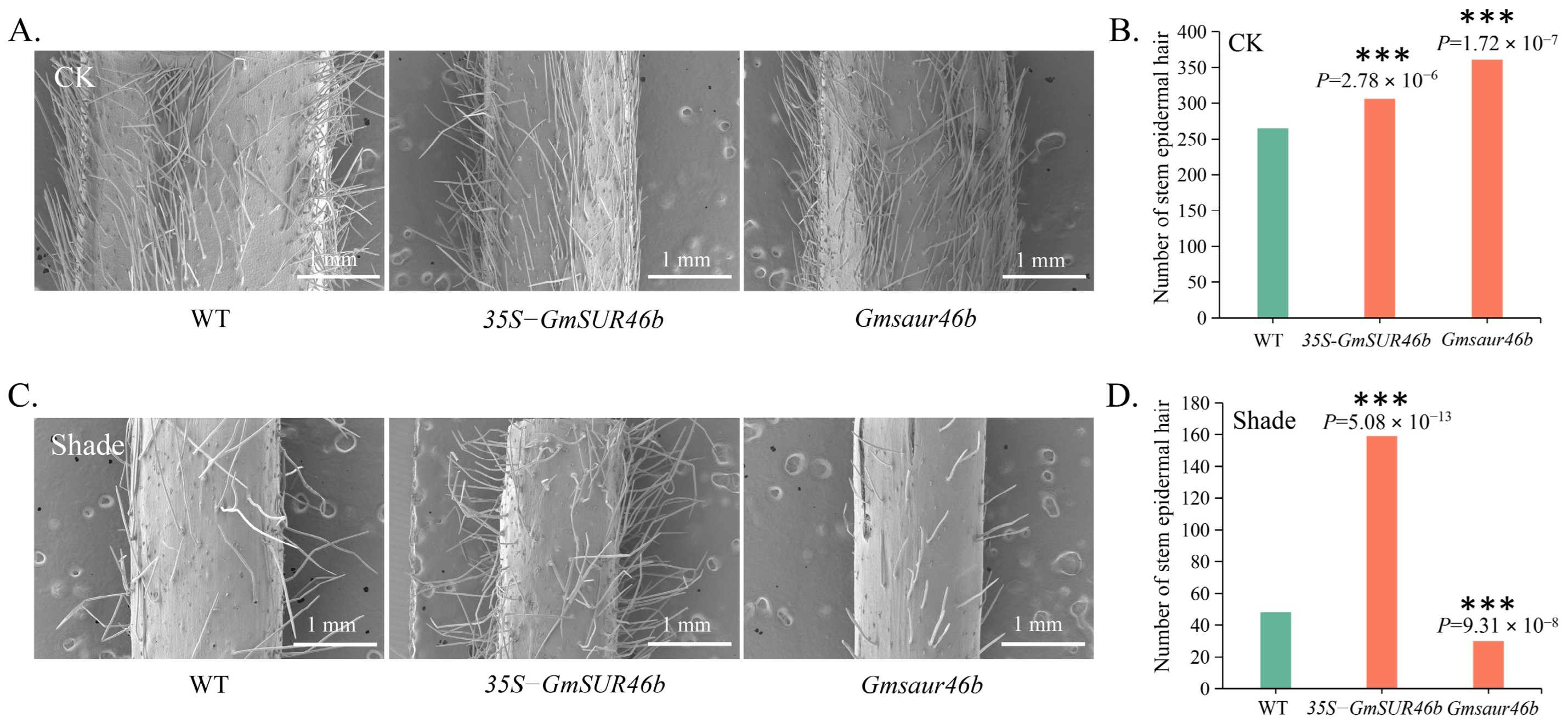
2.10. GmSAUR46b Regulates the Stem Trichome Density in a Light-Dependent Manner
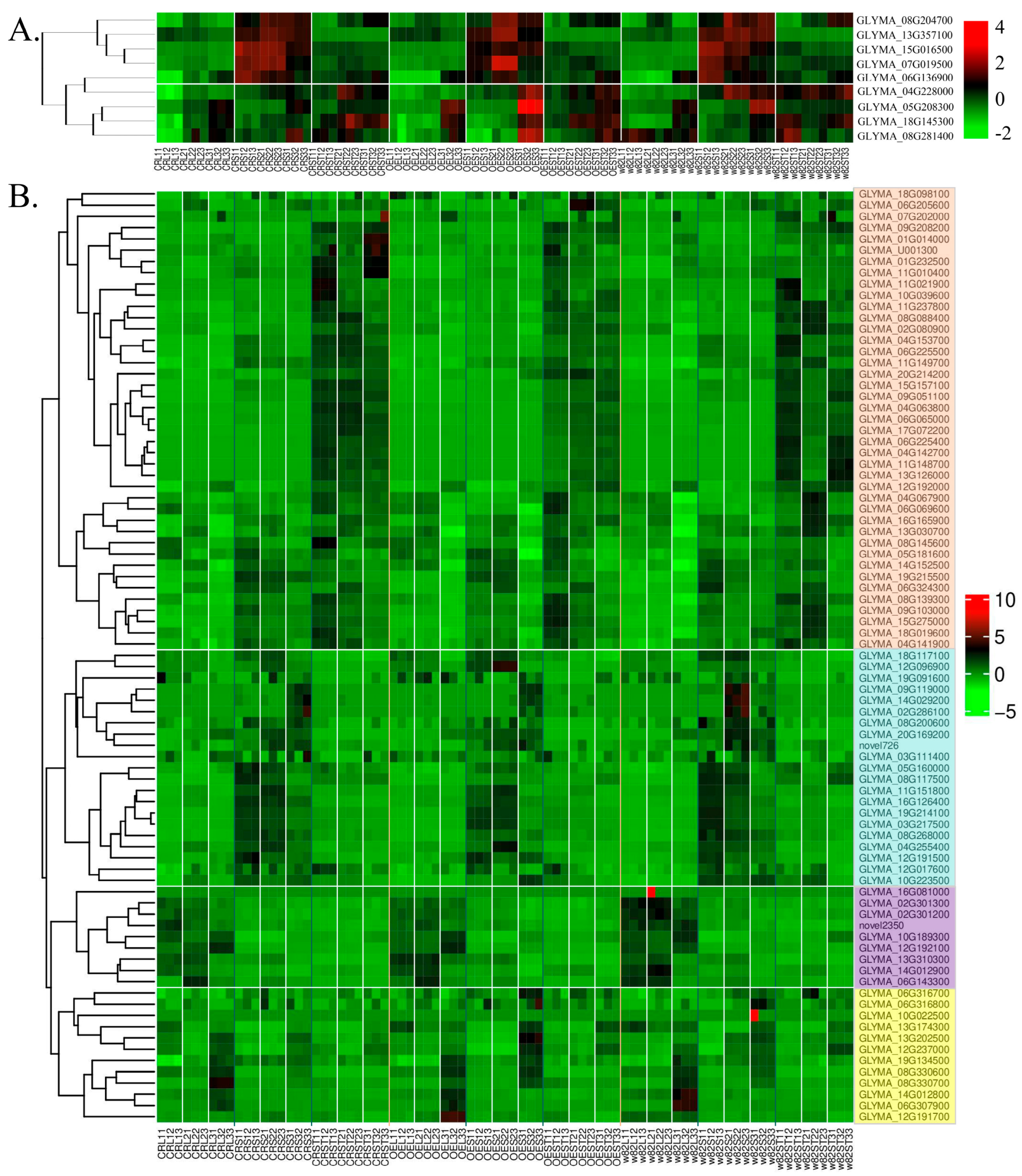
2.11. Differentially Expressed Genes Associated with Observed Morphological Phenotypes
3. Discussion
3.1. The Role of GmSAUR46b in Light-Responsive Soybean Development
3.2. Specific Regulation of Leaf Midrib Thickness by GmSAUR46b
3.3. Light-Dependent Regulation of Stem Trichome Density
3.4. Novel Transcripts and Functional Annotation
4. Materials and Methods
4.1. Construction of Gene Knockout Vector and Soybean Transformation
4.1.1. CRISPR-Cas9 Vector Construction for GmSAUR46b Knockout
4.1.2. Agrobacterium-Mediated Soybean Transformation for Knockout Lines
4.1.3. Construction of Overexpression Vector and Soybean Transformation
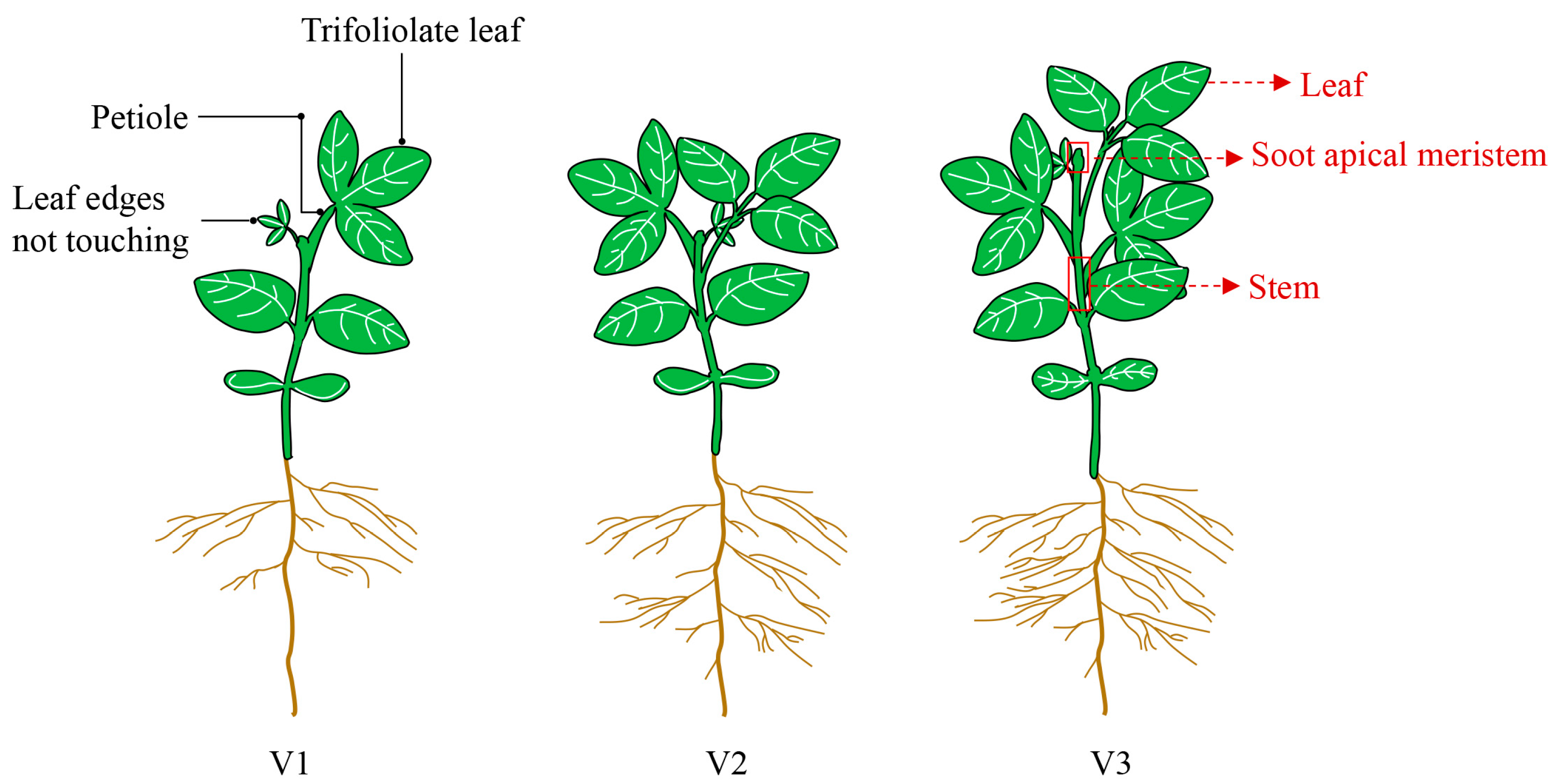
4.2. Plant Materials and Growth Conditions
4.3. Analysis of Gene Evolution, Structure and Conserved Motifs
4.4. Analysis of the Expression Patterns
4.5. RNA-Seq Sample Collection
4.6. Transcriptome Data Analysis
4.7. Analysis of Novel Genes and Novel Transcripts
4.8. Observation and Measurement of Leaf Tissue Thickness
4.9. SEM Preparation and Analysis Procedures
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fan, J.; Shen, Y.; Chen, C.; Chen, X.; Yang, X.; Liu, H.; Chen, R.; Liu, S.; Zhang, B.; Zhang, M.; et al. A large-scale integrated transcriptomic atlas for soybean organ development. Mol. Plant 2025, 18, 669–689. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Yang, S.; Wang, Q.; Yu, H.; Zhao, B.; Wu, T.; Tang, K.; Ma, J.; Yang, X.; Feng, X.; et al. Soybean GmHY2a encodes a phytochromobilin synthase that regulates internode length and flowering time. J. Exp. Bot. 2022, 73, 6646–6662. [Google Scholar] [CrossRef]
- Britz, S.J.; Sager, J.C. Photomorphogenesis and Photoassimilation in Soybean and Sorghum Grown under Broad Spectrum or Blue-Deficient Light Sources. Plant Physiol. 1990, 94, 448–454. [Google Scholar] [CrossRef]
- Cheng, B.; Wang, L.; Liu, R.; Wang, W.; Yu, R.; Zhou, T.; Ahmad, I.; Raza, A.; Jiang, S.; Xu, M.; et al. Shade-Tolerant Soybean Reduces Yield Loss by Regulating Its Canopy Structure and Stem Characteristics in the Maize-Soybean Strip Intercropping System. Front. Plant Sci. 2022, 13, 848893. [Google Scholar] [CrossRef]
- Mu, R.; Lyu, X.; Ji, R.; Liu, J.; Zhao, T.; Li, H.; Liu, B. GmBICs Modulate Low Blue Light-Induced Stem Elongation in Soybean. Front. Plant Sci. 2022, 13, 803122. [Google Scholar] [CrossRef]
- Wu, Y.; Chen, P.; Gong, W.; Gul, H.; Zhu, J.; Yang, F.; Wang, X.; Yong, T.; Liu, J.; Pu, T.; et al. Morphological and physiological variation of soybean seedlings in response to shade. Front. Plant Sci. 2022, 13, 1015414. [Google Scholar] [CrossRef]
- Lyu, X.; Cheng, Q.; Qin, C.; Li, Y.; Xu, X.; Ji, R.; Mu, R.; Li, H.; Zhao, T.; Liu, J.; et al. GmCRY1s modulate gibberellin metabolism to regulate soybean shade avoidance in response to reduced blue light. Mol. Plant 2021, 14, 298–314. [Google Scholar] [CrossRef]
- Sun, J.; Zheng, Y.; Guo, J.; Zhang, Y.; Liu, Y.; Tao, Y.; Wang, M.; Liu, T.; Liu, Y.; Li, X.; et al. GmGAMYB-BINDING PROTEIN 1 promotes small auxin-up RNA gene transcription to modulate soybean maturity and height. Plant Physiol. 2023, 193, 775–791. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Nan, H.; Kong, L.; Yue, L.; Yang, H.; Zhao, Q.; Fang, C.; Li, H.; Cheng, Q.; Lu, S.; et al. Soybean AP1 homologs control flowering time and plant height. J. Integr. Plant Biol. 2020, 62, 1868–1879. [Google Scholar] [CrossRef] [PubMed]
- Meng, Y.; Chen, F.; Shuai, H.; Luo, X.; Ding, J.; Tang, S.; Xu, S.; Liu, J.; Liu, W.; Du, J.; et al. Karrikins delay soybean seed germination by mediating abscisic acid and gibberellin biogenesis under shaded conditions. Sci. Rep. 2016, 6, 22073. [Google Scholar] [CrossRef]
- Yang, F.; Liu, Q.; Cheng, Y.; Feng, L.; Wu, X.; Fan, Y.; Raza, M.A.; Wang, X.; Yong, T.; Liu, W.; et al. Low red/far-red ratio as a signal promotes carbon assimilation of soybean seedlings by increasing the photosynthetic capacity. BMC Plant Biol. 2020, 20, 148. [Google Scholar] [CrossRef] [PubMed]
- Feng, L.; Raza, M.A.; Li, Z.; Chen, Y.; Khalid, M.H.B.; Du, J.; Liu, W.; Wu, X.; Song, C.; Yu, L.; et al. The Influence of Light Intensity and Leaf Movement on Photosynthesis Characteristics and Carbon Balance of Soybean. Front. Plant Sci. 2018, 9, 1952. [Google Scholar] [CrossRef] [PubMed]
- Yao, X.; Zhou, H.; Zhu, Q.; Li, C.; Zhang, H.; Wu, J.J.; Xie, F. Photosynthetic Response of Soybean Leaf to Wide Light-Fluctuation in Maize-Soybean Intercropping System. Front. Plant Sci. 2017, 8, 1695. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Chen, J.; Wang, Z.; Tan, T.; Li, S.; Li, J.; Wang, B.; Zhang, J.; Cheng, Y.; Wu, X.; et al. Soybean (Glycine max L. Merr.) seedlings response to shading: Leaf structure, photosynthesis and proteomic analysis. BMC Plant Biol. 2019, 19, 34. [Google Scholar] [CrossRef]
- Fan, Y.; Chen, J.; Cheng, Y.; Raza, M.A.; Wu, X.; Wang, Z.; Liu, Q.; Wang, R.; Wang, X.; Yong, T.; et al. Effect of shading and light recovery on the growth, leaf structure, and photosynthetic performance of soybean in a maize-soybean relay-strip intercropping system. PLoS ONE 2018, 13, e0198159. [Google Scholar] [CrossRef]
- Gong, W.; Qi, P.; Du, J.; Sun, X.; Wu, X.; Song, C.; Liu, W.; Wu, Y.; Yu, X.; Yong, T.; et al. Transcriptome analysis of shade-induced inhibition on leaf size in relay intercropped soybean. PLoS ONE 2014, 9, e98465. [Google Scholar] [CrossRef]
- Wu, Y.; Gong, W.; Yang, W. Shade Inhibits Leaf Size by Controlling Cell Proliferation and Enlargement in Soybean. Sci. Rep. 2017, 7, 9259. [Google Scholar] [CrossRef]
- Dias, A.P.S.; Li, X.; Harmon, P.F.; Harmon, C.L.; Yang, X.B. Effects of Shade Intensity and Duration on Asian Soybean Rust Caused by Phakopsora pachyrhizi. Plant Dis. 2011, 95, 485–489. [Google Scholar] [CrossRef]
- Li, G.; Wang, Q.; Lu, L.; Wang, S.; Chen, X.; Khan, M.H.U.; Zhang, Y.; Yang, S. Identification of the soybean small auxin upregulated RNA (SAUR) gene family and specific haplotype for drought tolerance. Biologia 2022, 77, 1197–1217. [Google Scholar] [CrossRef]
- Chen, L.; Yang, H.; Fang, Y.; Guo, W.; Chen, H.; Zhang, X.; Dai, W.; Chen, S.; Hao, Q.; Yuan, S.; et al. Overexpression of GmMYB14 improves high-density yield and drought tolerance of soybean through regulating plant architecture mediated by the brassinosteroid pathway. Plant Biotechnol. J. 2021, 19, 702–716. [Google Scholar] [CrossRef]
- Chen, Y.; Hao, X.; Cao, J. Small auxin upregulated RNA (SAUR) gene family in maize: Identification, evolution, and its phylogenetic comparison with Arabidopsis, rice, and sorghum. J. Integr. Plant Biol. 2014, 56, 133–150. [Google Scholar] [CrossRef]
- Zhang, H.; Yu, Z.; Yao, X.; Chen, J.; Chen, X.; Zhou, H.; Lou, Y.; Ming, F.; Jin, Y. Genome-wide identification and characterization of small auxin-up RNA (SAUR) gene family in plants: Evolution and expression profiles during normal growth and stress response. BMC Plant Biol. 2021, 21, 4. [Google Scholar] [CrossRef] [PubMed]
- Guo, G.; Yu, T.; Zhang, H.; Chen, M.; Dong, W.; Zhang, S.; Tang, X.; Liu, L.; Heng, W.; Zhu, L.; et al. Evidence That PbrSAUR72 Contributes to Iron Deficiency Tolerance in Pears by Facilitating Iron Absorption. Plants 2023, 12, 2173. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Lu, Z.; Zhai, L.; Li, N.; Yan, H. The Small Auxin-Up RNA SAUR10 Is Involved in the Promotion of Seedling Growth in Rice. Plants 2023, 12, 3880. [Google Scholar] [CrossRef]
- Qiu, T.; Qi, M.; Ding, X.; Zheng, Y.; Zhou, T.; Chen, Y.; Han, N.; Zhu, M.; Bian, H.; Wang, J. The SAUR41 subfamily of SMALL AUXIN UP RNA genes is abscisic acid inducible to modulate cell expansion and salt tolerance in Arabidopsis thaliana seedlings. Ann. Bot. 2020, 125, 805–819. [Google Scholar] [CrossRef]
- Stortenbeker, N.; Bemer, M. The SAUR gene family: The plant’s toolbox for adaptation of growth and development. J. Exp. Bot. 2019, 70, 17–27. [Google Scholar] [CrossRef]
- Muday, G.K.; Ren, H.; Park, M.Y.; Spartz, A.K.; Wong, J.H.; Gray, W.M. A subset of plasma membrane-localized PP2C.D phosphatases negatively regulate SAUR-mediated cell expansion in Arabidopsis. PLoS Genet. 2018, 14, e1007455. [Google Scholar]
- Spartz, A.K.; Ren, H.; Park, M.Y.; Grandt, K.N.; Lee, S.H.; Murphy, A.S.; Sussman, M.R.; Overvoorde, P.J.; Gray, W.M. SAUR Inhibition of PP2C-D Phosphatases Activates Plasma Membrane H+-ATPases to Promote Cell Expansion in Arabidopsis. Plant Cell 2014, 26, 2129–2142. [Google Scholar] [CrossRef] [PubMed]
- Wong, J.H.; Spartz, A.K.; Park, M.Y.; Du, M.; Gray, W.M. Mutation of a Conserved Motif of PP2C.D Phosphatases Confers SAUR Immunity and Constitutive Activity. Plant Physiol. 2019, 181, 353–366. [Google Scholar] [CrossRef]
- Du, M.; Spalding, E.P.; Gray, W.M. Rapid Auxin-Mediated Cell Expansion. Annu. Rev. Plant Biol. 2020, 71, 379–402. [Google Scholar] [CrossRef]
- Hur, Y.-S.; Oh, J.; Kim, N.; Kim, S.; Son, O.; Kim, J.; Um, J.-H.; Ji, Z.; Kim, M.-h.; Ko, J.-H.; et al. Arabidopsis transcription factor TCP13 promotes shade avoidance syndrome-like responses by directly targeting a subset of shade-responsive gene promoters. J. Exp. Bot. 2024, 75, 241–257. [Google Scholar] [CrossRef]
- Nagpal, P.; Reeves, P.H.; Wong, J.H.; Armengot, L.; Chae, K.; Rieveschl, N.B.; Trinidad, B.; Davidsdottir, V.; Jain, P.; Gray, W.M.; et al. SAUR63 stimulates cell growth at the plasma membrane. PLoS Genet. 2022, 18, e1010375. [Google Scholar] [CrossRef] [PubMed]
- Rovira, A.; Sentandreu, M.; Nagatani, A.; Leivar, P.; Monte, E. The Sequential Action of MIDA9/PP2C.D1, PP2C.D2, and PP2C.D5 Is Necessary to Form and Maintain the Hook After Germination in the Dark. Front. Plant Sci. 2021, 12, 636098. [Google Scholar] [CrossRef] [PubMed]
- Minami, A.; Takahashi, K.; Inoue, S.-i.; Tada, Y.; Kinoshita, T. Brassinosteroid Induces Phosphorylation of the Plasma Membrane H+-ATPase during Hypocotyl Elongation in Arabidopsis thaliana. Plant Cell Physiol. 2019, 60, 935–944. [Google Scholar] [CrossRef]
- Spartz, A.K.; Lor, V.S.; Ren, H.; Olszewski, N.E.; Miller, N.D.; Wu, G.; Spalding, E.P.; Gray, W.M. Constitutive Expression of Arabidopsis SMALL AUXIN UP RNA19 (SAUR19) in Tomato Confers Auxin-Independent Hypocotyl Elongation. Plant Physiol. 2017, 173, 1453–1462. [Google Scholar] [CrossRef]
- Kong, Y.; Zhu, Y.; Gao, C.; She, W.; Lin, W.; Chen, Y.; Han, N.; Bian, H.; Zhu, M.; Wang, J. Tissue-Specific Expression of SMALL AUXIN UP RNA41 Differentially Regulates Cell Expansion and Root Meristem Patterning in Arabidopsis. Plant Cell Physiol. 2013, 54, 609–621. [Google Scholar] [CrossRef]
- Hou, K.; Wu, W.; Gan, S.-S. SAUR36, a SMALL AUXIN UP RNA Gene, Is Involved in the Promotion of Leaf Senescence in Arabidopsis. Plant Physiol. 2013, 161, 1002–1009. [Google Scholar] [CrossRef]
- Xu, X.; Feng, G.; Li, P.; Yu, S.; Hao, F.; Nie, G.; Huang, L.; Zhang, X. Genome-wide association analysis reveals the function of DgSAUR71 in plant height improvement. BMC Plant Biol. 2025, 25, 240. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Liu, Y.; He, Y.; Yan, Y.; Yu, X.; Ali, M.; Pan, C.; Lu, G.; Murphy, A. Cytokinin-inducible response regulator SlRR6 controls plant height through gibberellin and auxin pathways in tomato. J. Exp. Bot. 2023, 74, 4471–4488. [Google Scholar] [CrossRef]
- Liu, R.; Wen, S.-S.; Sun, T.-T.; Wang, R.; Zuo, W.-T.; Yang, T.; Wang, C.; Hu, J.-J.; Lu, M.-Z.; Wang, L.-Q.; et al. PagWOX11/12a positively regulates the PagSAUR36 gene that enhances adventitious root development in poplar. J. Exp. Bot. 2022, 73, 7298–7311. [Google Scholar] [CrossRef]
- Li, M.; Liu, C.; Hepworth, S.R.; Ma, C.; Li, H.; Li, J.; Wang, S.-M.; Yin, H. SAUR15 interaction with BRI1 activates plasma membrane H+-ATPase to promote organ development of Arabidopsis. Plant Physiol. 2022, 189, 2454–2466. [Google Scholar] [CrossRef]
- Wen, Z.; Mei, Y.; Zhou, J.; Cui, Y.; Wang, D.; Wang, N.N. SAUR49 Can Positively Regulate Leaf Senescence by Suppressing SSPP in Arabidopsis. Plant Cell Physiol. 2020, 61, 644–658. [Google Scholar] [CrossRef]
- Gastaldi, V.; Lucero, L.E.; Ferrero, L.V.; Ariel, F.D.; Gonzalez, D.H. Class-I TCP Transcription Factors Activate the SAUR63 Gene Subfamily in Gibberellin-Dependent Stamen Filament Elongation. Plant Physiol. 2020, 182, 2096–2110. [Google Scholar] [CrossRef]
- Kathare, P.K.; Dharmasiri, S.; Dharmasiri, N. SAUR53 regulates organ elongation and apical hook development in Arabidopsis. Plant Signal. Behav. 2018, 13, e1514896. [Google Scholar] [CrossRef]
- He, S.-L.; Hsieh, H.-L.; Jauh, G.-Y. SMALL AUXIN UP RNA62/75 Are Required for the Translation of Transcripts Essential for Pollen Tube Growth. Plant Physiol. 2018, 178, 626–640. [Google Scholar] [CrossRef] [PubMed]
- Li, X.-M.; Zhai, H.-H.; An, X.-H.; Zhang, H.; Zhang, X.; Wang, P.; Chen, H.; Tian, Y. PpSAUR5 promotes plant growth by regulating lignin and hormone pathways. Front. Plant Sci. 2024, 15, 1291693. [Google Scholar] [CrossRef]
- Yin, C.; Sun, A.; Zhou, Y.; Liu, K.; Wang, P.; Ye, W.; Fang, Y.; Gibbs, D. The dynamics of H2A.Z on SMALL AUXIN UP RNAs regulate abscisic acid–auxin signaling crosstalk in Arabidopsis. J. Exp. Bot. 2023, 74, 4158–4168. [Google Scholar] [CrossRef] [PubMed]
- Bianchimano, L.; De Luca, M.B.; Borniego, M.B.; Iglesias, M.J.; Casal, J.J. Temperature regulation of auxin-related gene expression and its implications for plant growth. J. Exp. Bot. 2023, 74, 7015–7033. [Google Scholar] [CrossRef]
- Ferrero, L.V.; Gastaldi, V.; Ariel, F.D.; Viola, I.L.; Gonzalez, D.H. Class I TCP proteins TCP14 and TCP15 are required for elongation and gene expression responses to auxin. Plant Mol. Biol. 2021, 105, 147–159. [Google Scholar] [CrossRef]
- Yang, H.; Klopotek, Y.; Hajirezaei, M.R.; Zerche, S.; Franken, P.; Druege, U. Role of auxin homeostasis and response in nitrogen limitation and dark stimulation of adventitious root formation in petunia cuttings. Ann. Bot. 2019, 124, 1053–1066. [Google Scholar] [CrossRef] [PubMed]
- Sun, N.; Wang, J.; Gao, Z.; Dong, J.; He, H.; Terzaghi, W.; Wei, N.; Deng, X.W.; Chen, H. Arabidopsis SAURs are critical for differential light regulation of the development of various organs. Proc. Natl. Acad. Sci. USA 2016, 113, 6071–6076. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Li, Q.; Jiang, M.; Tian, H.; Khalid, M.H.B.; Wang, Y.; Yu, H. The Small Auxin-Up RNA 50 (SAUR50) Gene from Ammopiptanthus nanus Negatively Regulates Drought Tolerance. Plants 2024, 13, 2512. [Google Scholar] [CrossRef]
- Xu, K.; Lou, Q.; Wang, D.; Li, T.; Chen, S.; Li, T.; Luo, L.; Chen, L. Overexpression of a novel small auxin-up RNA gene, OsSAUR11, enhances rice deep rootedness. BMC Plant Biol. 2023, 23, 319. [Google Scholar] [CrossRef]
- Ma, S.; Hu, H.; Zhang, H.; Ma, F.; Gao, Z.; Li, X. Physiological response and transcriptome analyses of leguminous Indigofera bungeana Walp. to drought stress. PeerJ 2023, 11, e15440. [Google Scholar] [CrossRef]
- Chen, F.; Wang, R.-J.; Wu, C.-J.; Lin, M.; Yan, H.-W.; Xiang, Y. SAUR8, a small auxin-up RNA gene in poplar, confers drought tolerance to transgenic Arabidopsis plants. Gene 2022, 837, 146692. [Google Scholar] [CrossRef]
- Wong, J.H.; Klejchová, M.; Snipes, S.A.; Nagpal, P.; Bak, G.; Wang, B.; Dunlap, S.; Park, M.Y.; Kunkel, E.N.; Trinidad, B.; et al. SAUR proteins and PP2C.D phosphatases regulate H+-ATPases and K+ channels to control stomatal movements. Plant Physiol. 2021, 185, 256–273. [Google Scholar] [CrossRef]
- Li, M.; Chen, R.; Gu, H.; Cheng, D.; Guo, X.; Shi, C.; Li, L.; Xu, G.; Gu, S.; Wu, Z.; et al. Grape Small Auxin Upregulated RNA (SAUR) 041 Is a Candidate Regulator of Berry Size in Grape. Int. J. Mol. Sci. 2021, 22, 11818. [Google Scholar] [CrossRef]
- Ren, H.; Gray, W.M. SAUR Proteins as Effectors of Hormonal and Environmental Signals in Plant Growth. Mol. Plant 2015, 8, 1153–1164. [Google Scholar] [CrossRef] [PubMed]
- van Mourik, H.; van Dijk, A.D.J.; Stortenbeker, N.; Angenent, G.C.; Bemer, M. Divergent regulation of Arabidopsis SAUR genes: A focus on the SAUR10-clade. BMC Plant Biol. 2017, 17, 245. [Google Scholar] [CrossRef]
- Bai, M.-Y.; Shang, J.-X.; Oh, E.; Fan, M.; Bai, Y.; Zentella, R.; Sun, T.-P.; Wang, Z.-Y. Brassinosteroid, gibberellin and phytochrome impinge on a common transcription module in Arabidopsis. Nat. Cell Biol. 2012, 14, 810–817. [Google Scholar] [CrossRef] [PubMed]
- Stamm, P.; Kumar, P.P. Auxin and gibberellin responsive Arabidopsis SMALL AUXIN UP RNA36 regulates hypocotyl elongation in the light. Plant Cell Rep. 2013, 32, 759–769. [Google Scholar] [CrossRef]
- Lin, Z.; Zhu, P.; Gao, L.; Chen, X.; Li, M.; Wang, Y.; He, J.; Miao, Y.; Miao, R. Recent Advances in Understanding the Regulatory Mechanism of Plasma Membrane H+-ATPase through the Brassinosteroid Signaling Pathway. Plant Cell Physiol. 2024, 65, 1515–1529. [Google Scholar] [CrossRef] [PubMed]
- Oh, E.; Zhu, J.-Y.; Bai, M.-Y.; Arenhart, R.A.; Sun, Y.; Wang, Z.-Y. Cell elongation is regulated through a central circuit of interacting transcription factors in the Arabidopsis hypocotyl. eLife 2014, 3, e03031. [Google Scholar] [CrossRef]
- Wiesel, L.; Davis, J.L.; Milne, L.; Redondo Fernandez, V.; Herold, M.B.; Middlefell Williams, J.; Morris, J.; Hedley, P.E.; Harrower, B.; Newton, A.C.; et al. A transcriptional reference map of defence hormone responses in potato. Sci. Rep. 2015, 5, 15229. [Google Scholar] [CrossRef]
- Shi, Y.; Zhao, Y.; Yao, Q.; Liu, F.; Li, X.; Jin, X.; Zhang, Y.; Ahammed, G.J. Comparative Physiological and Transcriptomic Analyses Reveal Mechanisms of Exogenous Spermidine-Induced Tolerance to Low-Iron Stress in Solanum lycopersicum L. Antioxidants 2022, 11, 1260. [Google Scholar] [CrossRef]
- Kodaira, K.-S.; Qin, F.; Tran, L.-S.P.; Maruyama, K.; Kidokoro, S.; Fujita, Y.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Arabidopsis Cys2/His2 Zinc-Finger Proteins AZF1 and AZF2 Negatively Regulate Abscisic Acid-Repressive and Auxin-Inducible Genes under Abiotic Stress Conditions. Plant Physiol. 2011, 157, 742–756. [Google Scholar] [CrossRef]
- Nemhauser, J.L.; Hong, F.; Chory, J. Different Plant Hormones Regulate Similar Processes through Largely Nonoverlapping Transcriptional Responses. Cell 2006, 126, 467–475. [Google Scholar] [CrossRef]
- Roig-Villanova, I.; Bou-Torrent, J.; Galstyan, A.; Carretero-Paulet, L.; Portolés, S.; Rodríguez-Concepción, M.; Martínez-García, J.F. Interaction of shade avoidance and auxin responses: A role for two novel atypical bHLH proteins. EMBO J. 2007, 26, 4756–4767. [Google Scholar] [CrossRef]
- OuYang, F.; Mao, J.-F.; Wang, J.; Zhang, S.; Li, Y. Transcriptome Analysis Reveals that Red and Blue Light Regulate Growth and Phytohormone Metabolism in Norway Spruce [Picea abies (L.) Karst.]. PLoS ONE 2015, 10, e0127896. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.; Yan, H.; Luo, S.; Pan, F.; Wang, Y.; Xiang, Y. Genome-wide analysis of poplar SAUR gene family and expression profiles under cold, polyethylene glycol and indole-3-acetic acid treatments. Plant Physiol. Biochem. 2018, 128, 50–65. [Google Scholar] [PubMed]
- Wu, J.; Liu, S.; He, Y.; Guan, X.; Zhu, X.; Cheng, L.; Wang, J.; Lu, G. Genome-wide analysis of SAUR gene family in Solanaceae species. Gene 2012, 509, 38–50. [Google Scholar] [CrossRef]
- Fowler, S.; Thomashow, M.F. Arabidopsis Transcriptome Profiling Indicates That Multiple Regulatory Pathways Are Activated during Cold Acclimation in Addition to the CBF Cold Response Pathway. Plant Cell 2002, 14, 1675–1690. [Google Scholar] [CrossRef]
- Guo, Y.; Jiang, Q.; Hu, Z.; Sun, X.; Fan, S.; Zhang, H. Function of the auxin-responsive gene TaSAUR75 under salt and drought stress. Crop J. 2018, 6, 181–190. [Google Scholar] [CrossRef]
- Franklin, K.A.; Lee, S.H.; Patel, D.; Kumar, S.V.; Spartz, A.K.; Gu, C.; Ye, S.; Yu, P.; Breen, G.; Cohen, J.D.; et al. PHYTOCHROME-INTERACTING FACTOR 4 (PIF4) regulates auxin biosynthesis at high temperature. Proc. Natl. Acad. Sci. USA 2011, 108, 20231–20235. [Google Scholar] [CrossRef]
- Han, G.; Li, Y.; Yang, Z.; Wang, C.; Zhang, Y.; Wang, B. Molecular Mechanisms of Plant Trichome Development. Front. Plant Sci. 2022, 13, 910228. [Google Scholar] [CrossRef]
- Sun, C.; Wei, J.; Gu, X.; Wu, M.; Li, M.; Liu, Y.; An, N.; Wu, K.; Wu, S.; Wu, J.; et al. Different multicellular trichome types coordinate herbivore mechanosensing and defense in tomato. Plant Cell 2024, 36, 4952–4969. [Google Scholar] [CrossRef]
- Zheng, X.; Jian, Y.; Long, Q.; Luo, Y.; Xu, X.; Zhang, Q.; Cheng, Y.; Huang, B.; Qiu, D.; Li, Z.; et al. SlASR3 mediates crosstalk between auxin and jasmonic acid signaling to regulate trichome formation in tomato. Plant J. 2025, 121, e70053, Erratum in Plant J. 2025, 122, e70220. https://doi.org/10.1111/tpj.70220. [Google Scholar] [CrossRef]
- Chen, G.; Klinkhamer, P.G.L.; Escobar-Bravo, R.; Leiss, K.A. Type VI glandular trichome density and their derived volatiles are differently induced by jasmonic acid in developing and fully developed tomato leaves: Implications for thrips resistance. Plant Sci. 2018, 276, 87–98. [Google Scholar] [CrossRef] [PubMed]
- Ali, M.S.; Baek, K.H. Jasmonic Acid Signaling Pathway in Response to Abiotic Stresses in Plants. Int. J. Mol. Sci. 2020, 21, 621. [Google Scholar] [CrossRef] [PubMed]
- Marquis, V.; Smirnova, E.; Graindorge, S.; Delcros, P.; Villette, C.; Zumsteg, J.; Heintz, D.; Heitz, T. Broad-spectrum stress tolerance conferred by suppressing jasmonate signaling attenuation in Arabidopsis JASMONIC ACID OXIDASE mutants. Plant J. 2022, 109, 856–872. [Google Scholar] [CrossRef]
- Rehman, M.; Saeed, M.S.; Fan, X.; Salam, A.; Munir, R.; Yasin, M.U.; Khan, A.R.; Muhammad, S.; Ali, B.; Ali, I.; et al. The Multifaceted Role of Jasmonic Acid in Plant Stress Mitigation: An Overview. Plants 2023, 12, 3982. [Google Scholar] [CrossRef]
- Attia, H.; Alamer, K.H. Supplementation of Jasmonic acid Mitigates the Damaging Effects of Arsenic Stress on Growth, Photosynthesis and Nitrogen Metabolism in Rice. Rice 2024, 17, 31. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, Y.; Sano, R.; Wada, T.; Takabayashi, J.; Okada, K. Jasmonic acid control of GLABRA3 links inducible defense and trichome patterning in Arabidopsis. Development 2009, 136, 1039–1048. [Google Scholar] [CrossRef] [PubMed]
- Escobar-Bravo, R.; Klinkhamer, P.G.L.; Leiss, K.A. Induction of Jasmonic Acid-Associated Defenses by Thrips Alters Host Suitability for Conspecifics and Correlates with Increased Trichome Densities in Tomato. Plant Cell Physiol. 2017, 58, 622–634. [Google Scholar] [CrossRef]
- Li, J.; Yu, G.; Wang, X.; Guo, C.; Wang, Y.; Wang, X. Jasmonic acid plays an important role in mediating retrograde signaling under mitochondrial translational stress to balance plant growth and defense. Plant Commun. 2025, 6, 101133. [Google Scholar] [CrossRef]
- Rahman, M.M.; Mostofa, M.G.; Keya, S.S.; Ghosh, P.K.; Abdelrahman, M.; Anik, T.R.; Gupta, A.; Tran, L.-S.P. Jasmonic acid priming augments antioxidant defense and photosynthesis in soybean to alleviate combined heat and drought stress effects. Plant Physiol. Biochem. 2024, 206, 108193. [Google Scholar] [CrossRef]
- Tao, Y.; Huang, J.; Jing, H.K.; Shen, R.F.; Zhu, X.F. Jasmonic acid is involved in root cell wall phosphorus remobilization through the nitric oxide dependent pathway in rice. J. Exp. Bot. 2022, 73, 2618–2630. [Google Scholar] [CrossRef] [PubMed]
- Traw, M.B.; Bergelson, J. Interactive effects of jasmonic acid, salicylic acid, and gibberellin on induction of trichomes in Arabidopsis. Plant Physiol. 2003, 133, 1367–1375. [Google Scholar] [CrossRef]
- Li, S.; Cong, Y.; Liu, Y.; Wang, T.; Shuai, Q.; Chen, N.; Gai, J.; Li, Y. Optimization of Agrobacterium-Mediated Transformation in Soybean. Front. Plant Sci. 2017, 8, 246. [Google Scholar] [CrossRef]
- Zhong, H.; Li, C.; Yu, W.; Zhou, H.-P.; Lieber, T.; Su, X.; Wang, W.; Bumann, E.; Lunny Castro, R.M.; Jiang, Y.; et al. A fast and genotype-independent in planta Agrobacterium-mediated transformation method for soybean. Plant Commun. 2024, 5, 101063. [Google Scholar] [CrossRef]
- Hada, A.; Krishnan, V.; Punjabi, M.; Basak, N.; Pandey, V.; Jeevaraj, T.; Marathe, A.; Gupta, A.K.; Jolly, M.; Kumar, A.; et al. Refined glufosinate selection and its extent of exposure for improving the Agrobacterium-mediated transformation in Indian soybean (Glycine max) genotype JS-335. Plant Biotechnol. 2016, 33, 341–350. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hung, J.-H.; Weng, Z. Sequence Alignment and Homology Search with BLAST and ClustalW. Cold Spring Harb. Protoc. 2016, 11, 1–6. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular Evolutionary Genetics Analysis Using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Jin, J.; Guo, A.Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Bailey, T.L.; Williams, N.; Misleh, C.; Li, W.W. MEME: Discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006, 34, W369–W373. [Google Scholar] [CrossRef]
- Li, X.; Wu, Y.; Liu, Z.; Wei, H.; Wang, H.; Yu, S. Apical meristem transcriptome analysis identifies a role for the blue light receptor gene GhFKF1 in cotton architecture development. Crop J. 2024, 12, 1126–1136. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Schmutz, J.; Cannon, S.B.; Schlueter, J.; Ma, J.; Mitros, T.; Nelson, W.; Hyten, D.L.; Song, Q.; Thelen, J.J.; Cheng, J.; et al. Genome sequence of the palaeopolyploid soybean. Nature 2010, 463, 178–183. [Google Scholar] [CrossRef] [PubMed]
- Shimomura, M.; Kanamori, H.; Komatsu, S.; Namiki, N.; Mukai, Y.; Kurita, K.; Kamatsuki, K.; Ikawa, H.; Yano, R.; Ishimoto, M.; et al. The Glycine max cv. Enrei Genome for Improvement of Japanese Soybean Cultivars. Int. J. Genom. 2015, 2015, 358127. [Google Scholar] [CrossRef]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016, 11, 1650–1667. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L.; et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation 2021, 2, 100141. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. clusterProfiler: An R package for comparing biological themes among gene clusters. Omics 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef] [PubMed]

| Code | Number |
|---|---|
| i | 260 |
| j | 33,394 |
| o | 4204 |
| u | 3353 |
| x | 1543 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, X.; Liu, B.; Yang, Y.; Gou, H.; Du, H.; Chen, Y.; Yu, H.; Zhao, J.; Yuan, F. GmSAUR46b Integrates Light Signals to Regulate Leaf Midrib Thickness and Stem Trichome Density in Soybean. Int. J. Mol. Sci. 2025, 26, 9200. https://doi.org/10.3390/ijms26189200
Li X, Liu B, Yang Y, Gou H, Du H, Chen Y, Yu H, Zhao J, Yuan F. GmSAUR46b Integrates Light Signals to Regulate Leaf Midrib Thickness and Stem Trichome Density in Soybean. International Journal of Molecular Sciences. 2025; 26(18):9200. https://doi.org/10.3390/ijms26189200
Chicago/Turabian StyleLi, Xiao, Bei Liu, Yunhua Yang, Han Gou, Huan Du, Yuhao Chen, Huakun Yu, Jinming Zhao, and Fengjie Yuan. 2025. "GmSAUR46b Integrates Light Signals to Regulate Leaf Midrib Thickness and Stem Trichome Density in Soybean" International Journal of Molecular Sciences 26, no. 18: 9200. https://doi.org/10.3390/ijms26189200
APA StyleLi, X., Liu, B., Yang, Y., Gou, H., Du, H., Chen, Y., Yu, H., Zhao, J., & Yuan, F. (2025). GmSAUR46b Integrates Light Signals to Regulate Leaf Midrib Thickness and Stem Trichome Density in Soybean. International Journal of Molecular Sciences, 26(18), 9200. https://doi.org/10.3390/ijms26189200






