Monitoring of Allograft Adaptation After Kidney Transplantation in Pediatric Patients by Targeted Plasma Metabolomics
Abstract
1. Introduction
2. Results
2.1. Patient Characteristics
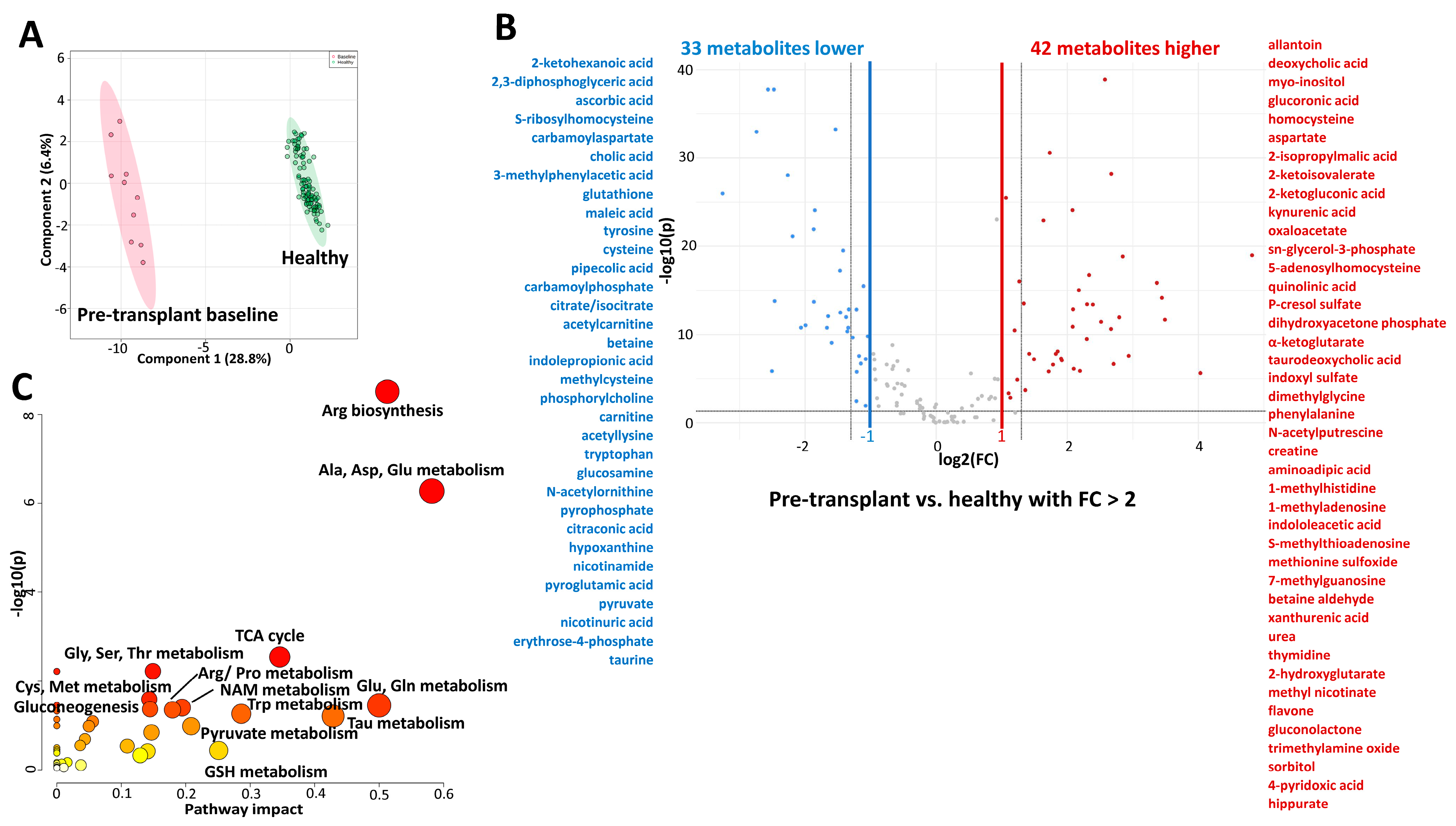
2.2. Comparison of Metabolic Profiles of Children with Kidney Failure (Pre-Transplant Baseline Timepoint) with Healthy Children
2.3. Comparison of Metabolic Profiles of Post-Transplant (Pre-Dose Timepoint) with Healthy Children
2.4. Pre-Transplant (Baseline) Versus Post-Transplant (Pre-Dose)
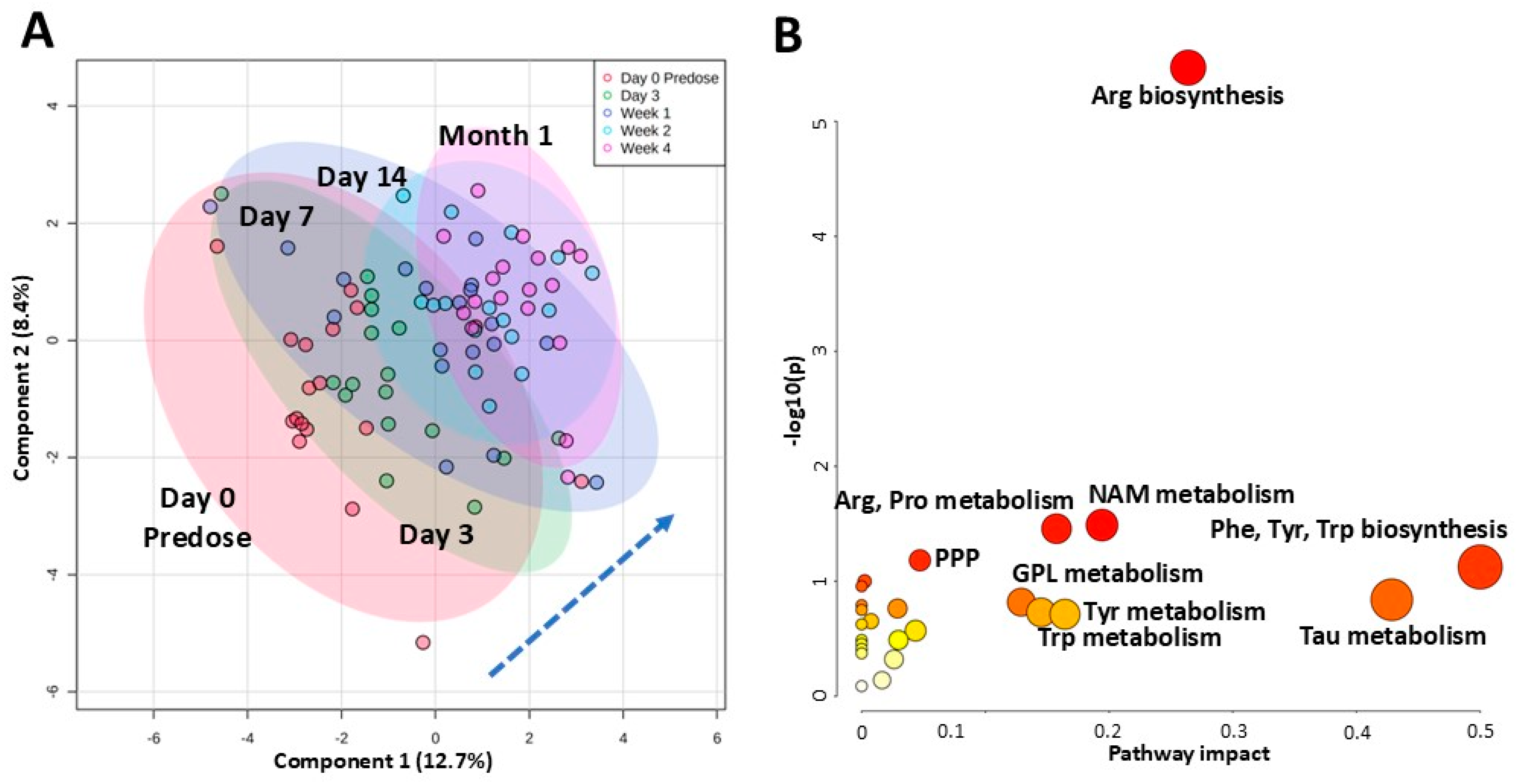
2.5. Changes Within the First Month Post Kidney Transplant and Start of Tacrolimus Regimen
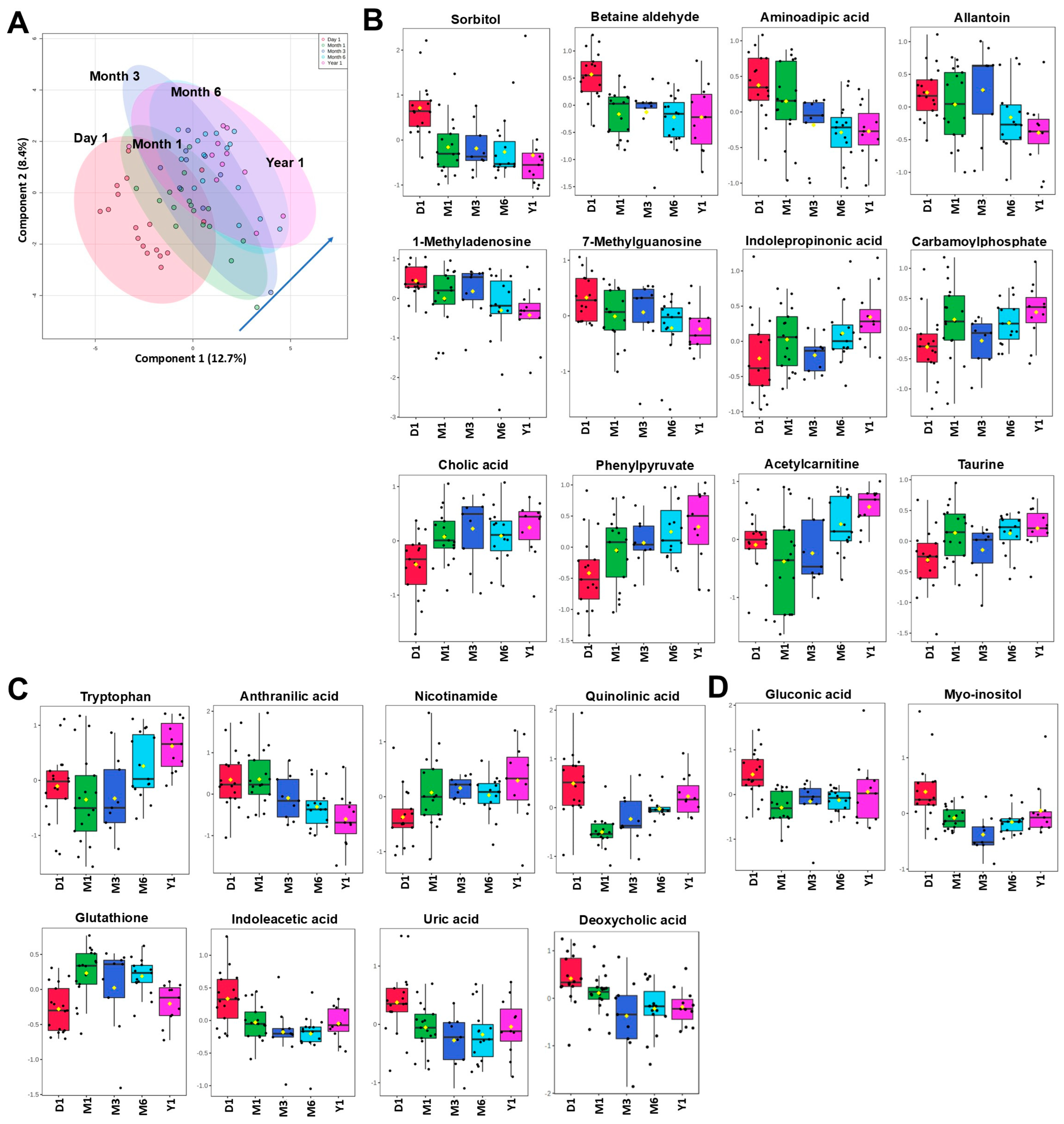
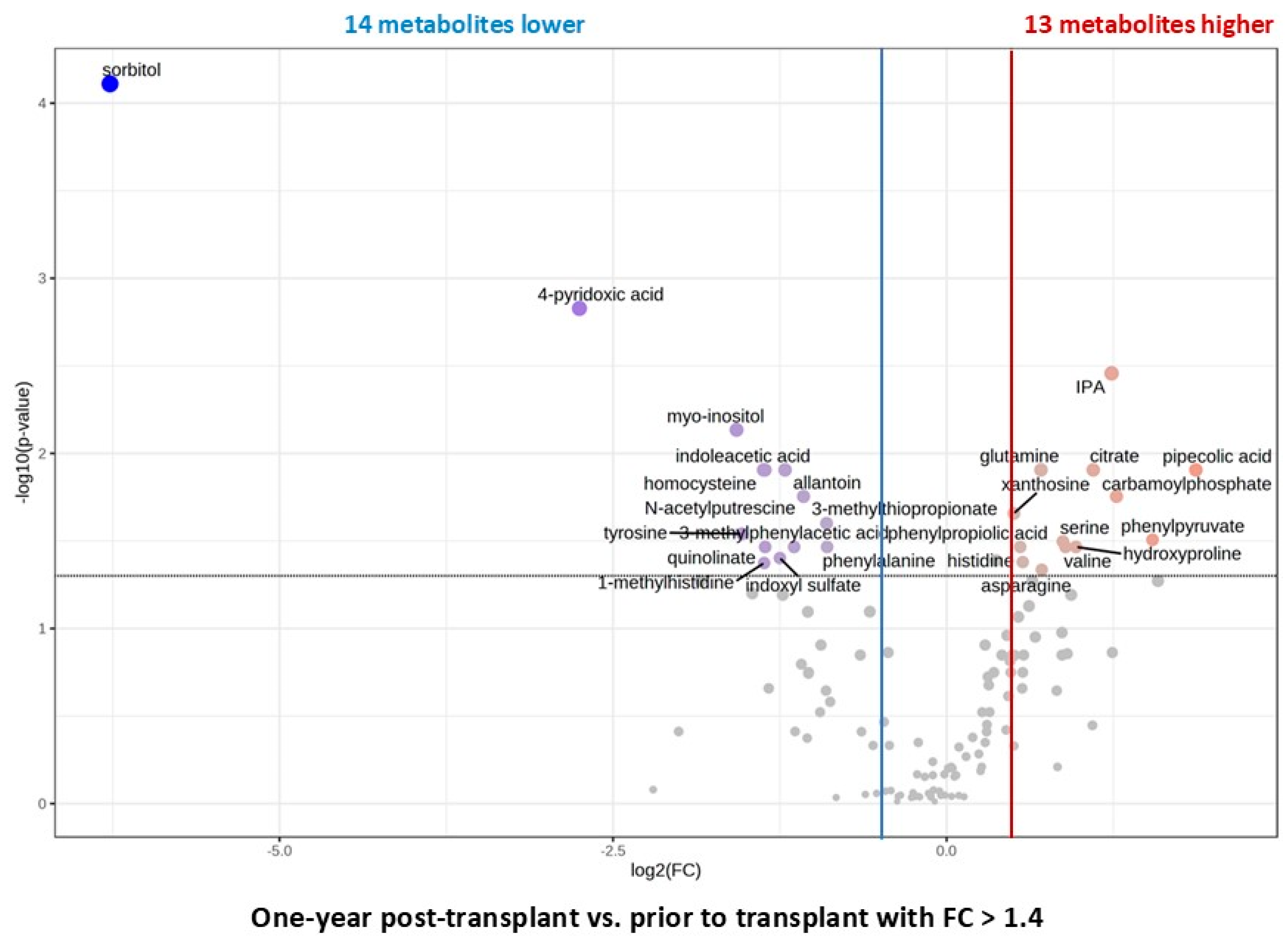
2.6. Changes Within the First-Year Post Kidney Transplant
2.7. Changes Within 2 to 4 Years Post Kidney Transplant and Comparison to Healthy Subjects
3. Discussion
Study Limitations
4. Materials and Methods
4.1. Trial Design
4.2. Healthy Participants
4.3. Targeted Metabolomics
4.4. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Suthanthiran, M.; Strom, T.B. Renal transplantation. N. Engl. J. Med. 1994, 331, 365–376. [Google Scholar] [CrossRef]
- Calne, R.Y.; Rolles, K.; White, D.J.; Thiru, S.; Evans, D.B.; McMaster, P.; Dunn, D.C.; Craddock, G.N.; Henderson, R.G.; Aziz, S.; et al. Cyclosporin A initially as the only immunosuppressant in 34 recipients of cadaveric organs: 32 kidneys, 2 pancreases, and 2 livers. Lancet 1979, 2, 1033–1036. [Google Scholar] [CrossRef] [PubMed]
- Starzl, T.E.; Todo, S.; Fung, J.; Demetris, A.J.; Venkataramman, R.; Jain, A. FK 506 for liver, kidney, and pancreas transplantation. Lancet 1989, 2, 1000–1004. [Google Scholar] [CrossRef]
- Hart, A.; Lentine, K.L.; Smith, J.M.; Miller, J.M.; Skeans, M.A.; Prentice, M.; Robinson, A.; Foutz, J.; Booker, S.E.; Israni, A.K.; et al. OPTN/SRTR 2019 Annual Data Report: Kidney. Am. J. Transplant. 2021, 21 (Suppl. S2), 21–137. [Google Scholar] [CrossRef]
- Meier-Kriesche, H.U.; Schold, J.D.; Kaplan, B. Long-term renal allograft survival: Have we made significant progress or is it time to rethink our analytic and therapeutic strategies? Am. J. Transplant. 2004, 4, 1289–1295. [Google Scholar] [CrossRef]
- Meier-Kriesche, H.U.; Schold, J.D.; Srinivas, T.R.; Kaplan, B. Lack of improvement in renal allograft survival despite a marked decrease in acute rejection rates over the most recent era. Am. J. Transplant. 2004, 4, 378–383. [Google Scholar] [CrossRef]
- Bia, M.; Adey, D.B.; Bloom, R.D.; Chan, L.; Kulkarni, S.; Tomlanovich, S. KDOQI US commentary on the 2009 KDIGO clinical practice guideline for the care of kidney transplant recipients. Am. J. Kidney Dis. 2010, 56, 189–218. [Google Scholar] [CrossRef]
- Kasiske, B.L.; Zeier, M.G.; Chapman, J.R.; Craig, J.C.; Ekberg, H.; Garvey, C.A.; Green, M.D.; Jha, V.; Josephson, M.A.; Kiberd, B.A.; et al. KDIGO clinical practice guideline for the care of kidney transplant recipients: A summary. Kidney Int. 2010, 77, 299–311. [Google Scholar] [CrossRef] [PubMed]
- Eckardt, K.-U.; Kasiske, B.L.; Zeier, M.G. Special Issue: KDIGO Clinical Practice Guideline for the Care of Kidney Transplant Recipients. Am. J. Transplant. 2009, 9 (Suppl. S3), S1–S155. [Google Scholar] [CrossRef]
- Furness, P.N.; Taub, N.; Convergence of European Renal Transplant Pathology Assessment Procedures (CERTPAP) Project. International variation in the interpretation of renal transplant biopsies: Report of the CERTPAP Project. Kidney Int. 2001, 60, 1998–2012. [Google Scholar] [CrossRef] [PubMed]
- Park, S.H.; Friedewald, J.J. Are borderline changes real rejection? Current viewpoints. Curr. Opin. Nephrol. Hypertens. 2020, 29, 656–662. [Google Scholar] [CrossRef]
- Schwarz, A.; Gwinner, W.; Hiss, M.; Radermacher, J.; Mengel, M.; Haller, H. Safety and adequacy of renal transplant protocol biopsies. Am. J. Transplant. 2005, 5, 1992–1996. [Google Scholar] [CrossRef]
- Banas, M.; Neumann, S.; Eiglsperger, J.; Schiffer, E.; Putz, F.J.; Reichelt-Wurm, S.; Kramer, B.K.; Pagel, P.; Banas, B. Identification of a urine metabolite constellation characteristic for kidney allograft rejection. Metabolomics 2018, 14, 116. [Google Scholar] [CrossRef]
- Kalantari, S.; Chashmniam, S.; Nafar, M.; Samavat, S.; Rezaie, D.; Dalili, N. A Noninvasive Urine Metabolome panel as potential biomarkers for diagnosis of t cell-mediated renal transplant rejection. OMICS 2020, 24, 140–147. [Google Scholar] [CrossRef] [PubMed]
- Kim, C.D.; Kim, E.Y.; Yoo, H.; Lee, J.W.; Ryu, D.H.; Noh, D.W.; Park, S.H.; Kim, Y.L.; Hwang, G.S.; Kwon, T.H. Metabonomic analysis of serum metabolites in kidney transplant recipients with cyclosporine A- or tacrolimus-based immunosuppression. Transplantation 2010, 90, 748–756. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.K.; Kim, B.K.; Jung, H.Y.; Cho, J.H.; Park, S.H.; Kim, Y.L.; Kim, S.Y.; Yoon, Y.R.; Chung, B.H.; Lee, S.H.; et al. Metabolomics study for identification of potential biomarkers of long-term survival in kidney transplantation recipients. Transplant. Proc. 2017, 49, 1005–1011. [Google Scholar] [CrossRef]
- Wishart, D.S. Metabolomics: A complementary tool in renal transplantation. Contrib. Nephrol. 2008, 160, 76–87. [Google Scholar] [CrossRef]
- Lentine, K.L.; Smith, J.M.; Lyden, G.R.; Miller, J.M.; Dolan, T.G.; Bradbrook, K.; Larkin, L.; Temple, K.; Handarova, D.K.; Weiss, S.; et al. OPTN/SRTR 2022 Annual Data Report: Kidney. Am. J. Transplant. 2024, 24, S19–S118. [Google Scholar] [CrossRef] [PubMed]
- Lees, H.J.; Swann, J.R.; Wilson, I.D.; Nicholson, J.K.; Holmes, E. Hippurate: The natural history of a mammalian-microbial cometabolite. J. Proteome Res. 2013, 12, 1527–1546. [Google Scholar] [CrossRef]
- Deltombe, O.; de Loor, H.; Glorieux, G.; Dhondt, A.; Van Biesen, W.; Meijers, B.; Eloot, S. Exploring binding characteristics and the related competition of different protein-bound uremic toxins. Biochimie 2017, 139, 20–26. [Google Scholar] [CrossRef]
- Mair, R.D.; Lee, S.; Plummer, N.S.; Sirich, T.L.; Meyer, T.W. Impaired Tubular Secretion of Organic Solutes in Advanced Chronic Kidney Disease. J. Am. Soc. Nephrol. 2021, 32, 2877–2884. [Google Scholar] [CrossRef] [PubMed]
- Stanimirova, I.; Banasik, M.; Zabek, A.; Dawiskiba, T.; Koscielska-Kasprzak, K.; Wojtowicz, W.; Krajewska, M.; Janczak, D.; Mlynarz, P. Serum metabolomics approach to monitor the changes in metabolite profiles following renal transplantation. Sci. Rep. 2020, 10, 17223. [Google Scholar] [CrossRef]
- Sun, B.; Wang, X.; Liu, X.; Wang, L.; Ren, F.; Wang, X.; Leng, X. Hippuric acid promotes renal fibrosis by disrupting redox homeostasis via facilitation of NRF2-KEAP1-CUL3 interactions in chronic kidney disease. Antioxidants 2020, 9, 783. [Google Scholar] [CrossRef]
- Ishii, N.; Ikenaga, H.; Ogawa, Z.; Aoki, Y.; Saruta, T.; Suga, T. Effects of renal sorbitol accumulation on urinary excretion of enzymes in hyperglycaemic rats. Ann. Clin. Biochem. 2001, 38, 391–398. [Google Scholar] [CrossRef]
- Klepacki, J.; Klawitter, J.; Klawitter, J.; Thurman, J.M.; Christians, U. A high-performance liquid chromatography-tandem mass spectrometry-based targeted metabolomics kidney dysfunction marker panel in human urine. Clin. Chim. Acta 2015, 446, 43–53. [Google Scholar] [CrossRef] [PubMed]
- Rosas-Rodriguez, J.A.; Valenzuela-Soto, E.M. Enzymes involved in osmolyte synthesis: How does oxidative stress affect osmoregulation in renal cells? Life Sci. 2010, 87, 515–520. [Google Scholar] [CrossRef]
- Busch, M.; Gobert, A.; Franke, S.; Ott, U.; Gerth, J.; Muller, A.; Stein, G.; Bitsch, R.; Wolf, G. Vitamin B6 metabolism in chronic kidney disease--relation to transsulfuration, advanced glycation and cardiovascular disease. Nephron Clin. Pract. 2010, 114, c38–c46. [Google Scholar] [CrossRef]
- Coburn, S.P.; Reynolds, R.D.; Mahuren, J.D.; Schaltenbrand, W.E.; Wang, Y.; Ericson, K.L.; Whyte, M.P.; Zubovic, Y.M.; Ziegler, P.J.; Costill, D.L.; et al. Elevated plasma 4-pyridoxic acid in renal insufficiency. Am. J. Clin. Nutr. 2002, 75, 57–64. [Google Scholar] [CrossRef]
- Kang, S.K.; Park, S.K. 2,3-diphosphoglyceric acid changes in uremia and during hemodialysis. Korean J. Intern. Med. 1986, 1, 86–91. [Google Scholar] [CrossRef]
- Balestri, P.L.; Berni, R.; Cupisti, A.; Giordani, R. An increase in bile acids in the serum of patients with chronic kidney failure. Minerva Med. 1995, 86, 207–209. [Google Scholar] [PubMed]
- Li, R.; Zeng, L.; Xie, S.; Chen, J.; Yu, Y.; Zhong, L. Targeted metabolomics study of serum bile acid profile in patients with end-stage renal disease undergoing hemodialysis. PeerJ 2019, 7, e7145. [Google Scholar] [CrossRef]
- Jimenez, F.; Monte, M.J.; El-Mir, M.Y.; Pascual, M.J.; Marin, J.J. Chronic renal failure-induced changes in serum and urine bile acid profiles. Dig. Dis. Sci. 2002, 47, 2398–2406. [Google Scholar] [CrossRef]
- Gai, Z.; Chu, L.; Hiller, C.; Arsenijevic, D.; Penno, C.A.; Montani, J.P.; Odermatt, A.; Kullak-Ublick, G.A. Effect of chronic renal failure on the hepatic, intestinal, and renal expression of bile acid transporters. Am. J. Physiol. Renal Physiol. 2014, 306, F130–F137. [Google Scholar] [CrossRef] [PubMed]
- Chu, L.; Zhang, K.; Zhang, Y.; Jin, X.; Jiang, H. Mechanism underlying an elevated serum bile acid level in chronic renal failure patients. Int. Urol. Nephrol. 2015, 47, 345–351. [Google Scholar] [CrossRef]
- Fan, C.Y.; Wang, M.X.; Ge, C.X.; Wang, X.; Li, J.M.; Kong, L.D. Betaine supplementation protects against high-fructose-induced renal injury in rats. J. Nutr. Biochem. 2014, 25, 353–362. [Google Scholar] [CrossRef] [PubMed]
- McGregor, D.O.; Dellow, W.J.; Robson, R.A.; Lever, M.; George, P.M.; Chambers, S.T. Betaine supplementation decreases post-methionine hyperhomocysteinemia in chronic renal failure. Kidney Int. 2002, 61, 1040–1046. [Google Scholar] [CrossRef]
- Bassi, R.; Niewczas, M.A.; Biancone, L.; Bussolino, S.; Merugumala, S.; Tezza, S.; D’Addio, F.; Ben Nasr, M.; Valderrama-Vasquez, A.; Usuelli, V.; et al. Metabolomic Profiling in Individuals with a Failing Kidney Allograft. PLoS ONE 2017, 12, e0169077. [Google Scholar] [CrossRef]
- Bergstrom, J.; Alvestrand, A.; Furst, P.; Lindholm, B. Sulphur amino acids in plasma and muscle in patients with chronic renal failure: Evidence for taurine depletion. J. Intern. Med. 1989, 226, 189–194. [Google Scholar] [CrossRef] [PubMed]
- Mor, A.; Kalaska, B.; Pawlak, D. Kynurenine Pathway in Chronic Kidney Disease: What’s Old, What’s New, and What’s Next? Int. J. Tryptophan Res. 2020, 13, 1178646920954882. [Google Scholar] [CrossRef]
- Zakrocka, I.; Zaluska, W. Kynurenine pathway in kidney diseases. Pharmacol. Rep. 2022, 74, 27–39. [Google Scholar] [CrossRef]
- Pawlak, K.; Domaniewski, T.; Mysliwiec, M.; Pawlak, D. The kynurenines are associated with oxidative stress, inflammation and the prevalence of cardiovascular disease in patients with end-stage renal disease. Atherosclerosis 2009, 204, 309–314. [Google Scholar] [CrossRef]
- Yilmaz, N.; Ustundag, Y.; Kivrak, S.; Kahvecioglu, S.; Celik, H.; Kivrak, I.; Huysal, K. Serum indoleamine 2,3 dioxygenase and tryptophan and kynurenine ratio using the UPLC-MS/MS method, in patients undergoing peritoneal dialysis, hemodialysis, and kidney transplantation. Ren. Fail. 2016, 38, 1300–1309. [Google Scholar] [CrossRef] [PubMed]
- Roager, H.M.; Licht, T.R. Microbial tryptophan catabolites in health and disease. Nat. Commun. 2018, 9, 3294. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.Y.; Lin, C.J.; Pan, H.C.; Lee, C.C.; Lu, S.C.; Hsieh, Y.T.; Huang, S.Y.; Huang, H.Y. Clinical association between the metabolite of healthy gut microbiota, 3-indolepropionic acid and chronic kidney disease. Clin. Nutr. 2019, 38, 2945–2948. [Google Scholar] [CrossRef]
- Tourountzis, T.; Lioulios, G.; Fylaktou, A.; Moysidou, E.; Papagianni, A.; Stangou, M. Microbiome in Chronic Kidney Disease. Life 2022, 12, 1513. [Google Scholar] [CrossRef]
- Snauwaert, E.; Holvoet, E.; Van Biesen, W.; Raes, A.; Glorieux, G.; Vande Walle, J.; Roels, S.; Vanholder, R.; Askiti, V.; Azukaitis, K.; et al. Uremic Toxin Concentrations are Related to Residual Kidney Function in the Pediatric Hemodialysis Population. Toxins 2019, 11, 235. [Google Scholar] [CrossRef]
- Castillo-Rodriguez, E.; Fernandez-Prado, R.; Esteras, R.; Perez-Gomez, M.V.; Gracia-Iguacel, C.; Fernandez-Fernandez, B.; Kanbay, M.; Tejedor, A.; Lazaro, A.; Ruiz-Ortega, M.; et al. Impact of Altered Intestinal Microbiota on Chronic Kidney Disease Progression. Toxins 2018, 10, 300. [Google Scholar] [CrossRef] [PubMed]
- Meijers, B.K.; Evenepoel, P. The gut-kidney axis: Indoxyl sulfate, p-cresyl sulfate and CKD progression. Nephrol. Dial. Transplant. 2011, 26, 759–761. [Google Scholar] [CrossRef]
- Kim, J.E.; Kim, H.E.; Park, J.I.; Cho, H.; Kwak, M.J.; Kim, B.Y.; Yang, S.H.; Lee, J.P.; Kim, D.K.; Joo, K.W.; et al. The association between gut microbiota and uremia of chronic kidney disease. Microorganisms 2020, 8, 907. [Google Scholar] [CrossRef]
- Tummalapalli, L.; Nadkarni, G.N.; Coca, S.G. Biomarkers for predicting outcomes in chronic kidney disease. Curr. Opin. Nephrol. Hypertens. 2016, 25, 480–486. [Google Scholar] [CrossRef]
- Wu, I.W.; Hsu, K.H.; Lee, C.C.; Sun, C.Y.; Hsu, H.J.; Tsai, C.J.; Tzen, C.Y.; Wang, Y.C.; Lin, C.Y.; Wu, M.S. p-Cresyl sulphate and indoxyl sulphate predict progression of chronic kidney disease. Nephrol. Dial. Transplant. 2011, 26, 938–947. [Google Scholar] [CrossRef]
- Vaziri, N.D.; Liu, S.M.; Lau, W.L.; Khazaeli, M.; Nazertehrani, S.; Farzaneh, S.H.; Kieffer, D.A.; Adams, S.H.; Martin, R.J. High amylose resistant starch diet ameliorates oxidative stress, inflammation, and progression of chronic kidney disease. PLoS ONE 2014, 9, e114881. [Google Scholar] [CrossRef]
- Alvarenga, L.; Saldanha, J.F.; Stockler-Pinto, M.B.; Fouque, D.; Soulage, C.O.; Mafra, D. Effects of resveratrol on inflammation and oxidative stress induced by the uremic toxin indoxyl sulfate in Murine macrophage-like RAW 264.7. Biochimie 2023, 213, 22–29. [Google Scholar] [CrossRef]
- Saranya, G.R.; Viswanathan, P. Gut microbiota dysbiosis in AKI to CKD transition. Biomed. Pharmacother. 2023, 161, 114447. [Google Scholar] [CrossRef]
- Moller, N.; Meek, S.; Bigelow, M.; Andrews, J.; Nair, K.S. The kidney is an important site for in vivo phenylalanine-to-tyrosine conversion in adult humans: A metabolic role of the kidney. Proc. Natl. Acad. Sci. USA 2000, 97, 1242–1246. [Google Scholar] [CrossRef]
- Riccio, S.; Valentino, M.S.; Passaro, A.P.; Izzo, M.; Guarino, S.; Miraglia Del Giudice, E.; Marzuillo, P.; Di Sessa, A. new insights from metabolomics in pediatric renal diseases. Children 2022, 9, 118. [Google Scholar] [CrossRef] [PubMed]
- Wen, D.; Zheng, Z.; Surapaneni, A.; Yu, B.; Zhou, L.; Zhou, W.; Xie, D.; Shou, H.; Avila-Pacheco, J.; Kalim, S.; et al. Metabolite profiling of CKD progression in the chronic renal insufficiency cohort study. JCI Insight 2022, 7, 16196. [Google Scholar] [CrossRef]
- Sood, M.M.; Murphy, M.S.Q.; Hawken, S.; Wong, C.A.; Potter, B.K.; Burns, K.D.; Tsampalieros, A.; Atkinson, K.M.; Chakraborty, P.; Wilson, K. Association between newborn metabolic profiles and pediatric kidney disease. Kidney Int. Rep. 2018, 3, 691–700. [Google Scholar] [CrossRef] [PubMed]
- Welch, G.N.; Loscalzo, J. Homocysteine and atherothrombosis. N. Engl. J. Med. 1998, 338, 1042–1050. [Google Scholar] [CrossRef]
- Friedman, A.N.; Rosenberg, I.H.; Selhub, J.; Levey, A.S.; Bostom, A.G. Hyperhomocysteinemia in renal transplant recipients. Am. J. Transplant. 2002, 2, 308–313. [Google Scholar] [CrossRef] [PubMed]
- Andrade, F.; Rodriguez-Soriano, J.; Prieto, J.A.; Aguirre, M.; Ariceta, G.; Lage, S.; Azcona, I.; Prado, C.; Sanjurjo, P.; Aldamiz-Echevarria, L. Methylation cycle, arginine-creatine pathway and asymmetric dimethylarginine in paediatric renal transplant. Nephrol. Dial. Transplant. 2011, 26, 328–336. [Google Scholar] [CrossRef]
- Andrade, F.; Rodriguez-Soriano, J.; Prieto, J.A.; Elorz, J.; Aguirre, M.; Ariceta, G.; Martin, S.; Sanjurjo, P.; Aldamiz-Echevarria, L. The arginine-creatine pathway is disturbed in children and adolescents with renal transplants. Pediatr. Res. 2008, 64, 218–222. [Google Scholar] [CrossRef]
- Arumugam, M.K.; Paal, M.C.; Donohue, T.M., Jr.; Ganesan, M.; Osna, N.A.; Kharbanda, K.K. Beneficial effects of betaine: A comprehensive review. Biology 2021, 10, 456. [Google Scholar] [CrossRef]
- Zhao, G.; He, F.; Wu, C.; Li, P.; Li, N.; Deng, J.; Zhu, G.; Ren, W.; Peng, Y. Betaine in inflammation: Mechanistic aspects and applications. Front. Immunol. 2018, 9, 1070. [Google Scholar] [CrossRef]
- Ozbay, L.A.; Moller, N.; Juhl, C.; Bjerre, M.; Carstens, J.; Rungby, J.; Jorgensen, K.A. The impact of calcineurin inhibitors on insulin sensitivity and insulin secretion: A randomized crossover trial in uraemic patients. Diabet. Med. 2012, 29, e440–e444. [Google Scholar] [CrossRef]
- Song, J.L.; Li, M.; Yan, L.N.; Yang, J.Y.; Yang, J.; Jiang, L. Higher tacrolimus blood concentration is related to increased risk of post-transplantation diabetes mellitus after living donor liver transplantation. Int. J. Surg. 2018, 51, 17–23. [Google Scholar] [CrossRef] [PubMed]
- Gelens, M.A.; Christiaans, M.H.; van Hooff, J.P. Glucose metabolism before and after conversion from cyclosporine microemulsion to tacrolimus in stable renal recipients. Nephrol. Dial. Transplant. 2008, 23, 701–706. [Google Scholar] [CrossRef]
- Santos, L.; Rodrigo, E.; Pinera, C.; Quintella, E.; Ruiz, J.C.; Fernandez-Fresnedo, G.; Palomar, R.; Gomez-Alamillo, C.; de Francisco, A.; Arias, M. New-onset diabetes after transplantation: Drug-related risk factors. Transplant. Proc. 2012, 44, 2585–2587. [Google Scholar] [CrossRef] [PubMed]
- Sato, T.; Inagaki, A.; Uchida, K.; Ueki, T.; Goto, N.; Matsuoka, S.; Katayama, A.; Haba, T.; Tominaga, Y.; Okajima, Y.; et al. Diabetes mellitus after transplant: Relationship to pretransplant glucose metabolism and tacrolimus or cyclosporine A-based therapy. Transplantation 2003, 76, 1320–1326. [Google Scholar] [CrossRef] [PubMed]
- Zheng, C.; Wang, C.; Zhang, T.; Li, D.; Ni, X.F.; Lin, J.H.; Sun, L.; Chen, B. Exploring the mechanism of skeletal muscle in a tacrolimus-induced posttransplantation diabetes mellitus model on gene expression profiles. J. Diabetes Res. 2020, 2020, 6542346. [Google Scholar] [CrossRef]
- Joncquel, M.; Labasque, J.; Demaret, J.; Bout, M.A.; Hamroun, A.; Hennart, B.; Tronchon, M.; Defevre, M.; Kim, I.; Kerckhove, A.; et al. Targeted metabolomics analysis suggests that tacrolimus alters protection against oxidative stress. Antioxidants 2023, 12, 1412. [Google Scholar] [CrossRef] [PubMed]
- Wee, H.N.; Liu, J.J.; Ching, J.; Kovalik, J.P.; Lim, S.C. The kynurenine pathway in acute kidney injury and chronic kidney disease. Am. J. Nephrol. 2021, 52, 771–787. [Google Scholar] [CrossRef]
- Minovic, I.; van der Veen, A.; van Faassen, M.; Riphagen, I.J.; van den Berg, E.; van der Ley, C.; Gomes-Neto, A.W.; Geleijnse, J.M.; Eggersdorfer, M.; Navis, G.J.; et al. Functional vitamin B6 status and long-term mortality in renal transplant recipients. Am. J. Clin. Nutr. 2017, 106, 1366–1374. [Google Scholar] [CrossRef] [PubMed]
- Wagner-Golbs, A.; Neuber, S.; Kamlage, B.; Christiansen, N.; Bethan, B.; Rennefahrt, U.; Schatz, P.; Lind, L. Effects of long-term storage at −80 degrees c on the human plasma metabolome. Metabolites 2019, 9, 99. [Google Scholar] [CrossRef]
- Gaspari, F.; Perico, N.; Remuzzi, G. Measurement of glomerular filtration rate. Kidney Int. Suppl. 1997, 63, S151–S154. [Google Scholar] [PubMed]
- Schwartz, G.J.; Munoz, A.; Schneider, M.F.; Mak, R.H.; Kaskel, F.; Warady, B.A.; Furth, S.L. New equations to estimate GFR in children with CKD. J. Am. Soc. Nephrol. 2009, 20, 629–637. [Google Scholar] [CrossRef]
- Davidson, J.A.; Pfeifer, Z.; Frank, B.; Tong, S.; Urban, T.T.; Wischmeyer, P.A.; Mourani, P.; Landeck, B.; Christians, U.; Klawitter, J. Metabolomic fingerprinting of infants undergoing cardiopulmonary bypass: Changes in metabolic pathways and association with mortality and cardiac intensive care unit length of stay. J. Am. Heart Assoc. 2018, 7, e010711. [Google Scholar] [CrossRef]
- Davidson, J.A.; Robison, J.; Khailova, L.; Frank, B.S.; Jaggers, J.; Ing, R.J.; Lawson, S.; Iguidbashian, J.; Ali, E.; Treece, A.; et al. Metabolomic profiling demonstrates evidence for kidney and urine metabolic dysregulation in a piglet model of cardiac surgery-induced acute kidney injury. Am. J. Physiol. Renal Physiol. 2022, 323, F20–F32. [Google Scholar] [CrossRef]
- Yuan, M.; Breitkopf, S.B.; Yang, X.; Asara, J.M. A positive/negative ion-switching, targeted mass spectrometry-based metabolomics platform for bodily fluids, cells, and fresh and fixed tissue. Nat. Protoc. 2012, 7, 872–881. [Google Scholar] [CrossRef]
- Pang, Z.; Zhou, G.; Ewald, J.; Chang, L.; Hacariz, O.; Basu, N.; Xia, J. Using MetaboAnalyst 5.0 for LC-HRMS spectra processing, multi-omics integration and covariate adjustment of global metabolomics data. Nat. Protoc. 2022, 17, 1735–1761. [Google Scholar] [CrossRef]

| Characteristic | Healthy | Predose (N = 23) | 1 Month (N = 15) | 6 Months (N = 18) | 12 Months (N = 15) | 24 Months (N = 10) |
|---|---|---|---|---|---|---|
| Age (years) | 12 ± 5 | 12 ± 4.3 | 11 ± 4.2 | 13 ± 4.1 | 12 ± 3.4 | 12 ± 2.7 |
| % Female | 53 | 48 | 60 | 39 | 33 | 30 |
| Race % White | 90 | 87 | 80 | 89 | 87 | 80 |
| Race % Other | 10 | 13 | 20 | 11 | 13 | 20 |
| Height (cm) | - | 138 ± 22 | 133 ± 21 | 140 ± 19 | 139 ± 18 | 137 ± 17 |
| Weight (kg) | - | 40 ± 19 | 36 ± 16 | 40 ± 14 | 40 ± 13 | 37 ± 13 |
| eGFR | - | 66 ± 30 | 79 ± 15 | 71 ± 19 | 78 ± 16 | 74 ± 22 |
| Creatinine (mg/dL) | 0.6 ± 0.1 | 1.5 ± 2.1 | 0.7 ± 0.2 | 0.9 ± 0.3 | 0.8 ± 0.2 | 0.8 ± 0.3 |
| Hypertension, treated | - | N = 11 | N = 6 | N = 8 | N = 7 | N = 3 |
| SBP (mmHg) | - | 122 ± 12 | 113 ± 9 | 108 ± 15 | 111 ± 18 | 111 ± 8.6 |
| BUN (mg/dL) | 13 ± 4.3 | 29 ± 25 | 20 ± 5.6 | 18 ± 6.3 | 17 ± 6.6 | 19 ± 6.9 |
| Bicarbonate (mmol/L) | 24 ± 3.4 | 23 ± 2.5 | 23± 2.4 | 24 ± 3.1 | 23 ± 2.5 | 23 ± 1.9 |
| Chloride (mmol/L) | 104 ± 2.2 | 108 ± 4.5 | 107 ± 3.8 | 106 ± 2.6 | 106 ± 3.3 | 106 ± 2.0 |
| Potassium (mmol/L) | 4.2 ± 0.4 | 4.0 ± 0.5 | 4.3 ± 0.5 | 4.4 ± 0.5 | 4.2 ± 0.3 | 4.2 ± 0.3 |
| Sodium (mmol/L) | 141 ± 2.3 | 138 ± 2.8 | 139 ± 2.7 | 140 ± 2.4 | 140 ± 2.7 | 139 ± 2.2 |
| Hematocrit (%) | 42 ± 4.3 * | 28 ± 3.4 * | 35 ± 4.6 | 35 ± 6.8 | 37 ± 3.9 | 40 ± 4.7 |
| Platelet (×109/L) | - | 210 ± 99 | 265 ± 43 | 289 ± 109 | 279 ± 63 | 278 ± 38 |
| WBC (×109/L) | 6.2 ± 3.9 | 6.0 ± 2.7 | 8.0 ± 2.4 | 4.6 ± 2.0 | 5.8 ± 1.9 | 6.7 ± 2.1 |
| Glucose (mg/dL) | 93 ± 19 | 86 ± 11 | 86 ± 14 | 91 ± 10 | 90 ± 13 | 81 ± 8.5 |
| FDR | Fold Change | |
|---|---|---|
| 1-methyladenosine | 1.34 × 10−6 | 4.57 |
| 1-methylhistidine | 9.60 × 10−16 | 4.52 |
| 2,3-diphosphoglyceric acid | 1.77 × 10−38 | 0.17 |
| 2-hydroxyglutarate | 6.16 × 10−29 | 6.37 |
| 2-isopropylmalic acid | 2.07 × 10−4 | 2.57 |
| 2-ketohexanoic acid | 1.09 × 10−33 | 0.15 |
| 2-ketoisovalerate | 1.56 × 10−8 | 2.67 |
| 3-methylphenylacetic acid | 1.66 × 10−11 | 0.24 |
| 3-methylthiopropionate | 1.02 × 10−5 | 1.86 |
| 4-pyridoxic acid | 6.95 × 10−15 | 10.9 |
| 5-hydroxyindoleacetic acid | 2.02 × 10−3 | 1.75 |
| 2-ketogluconic acid | 6.48 × 10−8 | 2.81 |
| 7-methylguanosine | 1.84 × 10−17 | 5.04 |
| acetyllysine | 1.09 × 10−12 | 0.39 |
| α-ketoglutarate | 8.63 × 10−8 | 3.78 |
| allantoin | 4.68 × 10−4 | 2.15 |
| aminoadipic acid | 7.82 × 10−7 | 4.28 |
| arginine | 7.01 × 10−8 | 0.52 |
| argininosuccinate | 1.63 × 10−10 | 0.48 |
| ascorbic acid | 1.43 × 10−6 | 0.18 |
| aspartate | 3.04 × 10−14 | 2.53 |
| betaine | 5.87 × 10−34 | 0.34 |
| betaine aldehyde | 3.99 × 10−14 | 5.24 |
| carbamoylaspartate | 8.98 × 10−29 | 0.21 |
| carbamoylphosphate | 1.71 × 10−11 | 0.31 |
| carnitine | 3.13 × 10−20 | 0.37 |
| citraconic acid | 1.46 × 10−13 | 0.43 |
| citrate | 1.77 × 10−38 | 0.18 |
| creatine | 1.37 × 10−13 | 4.25 |
| cysteine | 1.99 × 10−14 | 0.27 |
| deoxycholic acid | 1.48 × 10−3 | 2.19 |
| dihydroxyacetone phosphate | 5.54 × 10−8 | 3.75 |
| dimethylglycine | 8.01 × 10−25 | 4.23 |
| erythrose-4-phosphate | 6.08 × 10−8 | 0.47 |
| gluconic acid | 1.29 × 10−5 | 2.36 |
| gluconolactone | 1.45 × 10−19 | 7.19 |
| glucosamine | 1.42 × 10−13 | 0.40 |
| glutamate | 1.17 × 10−5 | 1.92 |
| glutathione | 9.14 × 10−12 | 0.25 |
| glycolic acid | 8.95 × 10−7 | 0.52 |
| hippurate | 2.38 × 10−6 | 16.3 |
| homocysteine | 9.63 × 10−17 | 2.41 |
| indoleacetic acid | 1.12 × 10−11 | 4.78 |
| indolepropionic acid | 2.68 × 10−26 | 0.36 |
| indoxyl sulfate | 1.57 × 10−13 | 4.10 |
| kynurenic acid | 1.55 × 10−6 | 3.29 |
| kynurenine | 1.17 × 10−3 | 1.79 |
| maleic acid | 1.18 × 10−22 | 0.27 |
| methionine sulfoxide | 3.68 × 10−14 | 4.93 |
| methyl nicotinate | 2.24 × 10−7 | 6.53 |
| methylcysteine | 6.06 × 10−18 | 0.36 |
| myo-inositol | 3.39 × 10−11 | 2.29 |
| N-acetylcarnitine | 9.10 × 10−10 | 0.33 |
| N-acetylphosphate | 1.26 × 10−5 | 0.53 |
| N-acetylputrescine | 1.32 × 10−11 | 4.23 |
| nicotinamide | 2.75 × 10−8 | 0.44 |
| nicotinuric acid | 1.21 × 10−2 | 0.47 |
| orotate | 9.23 × 10−4 | 1.62 |
| oxaloacetate | 2.63 × 10−31 | 3.32 |
| p-cresol sulfate | 2.04 × 10−18 | 3.71 |
| phenylalanine | 2.85 × 10−10 | 2.57 |
| phenyllactic acid | 1.95 × 10−3 | 1.56 |
| phenylpropiolic acid | 1.70 × 10−3 | 0.53 |
| phosphorylcholine | 3.28 × 10−13 | 0.36 |
| p-hydroxybenzoate | 1.58 × 10−8 | 0.51 |
| pyroglutamic acid | 1.99 × 10−7 | 0.45 |
| pyridoxal | 3.61 × 10−3 | 0.43 |
| pyruvate | 3.24 × 10−16 | 0.46 |
| quinolinic acid | 8.41 × 10−9 | 3.61 |
| S-adenosylhomocysteine | 1.58 × 10−8 | 3.54 |
| shikimate | 1.70 × 10−3 | 1.86 |
| S-methylthioadenosine | 3.42 × 10−10 | 4.92 |
| sn-glycerol-3-phosphate | 2.59 × 10−7 | 3.44 |
| sorbitol | 1.03 × 10−19 | 10.3 |
| S-ribosylhomocysteine | 1.62 × 10−14 | 0.18 |
| succinate | 1.19 × 10−2 | 1.46 |
| taurine | 4.50 × 10−5 | 0.59 |
| taurodeoxycholic acid | 3.89 × 10−6 | 3.89 |
| thymidine | 2.40 × 10−11 | 6.35 |
| trimethylamine oxide | 2.63 × 10−8 | 7.66 |
| tryptophan | 1.71 × 10−11 | 0.39 |
| tyrosine | 5.33 × 10−14 | 0.27 |
| urea | 1.25 × 10−39 | 5.96 |
| uric acid | 8.71 × 10−24 | 1.90 |
| xanthurenic acid | 3.71 × 10−12 | 5.72 |
| FDR | Fold Change 4-Week to Pre-Dose | |
|---|---|---|
| putrescine | 0.0141 | 0.25 |
| acetylcarnitine | 0.0141 | 0.29 |
| N-acetylornithine | 0.0141 | 0.31 |
| quinolinic acid | 0.0004 | 0.32 |
| deoxycholic acid | 0.0071 | 0.44 |
| gluconic acid | 0.0022 | 0.44 |
| trimethylamine oxide | 0.0124 | 0.45 |
| N-acetylglutamate | 0.0106 | 0.47 |
| guanidoacetic acid | 0.0141 | 0.52 |
| N-acetylputrescine | 0.0183 | 0.60 |
| phenylalanine | 0.0127 | 0.60 |
| phenylacetic acid | 0.0306 | 0.60 |
| fumarate | 0.0183 | 1.46 |
| ornithine | 0.0183 | 1.47 |
| citraconic acid | 0.0339 | 1.64 |
| taurine | 0.0287 | 1.67 |
| 2-ketohexanoic acid | 0.0156 | 1.68 |
| uridine | 0.0141 | 1.73 |
| proline | 0.0172 | 1.74 |
| carbamoylphosphate | 0.0438 | 1.85 |
| nicotinamide | 0.0424 | 1.95 |
| tyrosine | 0.0296 | 1.99 |
| glycerophosphocholine | 0.0141 | 2.06 |
| flavone | 0.0172 | 2.17 |
| sn-glycerol-3-phosphate | 0.0141 | 2.18 |
| anthranilic acid | 0.0047 | 2.26 |
| cholesteryl sulfate | 0.0028 | 2.32 |
| dihydroxybenzoic acid | 0.0053 | 2.43 |
| hypoxanthine | 0.0382 | 2.44 |
| pipecolic acid | 0.0295 | 2.54 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Klawitter, J.; Kirkpatrick, B.E.; Shillingburg, R.; Klawitter, J.; Wheeler, G.; Shokati, T.; Cadnapaphornchai, M.A.; Galinkin, J.L.; Thurman, J.M.; Christians, U. Monitoring of Allograft Adaptation After Kidney Transplantation in Pediatric Patients by Targeted Plasma Metabolomics. Int. J. Mol. Sci. 2025, 26, 9190. https://doi.org/10.3390/ijms26189190
Klawitter J, Kirkpatrick BE, Shillingburg R, Klawitter J, Wheeler G, Shokati T, Cadnapaphornchai MA, Galinkin JL, Thurman JM, Christians U. Monitoring of Allograft Adaptation After Kidney Transplantation in Pediatric Patients by Targeted Plasma Metabolomics. International Journal of Molecular Sciences. 2025; 26(18):9190. https://doi.org/10.3390/ijms26189190
Chicago/Turabian StyleKlawitter, Jelena, Bruce E. Kirkpatrick, Ryan Shillingburg, Jost Klawitter, Garrett Wheeler, Touraj Shokati, Melissa A. Cadnapaphornchai, Jeffrey L. Galinkin, Joshua M. Thurman, and Uwe Christians. 2025. "Monitoring of Allograft Adaptation After Kidney Transplantation in Pediatric Patients by Targeted Plasma Metabolomics" International Journal of Molecular Sciences 26, no. 18: 9190. https://doi.org/10.3390/ijms26189190
APA StyleKlawitter, J., Kirkpatrick, B. E., Shillingburg, R., Klawitter, J., Wheeler, G., Shokati, T., Cadnapaphornchai, M. A., Galinkin, J. L., Thurman, J. M., & Christians, U. (2025). Monitoring of Allograft Adaptation After Kidney Transplantation in Pediatric Patients by Targeted Plasma Metabolomics. International Journal of Molecular Sciences, 26(18), 9190. https://doi.org/10.3390/ijms26189190







