Integrated Analysis, Machine Learning, Molecular Docking and Dynamics of CDK1 Inhibitors in Epithelial Ovarian Cancer: A Multifaceted Approach Towards Targeted Therapy
Abstract
1. Introduction
2. Results
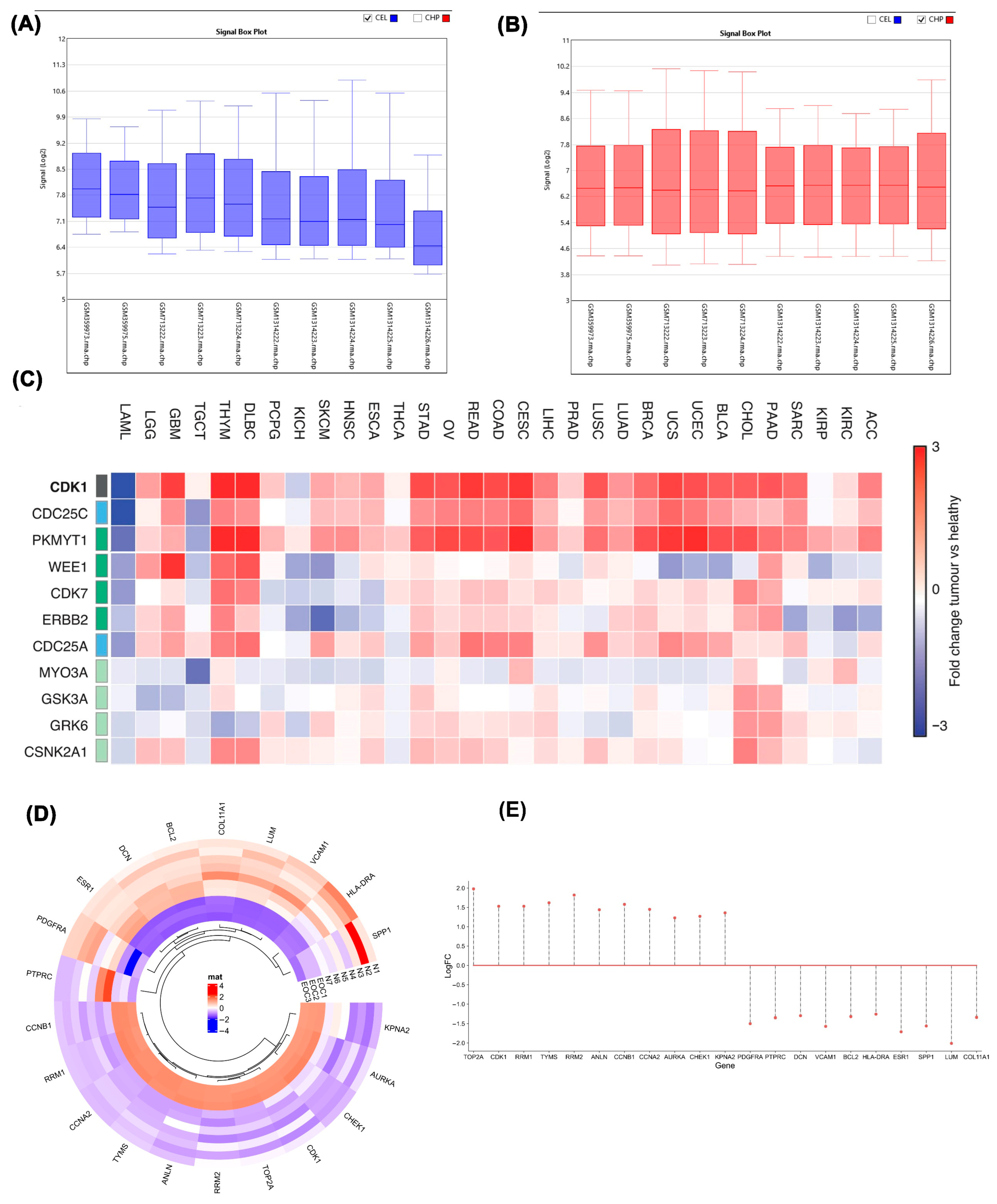
2.1. Microarray Data Analysis
2.2. Differential Expression Analysis of CDK1
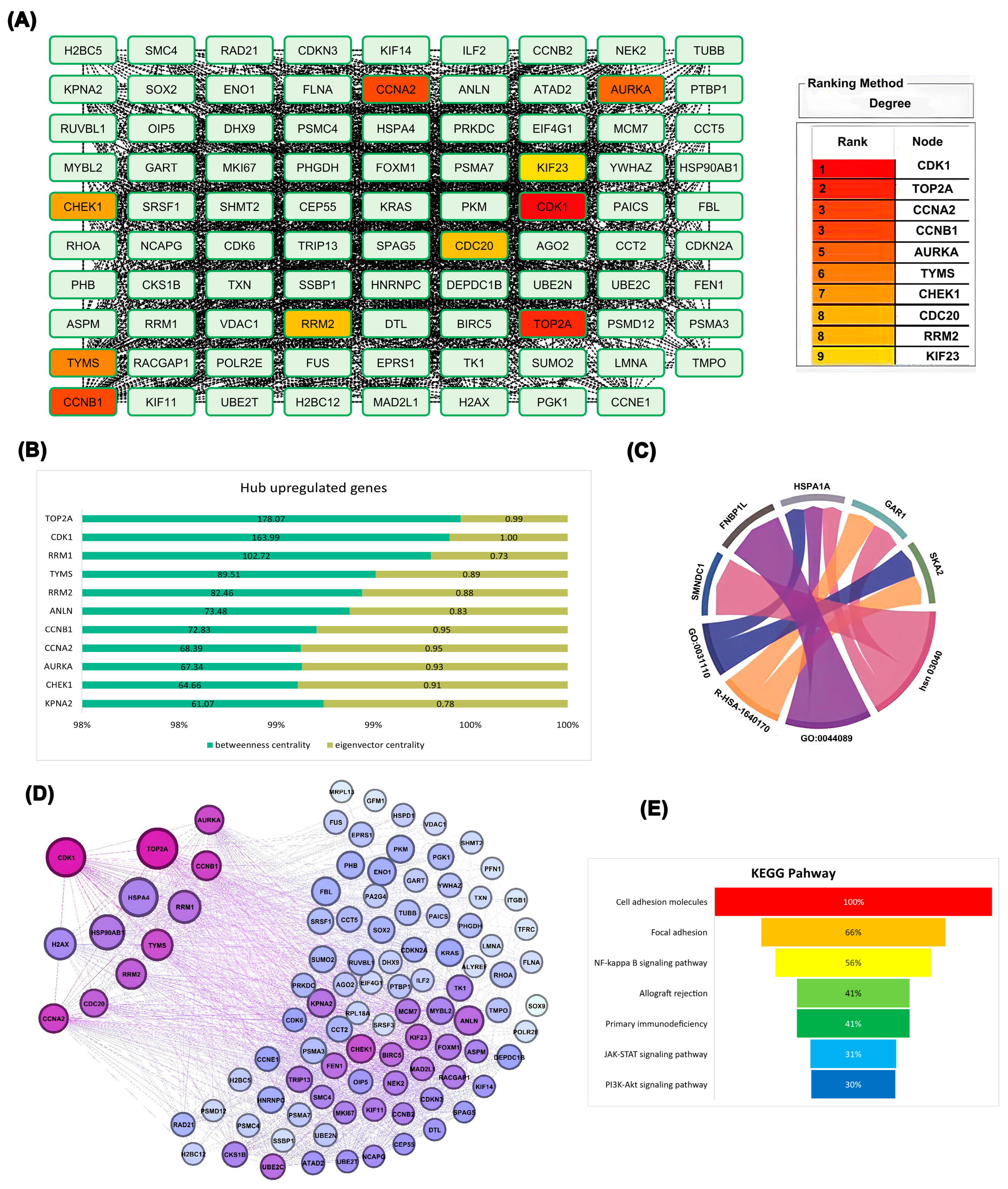
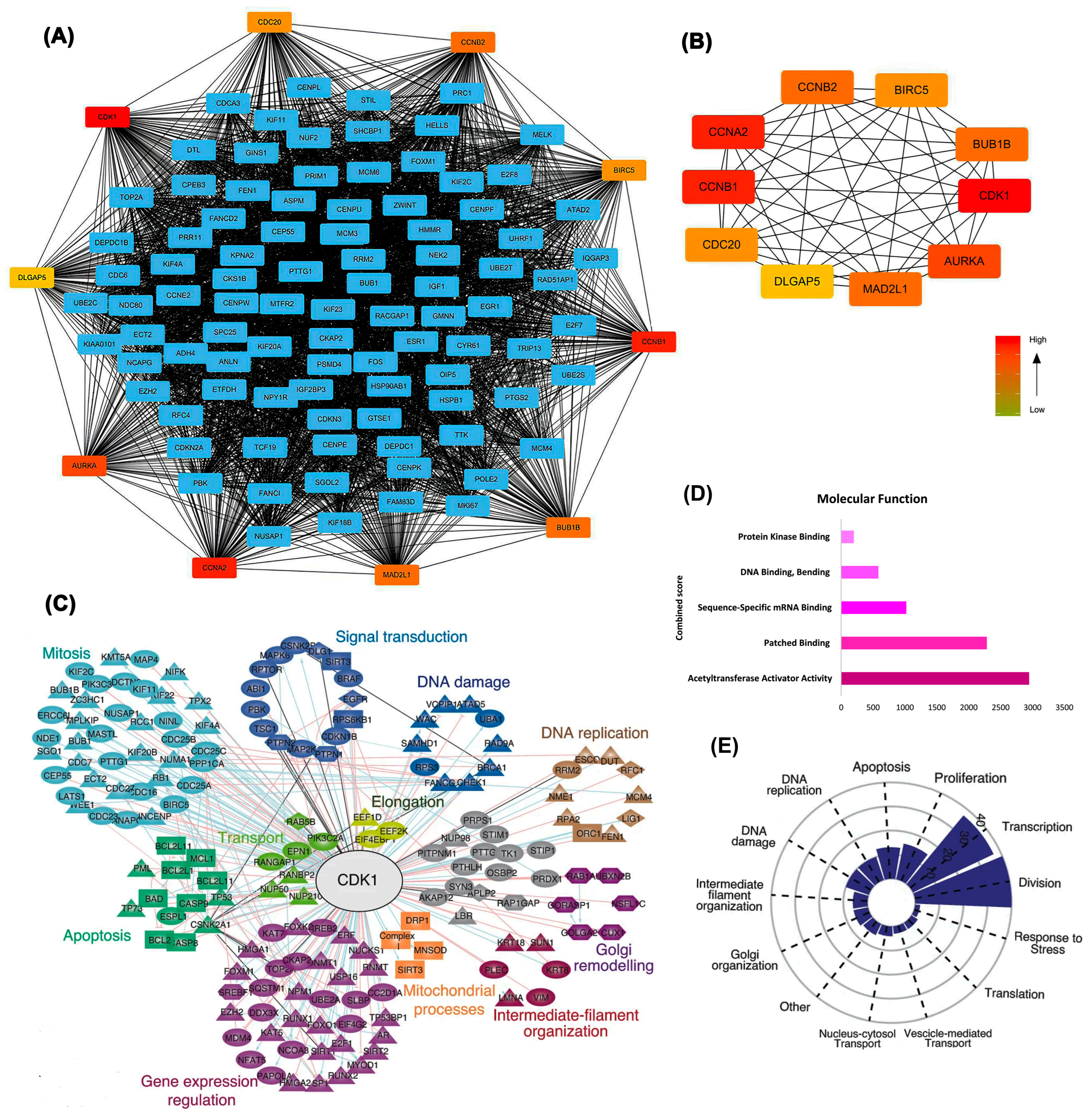
2.3. Protein–Protein Interaction Network Analysis
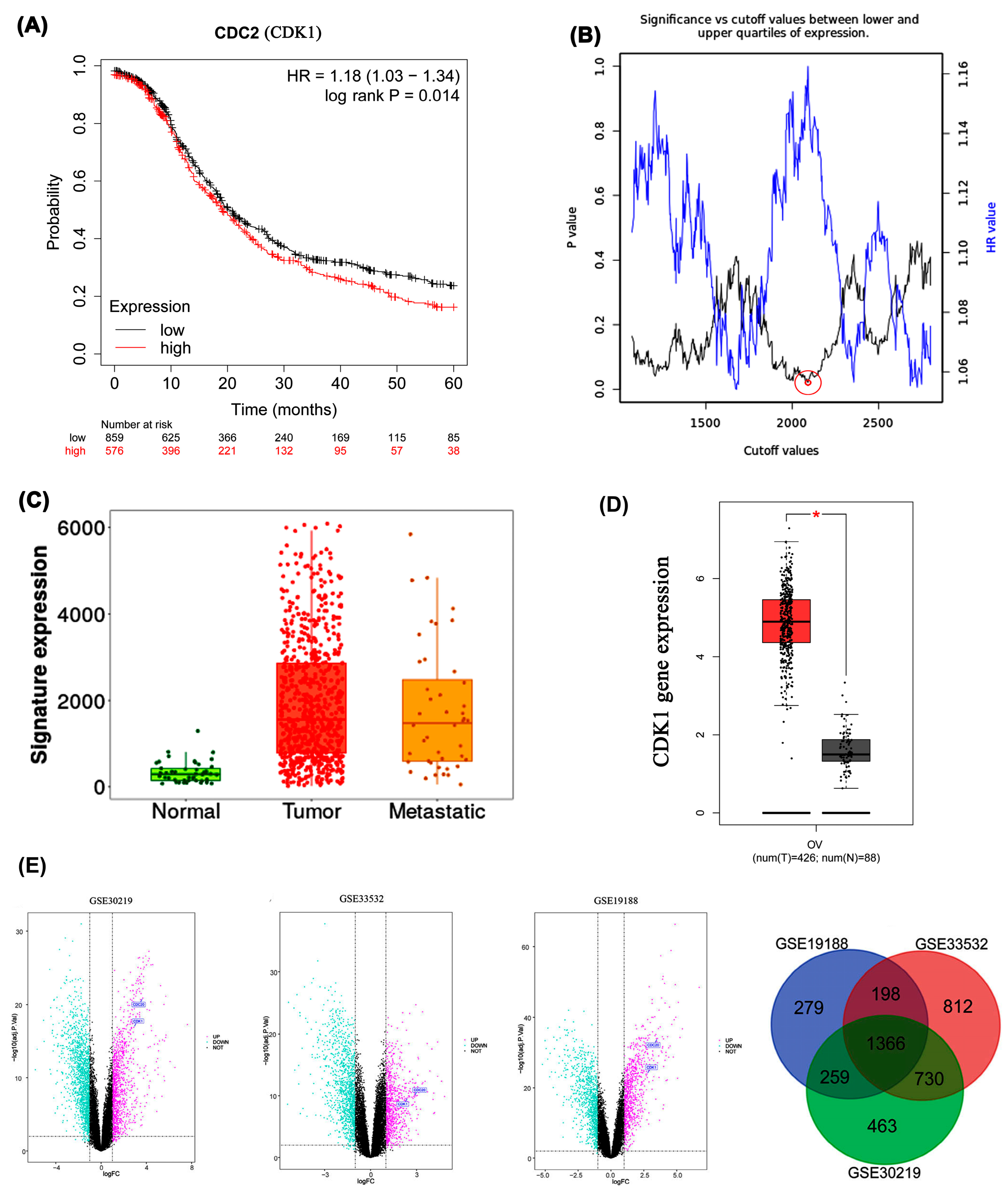
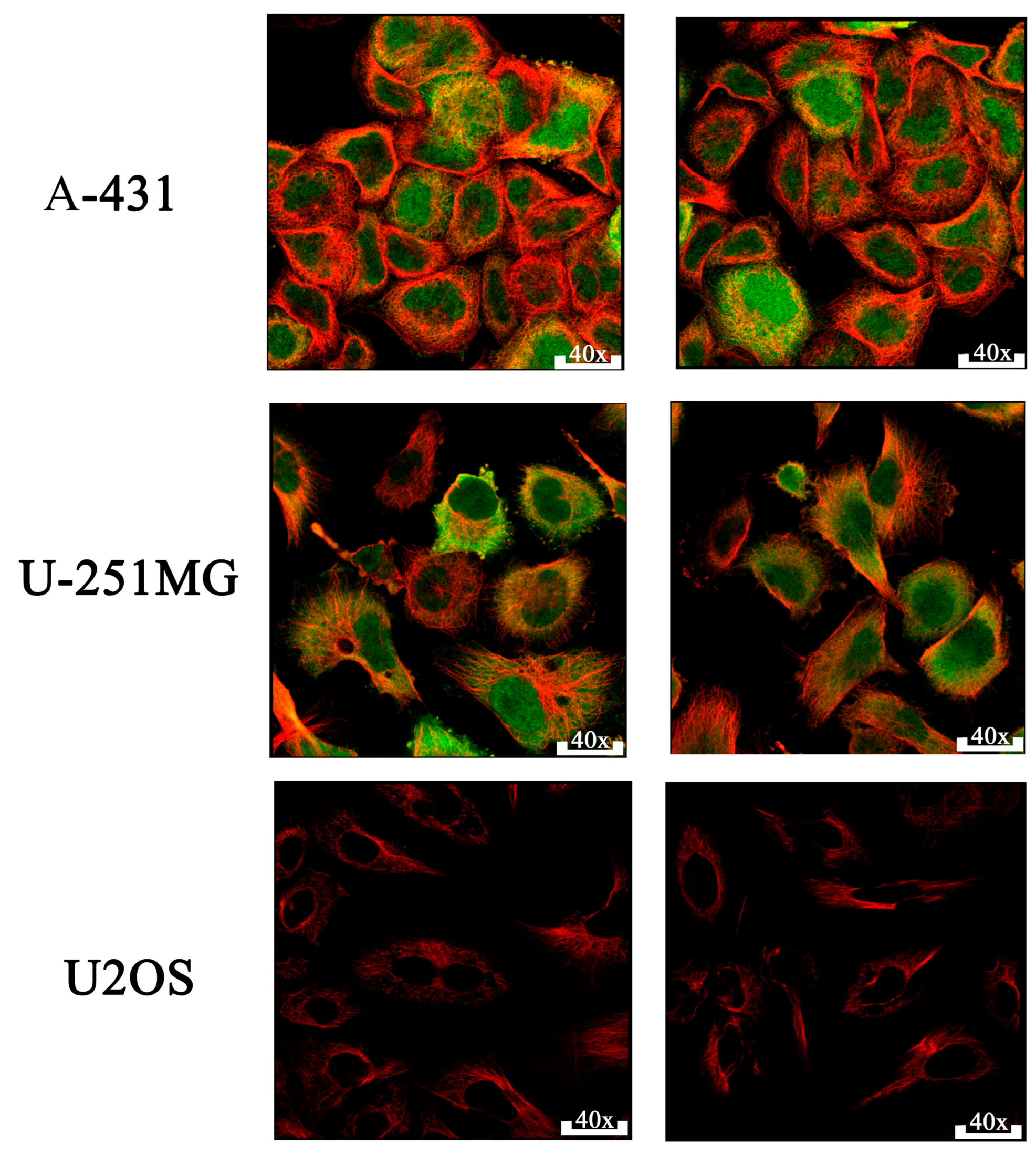
2.4. Proteomic and Survival Analyses
2.5. Molecular Docking Simulation
2.6. Molecular Dynamics Analysis
2.7. Pharmacokinetic Property Analysis
3. Discussion
4. Materials and Methods
4.1. Extraction and Processing of Microarray Data
4.2. Creation of Protein–Protein Interaction Network
4.3. Validation of CDK1 Gene
4.4. Machine Learning in CDK1 Gene Expression
4.5. Pharmacological Effects In Silico
4.6. Statistical Analysis
5. Conclusions
6. Limitations and Future Directions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jiang, Y.T.; Gong, T.T.; Zhang, J.Y.; Li, X.Q.; Gao, S.; Zhao, Y.H.; Wu, Q.J. Infertility and ovarian cancer risk: Evidence from nine prospective cohort studies. Int. J. Cancer 2020, 147, 2121–2130. [Google Scholar] [CrossRef]
- Torre, L.A.; Trabert, B.; DeSantis, C.E.; Miller, K.D.; Samimi, G.; Runowicz, C.D.; Gaudet, M.M.; Jemal, A.; Siegel, R.L. Ovarian cancer statistics, 2018. CA A Cancer J. Clin. 2018, 68, 284–296. [Google Scholar] [CrossRef]
- Sowamber, R.; Lukey, A.; Huntsman, D.; Hanley, G. Ovarian cancer: From precursor lesion identification to population-based prevention programs. Curr. Oncol. 2023, 30, 10179–10194. [Google Scholar] [CrossRef] [PubMed]
- Ghose, A.; Bolina, A.; Mahajan, I.; Raza, S.A.; Clarke, M.; Pal, A.; Sanchez, E.; Rallis, K.S.; Boussios, S. Hereditary ovarian cancer: Towards a cost-effective prevention strategy. Int. J. Environ. Res. Public Health 2022, 19, 12057. [Google Scholar] [CrossRef]
- Pavlidis, N.; Rassy, E.; Vermorken, J.B.; Assi, T.; Kattan, J.; Boussios, S.; Smith-Gagen, J. The outcome of patients with serous papillary peritoneal cancer, fallopian tube cancer, and epithelial ovarian cancer by treatment eras: 27 years data from the SEER registry. Cancer Epidemiol. 2021, 75, 102045. [Google Scholar] [CrossRef] [PubMed]
- Hyriavenko, N.; Lyndin, M.; Sikora, V.; Chyzhma, R.; Lyndina, Y.; Sikora, K.; Awuah, W.A.; Romaniuk, A. Neuroendocrine Tumor of the Fallopian Tube and Serous Adenocarcinoma of the Ovary: Multicentric Primary Tumors. Turk. J. Pathol. 2023, 39, 161–166. [Google Scholar] [CrossRef] [PubMed]
- Yousefi, M.; Dehghani, S.; Nosrati, R.; Ghanei, M.; Salmaninejad, A.; Rajaie, S.; Hasanzadeh, M.; Pasdar, A. Current insights into the metastasis of epithelial ovarian cancer-hopes and hurdles. Cell. Oncol. 2020, 43, 515–538. [Google Scholar] [CrossRef]
- Wang, B.; Ramazzotti, D.; De Sano, L.; Zhu, J.; Pierson, E.; Batzoglou, S. SIMLR: A tool for large-scale genomic analyses by multi-kernel learning. Proteomics 2018, 18, 1700232. [Google Scholar] [CrossRef]
- Della Corte, L.; Conte, C.; Palumbo, M.; Guerra, S.; Colacurci, D.; Riemma, G.; De Franciscis, P.; Giampaolino, P.; Fagotti, A.; Bifulco, G. Hyperthermic Intraperitoneal Chemotherapy (HIPEC): New approaches and controversies on the treatment of Advanced epithelial ovarian Cancer—Systematic review and Meta-analysis. J. Clin. Med. 2023, 12, 7012. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Yang, D.; Yu, H.-F.; Jin, J.; Nie, Y.; Zhang, S.; Ren, W.; Ge, Z.; Zhang, Z.; Ma, X. Copper is essential for cyclin B1-mediated CDK1 activation. Nat. Commun. 2025, 16, 2288. [Google Scholar] [CrossRef]
- Almalki, S.G. The pathophysiology of the cell cycle in cancer and treatment strategies using various cell cycle checkpoint inhibitors. Pathol. Res. Pr. 2023, 251, 154854. [Google Scholar] [CrossRef]
- Zheng, X.F.; Sarkar, A.; Lotana, H.; Syed, A.; Nguyen, H.; Ivey, R.G.; Kennedy, J.J.; Whiteaker, J.R.; Tomasik, B.; Huang, K.; et al. CDK5-cyclin B1 regulates mitotic fidelity. Nature 2024, 633, 932–940. [Google Scholar] [CrossRef]
- Liu, Y.; Gu, X.; Xuan, M.; Lou, N.; Fu, L.; Li, J.; Xue, C. Notch signaling in digestive system cancers: Roles and therapeutic prospects. Cell. Signal. 2024, 124, 111476. [Google Scholar] [CrossRef] [PubMed]
- Farahmand, S.; SamadiAfshar, S.; Khalili, M.; Haji Hosseini, R. Therapeutic implications of Epirubicin-induced miRNA-22 and miRNA-331 upregulation on cell viability and metastatic potential in triple-negative breast cancer. Hum. Gene 2025, 44, 201396. [Google Scholar] [CrossRef]
- Baliyan, D.; Sharma, R.; Goyal, S.; Chhabra, R.; Singh, B. Phytochemical strategies in glioblastoma therapy: Mechanisms, efficacy, and future perspectives. Biochim. et Biophys. Acta (BBA)-Mol. Basis Dis. 2025, 1871, 167647. [Google Scholar] [CrossRef]
- Ma, Y.; Patterson, B.; Zhu, L. Biased signaling in GPCRs: Structural insights and implications for drug development. Pharmacol. Ther. 2025, 266, 108786. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Fan, Y.; Zhao, Z.; Zhang, X.; Tucker, K.; Staley, A.; Suo, H.; Sun, W.; Shen, X.; Deng, B.; et al. Inhibition of CDK1 by RO-3306 Exhibits Anti-Tumorigenic Effects in Ovarian Cancer Cells and a Transgenic Mouse Model of Ovarian Cancer. Int. J. Mol. Sci. 2023, 24, 12375. [Google Scholar] [CrossRef]
- Liu, Y.; Xiong, Y. HADHA promotes ovarian cancer outgrowth via up-regulating CDK1. Cancer Cell Int. 2023, 23, 283. [Google Scholar] [CrossRef]
- Yang, L.; Xie, H.-J.; Li, Y.-Y.; Wang, X.; Liu, X.-X.; Mai, J. Molecular mechanisms of platinum-based chemotherapy resistance in ovarian cancer. Oncol. Rep. 2022, 47, 1–11. [Google Scholar] [CrossRef]
- Alme, C.; Pirim, H.; Akbulut, Y. Machine learning approaches for predicting craniofacial anomalies with graph neural networks. Comput. Biol. Chem. 2025, 115, 108294. [Google Scholar] [CrossRef]
- Wang, J.; Zheng, F.; Wang, D.; Yang, Q. Regulation of ULK1 by WTAP/IGF2BP3 axis enhances mitophagy and progression in epithelial ovarian cancer. Cell Death Dis. 2024, 15, 97. [Google Scholar] [CrossRef]
- Shen, J.; Gong, X.; Tan, S.; Zhang, Y.; Xia, R.; Xu, S.; Wang, S.; Zhou, H.; Jiang, Y.; Zhao, T.; et al. CDK1 Acts as a Prognostic Biomarker Associated with Immune Infiltration in Pan-Cancer, Especially in Gastrointestinal Tumors. Curr. Med. Chem. 2025, 32, 4836–4857. [Google Scholar] [CrossRef]
- Che Ismail, C.L.; Yusof, N.Y.; Mat Lazim, N.; Sarina, S.; Alwi, Z.B.; Abdullah, B. Exploring nasopharyngeal carcinoma genetics: Bioinformatics insights into pathways and gene associations. Med. J. Malays. 2024, 79, 615–625. [Google Scholar] [PubMed]
- Matthews, H.K.; Bertoli, C.; de Bruin, R.A. Cell cycle control in cancer. Nat. Rev. Mol. Cell Biol. 2022, 23, 74–88. [Google Scholar] [CrossRef] [PubMed]
- Xiao, C.; Gong, J.; Jie, Y.; Liang, W.; Tai, Y.; Qin, W.; Lu, T.; Chong, Y.; Hei, Z.; Hu, B.; et al. E2F1-mediated Up-regulation of NCAPG Promotes Hepatocellular Carcinoma Development by Inhibiting Pyroptosis. J. Clin. Transl. Hepatol. 2024, 12, 25–35. [Google Scholar] [CrossRef] [PubMed]
- Javed, A.; Özduman, G.; Altun, S.; Duran, D.; Yerli, D.; Özar, T.; Şimşek, F.; Korkmaz, K.S. Mitotic Kinase Inhibitors as Therapeutic Interventions for Prostate Cancer: Evidence from In Vitro Studies. Endocr. Metab. Immune Disord.-Drug Targets 2023, 23, 1699–1712. [Google Scholar] [CrossRef]
- Aliyuda, F.; Moschetta, M.; Ghose, A.; Sofia Rallis, K.; Sheriff, M.; Sanchez, E.; Rassy, E.; Boussios, S. Advances in ovarian cancer treatment beyond PARP inhibitors. Curr. Cancer Drug Targets 2023, 23, 433–446. [Google Scholar] [CrossRef]
- Qin, G.; Tu, X.; Li, H.; Cao, P.; Chen, X.; Song, J.; Han, H.; Li, Y.; Guo, B.; Yang, L. Long noncoding RNA p53-stabilizing and activating RNA promotes p53 signaling by inhibiting heterogeneous nuclear ribonucleoprotein K deSUMOylation and suppresses hepatocellular carcinoma. Hepatology 2020, 71, 112–129. [Google Scholar] [CrossRef] [PubMed]
- Ghandikota, S.K.; Jegga, A.G. Application of artificial intelligence and machine learning in drug repurposing. In Progress in Molecular Biology and Translational Science; Elsevier: Amsterdam, The Netherlands, 2024; Volume 205, pp. 171–211. [Google Scholar] [CrossRef]
- Zhang, R.; Shi, H.; Ren, F.; Zhang, M.; Ji, P.; Wang, W.; Liu, C. The aberrant upstream pathway regulations of CDK1 protein were implicated in the proliferation and apoptosis of ovarian cancer cells. J. Ovarian Res. 2017, 10, 60. [Google Scholar] [CrossRef] [PubMed]
- Cedeno-Rosario, L.; Honda, D.; Sunderland, A.M.; Lewandowski, M.D.; Taylor, W.R.; Chadee, D.N. Phosphorylation of mixed lineage kinase MLK3 by cyclin-dependent kinases CDK1 and CDK2 controls ovarian cancer cell division. J. Biol. Chem. 2022, 298, 102263. [Google Scholar] [CrossRef]
- Schmidt, M.; Rohe, A.; Platzer, C.; Najjar, A.; Erdmann, F.; Sippl, W. Regulation of G2/M transition by inhibition of WEE1 and PKMYT1 kinases. Molecules 2017, 22, 2045. [Google Scholar] [CrossRef]
- Roskoski Jr, R. Cyclin-dependent protein serine/threonine kinase inhibitors as anticancer drugs. Pharmacol. Res. 2019, 139, 471–488. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zhi, X.; Shen, X.; Chen, C.; Yuan, L.; Dong, X.; Zhu, C.; Yao, L.; Chen, M. Depletion of UBE2C reduces ovarian cancer malignancy and reverses cisplatin resistance via downregulating CDK1. Biochem. Biophys. Res. Commun. 2020, 523, 434–440. [Google Scholar] [CrossRef] [PubMed]
- Soni, D.V.; Jacobberger, J.W. Inhibition of cdk1 by alsterpaullone and thioflavopiridol correlates with increased transit time from mid G2 through prophase. Cell Cycle 2004, 3, 347–355. [Google Scholar] [CrossRef]
- Senturk, A.; Sahin, A.T.; Armutlu, A.; Kiremit, M.C.; Acar, O.; Erdem, S.; Bagbudar, S.; Esen, T.; Ozlu, N. Quantitative Phosphoproteomics Analysis Uncovers PAK2-and CDK1-Mediated Malignant Signaling Pathways in Clear Cell Renal Cell Carcinoma. Mol. Cell. Proteom. 2022, 21, 100417. [Google Scholar] [CrossRef]
- Rajgopal, A.; Young, D.W.; Mujeeb, K.A.; Stein, J.L.; Lian, J.B.; Van Wijnen, A.J.; Stein, G.S. Mitotic control of RUNX2 phosphorylation by both CDK1/cyclin B kinase and PP1/PP2A phosphatase in osteoblastic cells. J. Cell. Biochem. 2007, 100, 1509–1517. [Google Scholar] [CrossRef]
- Coley, H.; Safuwan, N.A.M.; Chivers, P.; Papacharalbous, E.; Giannopoulos, T.; Butler-Manuel, S.; Madhuri, K.; Lovell, D.; Crook, T. The cyclin-dependent kinase inhibitor p57Kip2 is epigenetically regulated in carboplatin resistance and results in collateral sensitivity to the CDK inhibitor seliciclib in ovarian cancer. Br. J. Cancer 2012, 106, 482–489. [Google Scholar] [CrossRef][Green Version]
- Chen, J.; Jia, X.; Li, Z.; Song, W.; Jin, C.; Zhou, M.; Xie, H.; Zheng, S.; Song, P. Targeting WEE1 by adavosertib inhibits the malignant phenotypes of hepatocellular carcinoma. Biochem. Pharmacol. 2021, 188, 114494. [Google Scholar] [CrossRef]
- Noguchi, R.; Yoshimatsu, Y.; Ono, T.; Sei, A.; Motoi, N.; Yatabe, Y.; Yoshida, Y.; Watanabe, S.; Kondo, T. Establishment and characterization of NCC-DMM1-C1, a novel patient-derived cell line of desmoplastic malignant pleural mesothelioma. Oncol. Lett. 2022, 23, 1–9. [Google Scholar] [CrossRef]
- Zhu, J.; Lin, S.; Zou, X.; Chen, X.; Liu, Y.; Yang, X.; Gao, J.; Zhu, H. Mechanisms of autophagy and endoplasmic reticulum stress in the reversal of platinum resistance of epithelial ovarian cancer cells by naringin. Mol. Biol. Rep. 2023, 50, 6457–6468. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Rodriguez, L.; Cianciaruso, C.; Bill, R.; Trefny, M.P.; Klar, R.; Kirchhammer, N.; Buchi, M.; Festag, J.; Michel, S.; Kohler, R.H. Dual TLR9 and PD-L1 targeting unleashes dendritic cells to induce durable antitumor immunity. J. Immunother. Cancer 2023, 11, e006714, Erratum in J. Immunother. Cancer 2023, 11, e006714corr1. [Google Scholar] [CrossRef]
- Xiaowei, W.; Tong, L.; Yanjun, Q.; Lili, F. PTH2R is related to cell proliferation and migration in ovarian cancer: A multi-omics analysis of bioinformatics and experiments. Cancer Cell Int. 2022, 22, 148. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Gable, A.L.; Fang, T.; Doncheva, N.T.; Pyysalo, S. The STRING database in 2023: Protein–protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023, 51, D638–D646. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.; Li, C.; Kang, B.; Gao, G.; Li, C.; Zhang, Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017, 45, W98–W102. [Google Scholar] [CrossRef]
- Bartha, Á.; Győrffy, B. TNMplot. com: A web tool for the comparison of gene expression in normal, tumor and metastatic tissues. Int. J. Mol. Sci. 2021, 22, 2622. [Google Scholar] [CrossRef] [PubMed]
- de Bruijn, I.; Kundra, R.; Mastrogiacomo, B.; Tran, T.N.; Sikina, L.; Mazor, T.; Li, X.; Ochoa, A.; Zhao, G.; Lai, B. Analysis and visualization of longitudinal genomic and clinical data from the AACR project GENIE biopharma collaborative in cBioPortal. Cancer Res. 2023, 83, 3861–3867. [Google Scholar] [CrossRef] [PubMed]
- Győrffy, B. Integrated analysis of public datasets for the discovery and validation of survival-associated genes in solid tumors. Innovation 2024, 5, 100625. [Google Scholar] [CrossRef]
- Thareja, P.; Chhillar, R.S.; Dalal, S.; Simaiya, S.; Lilhore, U.K.; Alroobaea, R.; Alsafyani, M.; Baqasah, A.M.; Algarni, S. Intelligence model on sequence-based prediction of PPI using AISSO deep concept with hyperparameter tuning process. Sci. Rep. 2024, 14, 21797. [Google Scholar] [CrossRef]
- Burley, S.K.; Bhikadiya, C.; Bi, C.; Bittrich, S.; Chao, H.; Chen, L.; Craig, P.A.; Crichlow, G.V.; Dalenberg, K.; Duarte, J.M. RCSB Protein Data Bank (RCSB. org): Delivery of experimentally-determined PDB structures alongside one million computed structure models of proteins from artificial intelligence/machine learning. Nucleic Acids Res. 2023, 51, D488–D508. [Google Scholar] [CrossRef]
- Farahmand, S.; SamadiAfshar, S.; Hosseini, L. TA-Cloning for Diabetes Treatment: Expressing Corynebacterium Malic Enzyme Gene in E. coli. Curr. Microbiol. 2024, 81, 167. [Google Scholar] [CrossRef]
- Hatami, S.; Sirous, H.; Mahnam, K.; Najafipour, A.; Fassihi, A. Preparing a database of corrected protein structures important in cell signaling pathways. Res. Pharm. Sci. 2023, 18, 67. [Google Scholar] [CrossRef] [PubMed]
- Samadiafshar, S.; Nikakhtar, A.; Samadiafshar, S.; Garmsiri, N.; Garmsiri, F.; Azizi, R.; Farahmand, S. In silico evaluation of microRNAs in kidney renal clear cell carcinoma and drugs effects in increasing apoptosis by docking. Acta Cell Biol. 2025, 1, 11–16. [Google Scholar]
- Yang, J.; Roy, A.; Zhang, Y. Protein–ligand binding site recognition using complementary binding-specific substructure comparison and sequence profile alignment. Bioinformatics 2013, 29, 2588–2595. [Google Scholar] [CrossRef]
- Handsel, J.; Matthews, B.; Knight, N.J.; Coles, S.J. Translating the InChI: Adapting neural machine translation to predict IUPAC names from a chemical identifier. J. Cheminform. 2021, 13, 79. [Google Scholar] [CrossRef]
- Eberhardt, J.; Santos-Martins, D.; Tillack, A.F.; Forli, S. AutoDock Vina 1.2.0: New docking methods, expanded force field, and python bindings. J. Chem. Inf. Model. 2021, 61, 3891–3898. [Google Scholar] [CrossRef]
- Dallakyan, S.; Olson, A.J. Small-molecule library screening by docking with PyRx. Chem. Biol. Methods Protoc. 2015, 1263, 243–250. [Google Scholar] [CrossRef]
- Adasme, M.F.; Linnemann, K.L.; Bolz, S.N.; Kaiser, F.; Salentin, S.; Haupt, V.J.; Schroeder, M. PLIP 2021: Expanding the scope of the protein–ligand interaction profiler to DNA and RNA. Nucleic Acids Res. 2021, 49, W530–W534. [Google Scholar] [CrossRef]
- Yang, H.; Lou, C.; Sun, L.; Li, J.; Cai, Y.; Wang, Z.; Li, W.; Liu, G.; Tang, Y. admetSAR 2.0: Web-service for prediction and optimization of chemical ADMET properties. Bioinformatics 2019, 35, 1067–1069. [Google Scholar] [CrossRef]
- Azad, H.; Akbar, M.Y.; Sarfraz, J.; Haider, W.; Ghazanfar, S. Simulation studies to identify high-affinity probiotic peptides for inhibiting PAK1 gastric cancer protein: A comparative approach. Comput. Biol. Chem. 2025, 115, 108345. [Google Scholar] [CrossRef]
- SamadiAfshar, S.; NikAkhtar, A.; SamadiAfshar, S.; Farahmand, S. Antibacterial Property of Silver Nanoparticles Green Synthesized from Stachys schtschegleevii Plant Extract on Urinary Tract Infection Bacteria. Curr. Microbiol. 2024, 81, 135. [Google Scholar] [CrossRef] [PubMed]
- Farahmand, S.; SamadiAfshar, S.; Shahmoradi, N.A. Electrospun PCL/PVP Nanofibers Meshes, A Novel Bisabolol Delivery System for Antidermatophytic Treatment. J. Pharm. Innov. 2025, 20, 25. [Google Scholar] [CrossRef]


| Prediction | CDK1 (4y72) | WEE1 (8bju) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Drug | Binding Affinity (kcal/mol) | Binding Site | Hydrophobic Interactions | Hydrogen Bonds | Salt Bridges | Binding Affinity (kcal/mol) | Binding Site | Hydrophobic Interactions | Hydrogen Bonds | Salt Bridges | |
| Adavosertib | * | * | * | * | * | −11 | ASN376/CYS379 | 8 | 4 | 1 | |
| Alsterpaullone | −10.9 | TYR 15/ASP 86 | 8 | 0 | 0 | −10.4 | CYS379 | 8 | 2 | 0 | |
| Avotaciclib | −9.3 | TYR 15/GLN 132 | 5 | 3 | 1 | −8.7 | ASN376/CYS379 | 4 | 5 | 0 | |
| Fostamatinib | −12.5 | TYR 15 | 7 | 6 | 0 | −7.5 | ASN376 | 1 | 4 | 2 | |
| Olomoucine | −8.5 | TYR 15/GLN 132 | 6 | 4 | 1 | * | * | * | * | * | |
| Seliciclib | −8.7 | TYR 15/GLN 132 | 10 | 4 | 0 | * | * | * | * | * | |
| Naringin | −10.6 | TYR 15/GLN 132 | 6 | 8 | 0 | −9.6 | ASN376/CYS379 | 5 | 5 | 1 | |
| Name of the Drug | Admet SAR | |||||||
|---|---|---|---|---|---|---|---|---|
| Subcellular Localization | AlogP | Molecular Weight | Blood–Brain Barrier | Human Oral Bioavailability | Nephrotoxicity | Hepatotoxicity | Ames Mutagenesis | |
| Adavosertib | Mitochondria | 2.89 | 500.607 | + | 0.6429(+) | 0.7773(−) | 0.7075(+) | 0.54(−) |
| Alsterpaullone | Mitochondria | 3.24 | 293.276 | + | 0.7143(+) | 0.5739(+) | 0.7875(+) | 0.88(+) |
| Avotaciclib | Mitochondria | 0.87 | 281.279 | + | 0.5571(+) | 0.4864(+) | 0.6125(+) | 0.5(−) |
| Fostamatinib | Mitochondria | 3.09 | 580.459 | − | 0.5571(+) | 0.7326(+) | 0.6677(+) | 0.53(−) |
| Olomoucine | Nucleus | 1.36 | 298.343 | + | 0.5714(−) | 0.5939(−) | 0.5587(+) | 0.58(+) |
| Seliciclib | Lysosomes | 3.2 | 354.449 | + | 0.5143(+) | 0.7729(−) | 0.5538(−) | 0.59(−) |
| Naringin | Mitochondria | −1.17 | 580.54 | − | 0.9857(−) | 0.6977(−) | 0.8750(−) | 0.61(−) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Masoudi, M.; Samadiafshar, S.; Azizi, H.; Skutella, T. Integrated Analysis, Machine Learning, Molecular Docking and Dynamics of CDK1 Inhibitors in Epithelial Ovarian Cancer: A Multifaceted Approach Towards Targeted Therapy. Int. J. Mol. Sci. 2025, 26, 9168. https://doi.org/10.3390/ijms26189168
Masoudi M, Samadiafshar S, Azizi H, Skutella T. Integrated Analysis, Machine Learning, Molecular Docking and Dynamics of CDK1 Inhibitors in Epithelial Ovarian Cancer: A Multifaceted Approach Towards Targeted Therapy. International Journal of Molecular Sciences. 2025; 26(18):9168. https://doi.org/10.3390/ijms26189168
Chicago/Turabian StyleMasoudi, Mahla, Saber Samadiafshar, Hossein Azizi, and Thomas Skutella. 2025. "Integrated Analysis, Machine Learning, Molecular Docking and Dynamics of CDK1 Inhibitors in Epithelial Ovarian Cancer: A Multifaceted Approach Towards Targeted Therapy" International Journal of Molecular Sciences 26, no. 18: 9168. https://doi.org/10.3390/ijms26189168
APA StyleMasoudi, M., Samadiafshar, S., Azizi, H., & Skutella, T. (2025). Integrated Analysis, Machine Learning, Molecular Docking and Dynamics of CDK1 Inhibitors in Epithelial Ovarian Cancer: A Multifaceted Approach Towards Targeted Therapy. International Journal of Molecular Sciences, 26(18), 9168. https://doi.org/10.3390/ijms26189168








