Regulation of NR4A2 Gene Expression and Its Importance in Neurodegenerative and Psychiatric Diseases
Abstract
1. Introduction
2. Methods
3. NR4A2 Protein
4. NR4A2 Gene Expression
5. Expression Regulation NR4A2
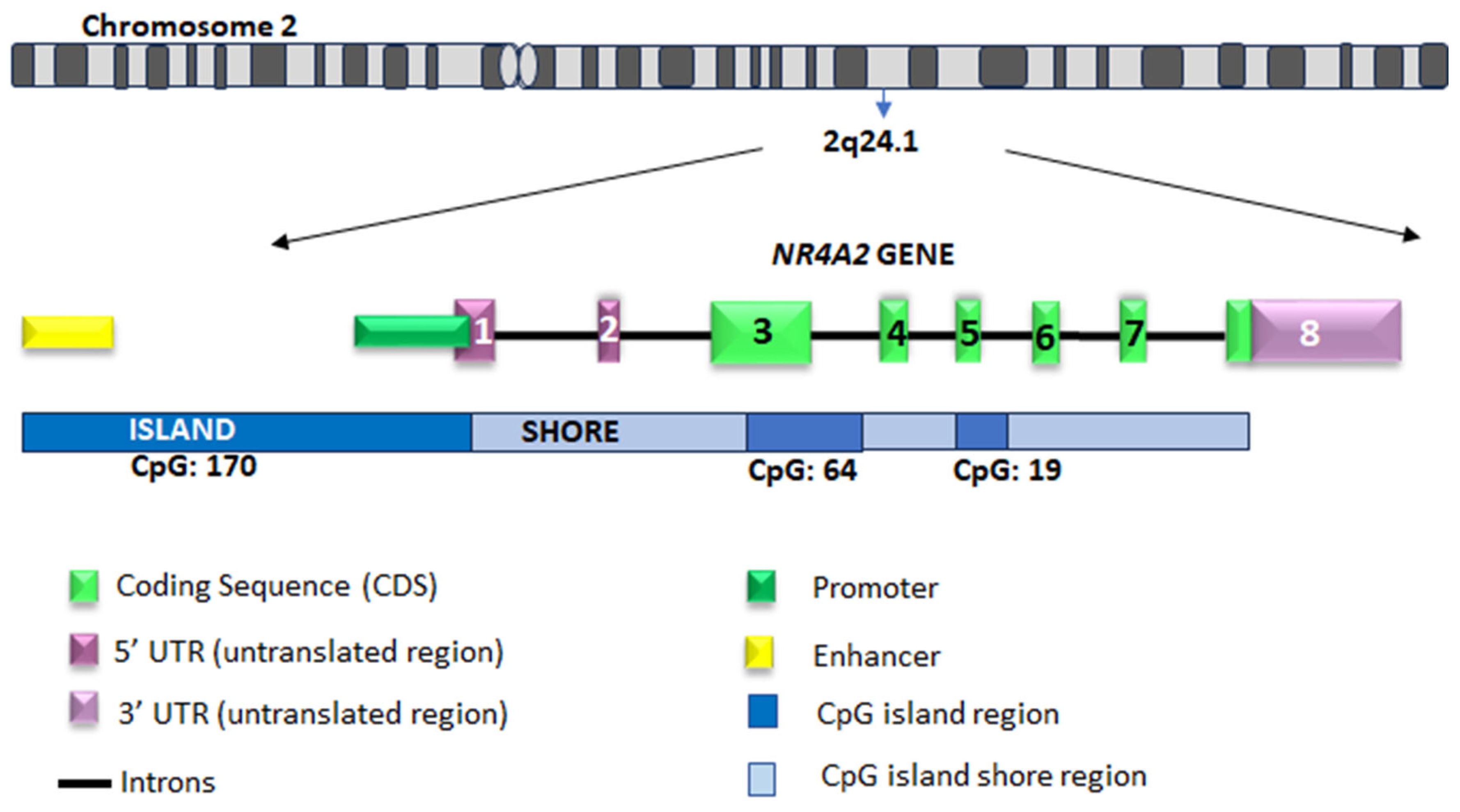
5.1. NR4A2 Gene and Cis Regulatory Elements (Promoter)
5.1.1. NR4A2 Gene
5.1.2. Promoter Region
5.1.3. NR4A2 Genetic Variants
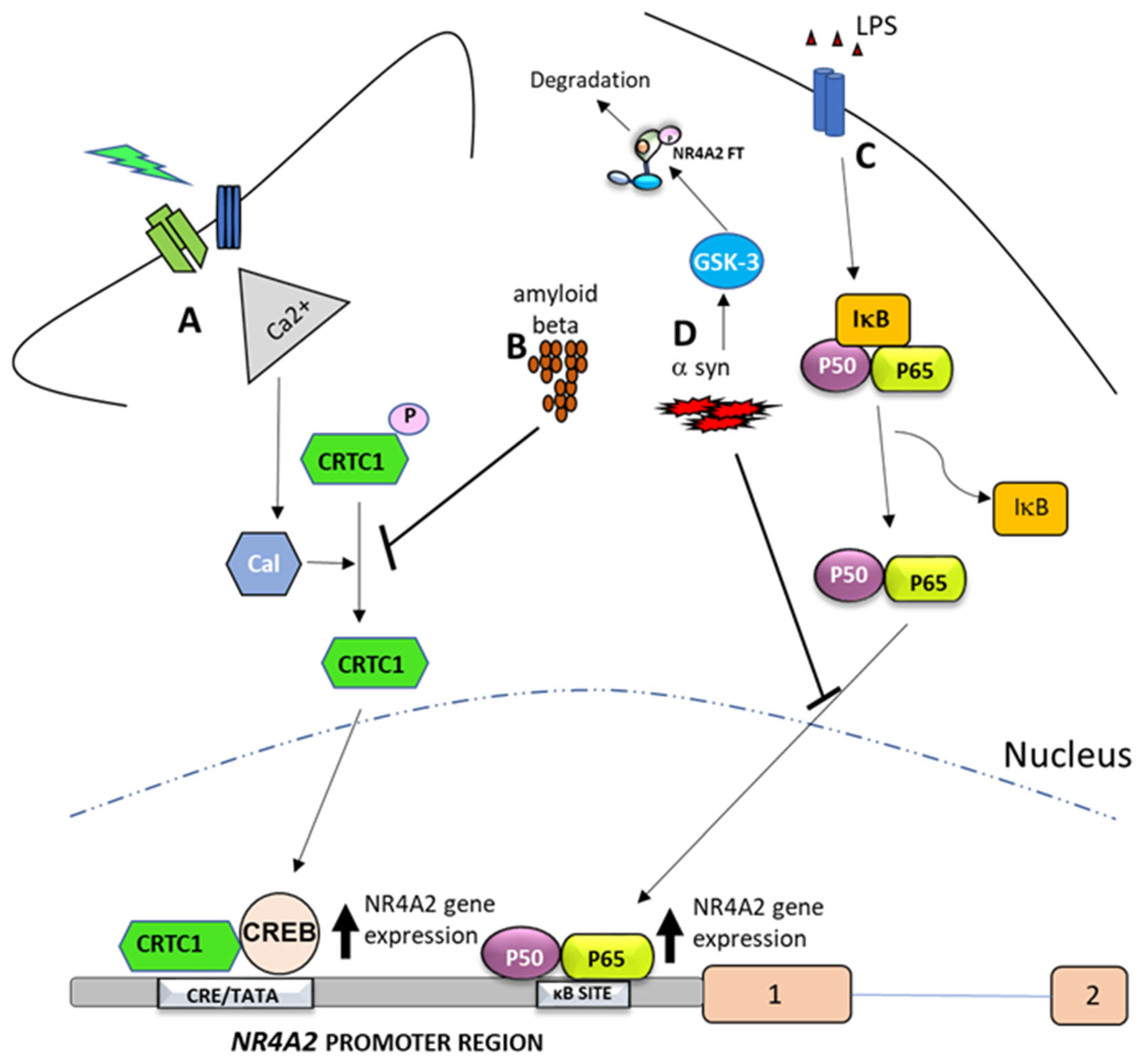
5.2. Transcription Factors Regulating NR4A2 Gene Expression
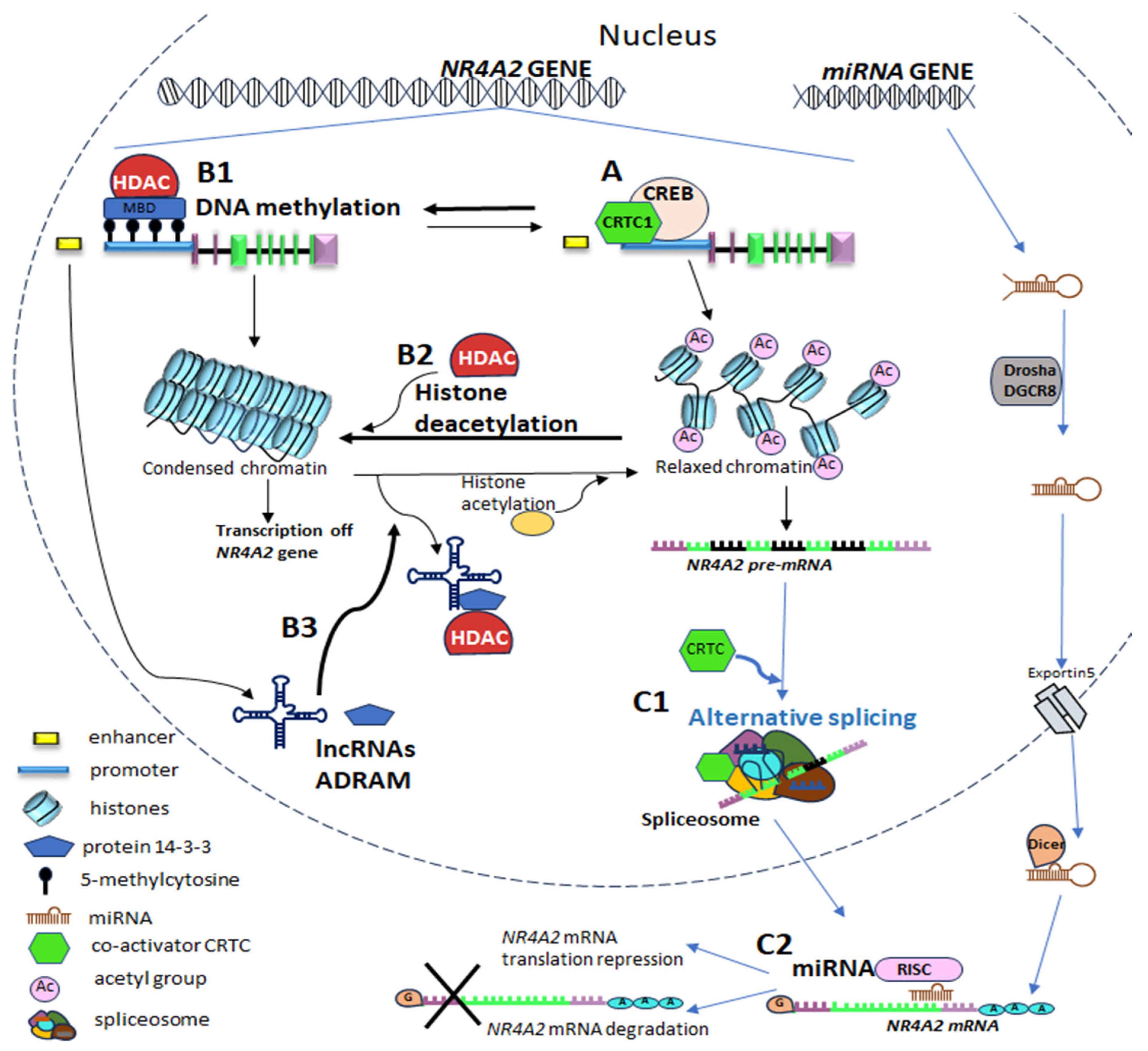
5.3. Epigenetic Mechanisms in NRA42 Gene Expression Regulation
5.3.1. DNA Methylation
5.3.2. NR4A2 Genetic Expression Through Histone Modification
5.3.3. Epigenetic Regulation of NR4A2 Gene Expression by Non-Coding RNA
5.4. Post-Transcriptional Mechanisms in NRA42 Gene Expression Regulation
5.4.1. Alternative Splicing of NR4A2 Pre-RNA and Its Regulation by CRTC
5.4.2. NR4A2 Gene Expression Through MicroRNAs
6. Dysregulation of the Expression of the NR4A2 Gene in Animal Models and in Clinical Studies of Neurological and Psychiatric Disorders
6.1. Parkinson′s Disease
6.2. Alzheimer′s Disease
6.3. Neurodevelopmental Disorders
6.4. Schizophrenia
6.5. Major Depressive Disorder
6.6. Substance Use Disorders
7. Therapeutic Potential of Modulating NR4A2 Gene Expression in Neurodegenerative Diseases and Psychiatric Disorders
8. Discussion
9. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| Aβ | amyloid β |
| AD | Alzheimer′s disease |
| ADHD | Attention-deficit hyperactivity disorder |
| AF1 | Ligand-independent activation function domain 1 |
| AF2 | Ligand-dependent activation function domain 2 |
| ASD | Autism spectrum disorder |
| BDNF | Brain-derived neurotrophic factor |
| CNS | Central nervous system |
| CP | Cognitively preserved |
| CI | Cognitive impaired |
| CNVs | Copy number variants |
| CREs | Cis-regulatory elements |
| CREB | cAMP response element-binding protein |
| CRTC | CREB-regulated transcription co-activators |
| CRTC1 | CREB regulated transcription co-activator 1 |
| CTCF | CCCTC-binding factor |
| DAergic | Dopaminergic neurons |
| DBD | DNA-binding domain |
| DMR | Differential methylation region |
| DR | Dorsal raphe nucleus |
| DSM-5-TR | Diagnostic and Statistical Manual of Mental Disorders |
| eRNA | Enhancer-derived RNA |
| GSK-3 | Glycogen synthase kinase 3 |
| HDAC | Histone deacetylases |
| HDAC 3 | Histone deacetylases 3 |
| IEG | Immediate early gene |
| ID | Intellectual disability |
| LBD | Ligand-binding site |
| LBD | Ligand-binding domain |
| LD | Linkage disequilibrium |
| lncRNAs | long non-coding RNAs |
| MDD | Major depressive disorder |
| mPFC | Medial prefrontal cortex |
| 5mC | 5-methylcytosine |
| miRNAs | microRNAs |
| miRNA-132 | microRNA 132 |
| miRNA-145-5p | microRNA-145-5p |
| miRNA-34 | microRNA-34 |
| mRNA | Messenger ribonucleic acid |
| MHb | Medial habenula |
| NAc | Nucleus accumbens |
| ncRNAs | Non-coding RNAs |
| NDDs | Neurodevelopmental disorders |
| NF-κB | Nuclear Factor Kappa B |
| NR4A2 | Nuclear receptor subfamily 4 group A member 2 |
| NR4A2 mRNA | messenger RNA transcript of NR4A2 |
| PD | Parkinson’s disease |
| PFC | Prefrontal cortex |
| piRNAs | Piwi-interacting RNAs |
| 5-HT | Serotonin |
| SZ | Schizophrenia |
| SNVs | Single nucleotide variants |
| SUDs | Substance use disorders |
| TF | Transcription factor |
| UTR | 5′-untranslated region |
| WHO | World Health Organization |
References
- Dodat, F.; Mader, S.; Lévesque, D. Minireview: What is Known about SUMOylation Among NR4A Family Members? J. Mol. Biol. 2021, 433, 167212. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Calvo, R.; Tajes, M.; Vázquez-Carrera, M. The NR4A subfamily of nuclear receptors: Potential new therapeutic targets for the treatment of inflammatory diseases. Expert. Opin. Ther. Targets 2017, 21, 291–304. [Google Scholar] [CrossRef]
- Odagiu, L.; May, J.; Boulet, S.; Baldwin, T.A.; Labrecque, N. Role of the Orphan Nuclear Receptor NR4A Family in T-Cell Biology. Front. Endocrinol. 2021, 11, 624122. [Google Scholar] [CrossRef]
- Herring, J.A.; Elison, W.S.; Tessem, J.S. Function of Nr4a Orphan Nuclear Receptors in Proliferation, Apoptosis and Fuel Utilization Across Tissues. Cells 2019, 8, 1373. [Google Scholar] [CrossRef]
- Safe, S. Natural products and synthetic analogs as selective orphan nuclear receptor 4A (NR4A) modulators. Histol. Histopathol. 2024, 39, 543–556. [Google Scholar] [CrossRef] [PubMed]
- Hashida, R.; Kawabata, T. Structural Perspective of NR4A Nuclear Receptor Family and Their Potential Endogenous Ligands. Biol. Pharm. Bull. 2024, 47, 580–590. [Google Scholar] [CrossRef]
- Phelan, D.E.; Shigemura, M.; Aldhafiri, S.; Mota, C.; Hall, T.J.; Sznajder, J.I.; Murphy, E.P.; Crean, D.; Cummins, E.P. Transcriptional Profiling of Monocytes Deficient in Nuclear Orphan Receptors NR4A2 and NR4A3 Reveals Distinct Signalling Roles Related to Antigen Presentation and Viral Response. Front. Immunol. 2021, 12, 676644. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Wang, Q.; Liu, W.; Liu, F.; Ji, A.; Li, Y. The Orphan Nuclear Receptor 4A1: A Potential New Therapeutic Target for Metabolic Diseases. J. Diabetes Res. 2018, 2018, 9363461. [Google Scholar] [CrossRef]
- Chen, L.; Fan, F.; Wu, L.; Zhao, Y. The nuclear receptor 4A family members: Mediators in human disease and autophagy. Cell Mol. Biol. Lett. 2020, 25, 48. [Google Scholar] [CrossRef]
- Zhao, S.; Wang, X.; Huang, F.; Zhou, Y.; Meng, D.; Zhao, D.; Wang, J.; Zhang, H.; Wu, L.; Zhang, Y.; et al. A role of NR4A2 in Graves’ disease: Regulation of Th17/Treg. Endocrine 2024, 83, 432–441. [Google Scholar] [CrossRef]
- Alavian, K.N.; Jeddi, S.; Naghipour, S.I.; Nabili, P.; Licznerski, P.; Tierney, T.S. The lifelong maintenance of mesencephalic dopaminergic neurons by Nurr1 and engrailed. J. Biomed. Sci. 2014, 21, 27. [Google Scholar] [CrossRef]
- Speranza, L.; di Porzio, U.; Viggiano, D.; de Donato, A.; Volpicelli, F. Dopamine: The Neuromodulator of Long-Term Synaptic Plasticity, Reward and Movement Control. Cells 2021, 10, 735. [Google Scholar] [CrossRef]
- Català-Solsona, J.; Miñano-Molina, A.J.; Rodríguez-Álvarez, J. Nr4a2 Transcription Factor in Hippocampal Synaptic Plasticity, Memory and Cognitive Dysfunction: A Perspective Review. Front. Mol. Neurosci. 2021, 14, 786226. [Google Scholar] [CrossRef] [PubMed]
- Jeon, S.G.; Yoo, A.; Chun, D.W.; Hong, S.B.; Chung, H.; Kim, J.I.; Moon, M. The Critical Role of Nurr1 as a Mediator and Therapeutic Target in Alzheimer’s Disease-related Pathogenesis. Aging Dis. 2020, 11, 705–724. [Google Scholar] [CrossRef]
- Montarolo, F.; Martire, S.; Perga, S.; Bertolotto, A. NURR1 Impairment in Multiple Sclerosis. Int. J. Mol. Sci. 2019, 20, 4858. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Shen, J. Machine learning-based predictive models and drug prediction for schizophrenia in multiple programmed cell death patterns. Front. Mol. Neurosci. 2023, 16, 1123708. [Google Scholar] [CrossRef]
- Huggett, S.B.; Stallings, M.C. Genetic Architecture and Molecular Neuropathology of Human Cocaine Addiction. J. Neurosci. 2020, 40, 5300–5313. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.; Gupta, A.; Zech, M.; Sigafoos, A.N.; Clark, K.J.; Dincer, Y.; Wagner, M.; Humberson, J.B.; Green, S.; van Gassen, K.; et al. De novo variants of NR4A2 are associated with neurodevelopmental disorder and epilepsy. Genet. Med. 2020, 22, 1413–1417. [Google Scholar] [CrossRef]
- Jakaria, M.; Haque, M.E.; Cho, D.Y.; Azam, S.; Kim, I.S.; Choi, D.K. Molecular Insights into NR4A2(Nurr1): An Emerging Target for Neuroprotective Therapy Against Neuroinflammation and Neuronal Cell Death. Mol. Neurobiol. 2019, 56, 5799–5814. [Google Scholar] [CrossRef]
- Jafari, H.; Hussain, S.; Campbell, M.J. Nuclear Receptor Coregulators in Hormone-Dependent Cancers. Cancers 2022, 14, 2402. [Google Scholar] [CrossRef]
- Talukdar, P.D.; Chatterji, U. Transcriptional co-activators: Emerging roles in signaling pathways and potential therapeutic targets for diseases. Signal. Transduct. Target Ther. 2023, 8, 427. [Google Scholar] [CrossRef]
- Li, F.; Song, C.; Zhang, Y.; Wu, D. Structural overview and perspectives of the nuclear receptors, a major family as the direct targets for small-molecule drugs. Acta Biochim. Biophys. Sin. 2022, 54, 12–24. [Google Scholar] [CrossRef]
- Weikum, E.R.; Liu, X.; Ortlund, E.A. The nuclear receptor superfamily: A structural perspective. Protein Sci. 2018, 27, 1876–1892. [Google Scholar] [CrossRef]
- de Vera, I.M.S.; Munoz-Tello, P.; Zheng, J.; Dharmarajan, V.; Marciano, D.P.; Matta-Camacho, E.; Giri, P.K.; Shang, J.; Hughes, T.S.; Rance, M.; et al. Defining a Canonical Ligand-Binding Pocket in the Orphan Nuclear Receptor Nurr1. Structure 2019, 27, 66–77.e5. [Google Scholar] [CrossRef]
- Scheepstra, M.; Andrei, S.A.; de Vries, R.M.J.M.; Meijer, F.A.; Ma, J.N.; Burstein, E.S.; Olsson, R.; Ottmann, C.; Milroy, L.G.; Brunsveld, L. Ligand Dependent Switch from RXR Homo- to RXR-NURR1 Heterodimerization. ACS Chem. Neurosci. 2017, 8, 2065–2077. [Google Scholar] [CrossRef]
- Eells, J.B.; Wilcots, J.; Sisk, S.; Guo-Ross, S.X. NR4A gene expression is dynamically regulated in the ventral tegmental area dopamine neurons and is related to expression of dopamine neurotransmission genes. J. Mol. Neurosci. 2012, 46, 545–553. [Google Scholar] [CrossRef][Green Version]
- Murphy, E.P.; Crean, D. Molecular Interactions between NR4A Orphan Nuclear Receptors and NF-κB Are Required for Appropriate Inflammatory Responses and Immune Cell Homeostasis. Biomolecules 2015, 5, 1302–1318. [Google Scholar] [CrossRef]
- Kadkhodaei, B.; Alvarsson, A.; Schintu, N.; Ramsköld, D.; Volakakis, N.; Joodmardi, E.; Yoshitake, T.; Kehr, J.; Decressac, M.; Björklund, A.; et al. Transcription factor Nurr1 maintains fiber integrity and nuclear-encoded mitochondrial gene expression in dopamine neurons. Proc. Natl. Acad. Sci. USA 2013, 110, 2360–2365. [Google Scholar] [CrossRef] [PubMed]
- Al-Nusaif, M.; Yang, Y.; Li, S.; Cheng, C.; Le, W. The role of NURR1 in metabolic abnormalities of Parkinson’s disease. Mol. Neurodegener. 2022, 17, 46. [Google Scholar] [CrossRef] [PubMed]
- García-Yagüe, Á.J.; Cuadrado, A. Mechanisms of NURR1 Regulation: Consequences for Its Biological Activity and Involvement in Pathology. Int. J. Mol. Sci. 2023, 24, 12280. [Google Scholar] [CrossRef] [PubMed]
- García-Yagüe, Á.J.; Rada, P.; Rojo, A.I.; Lastres-Becker, I.; Cuadrado, A. Nuclear import and export signals control the subcellular localization of Nurr1 protein in response to oxidative stress. J. Biol. Chem. 2013, 88, 5506–5517. [Google Scholar] [CrossRef]
- Bahrami, S.; Drabløs, F. Gene regulation in the immediate-early response process. Adv. Biol. Regul. 2016, 62, 37–49. [Google Scholar] [CrossRef]
- Salery, M.; Godino, A.; Nestler, E.J. Drug-activated cells: From immediate early genes to neuronal ensembles in addiction. Adv. Pharmacol. 2021, 90, 173–216. [Google Scholar] [CrossRef]
- Yap, E.L.; Greenberg, M.E. Activity-Regulated Transcription: Bridging the Gap between Neural Activity and Behavior. Neuron 2018, 100, 330–348. [Google Scholar] [CrossRef]
- Meenakshi, P.; Kumar, S.; Balaji, J. In vivo imaging of immediate early gene expression dynamics segregates neuronal ensemble of memories of dual events. Mol. Brain 2021, 14, 102. [Google Scholar] [CrossRef]
- Bisagno, V.; Cadet, J.L. Expression of immediate early genes in brain reward circuitries: Differential regulation by psychostimulant and opioid drugs. Neurochem. Int. 2019, 124, 10–18. [Google Scholar] [CrossRef]
- Fukuchi, M.; Tsuda, M. Convergence of neurotransmissions at synapse on IEG regulation in nucleus. Front. Biosci. 2017, 22, 1052–1072. [Google Scholar] [CrossRef]
- The Human Protein Atlas Version 23. Available online: https://www.proteinatlas.org/search/nr4a2 (accessed on 7 August 2025).
- Sjöstedt, E.; Zhong, W.; Fagerberg, L.; Karlsson, M.; Mitsios, N.; Adori, C.; Oksvold, P.; Edfors, F.; Limiszewska, A.; Hikmet, F.; et al. An atlas of the protein-coding genes in the human, pig, and mouse brain. Science 2020, 367, eaay5947. [Google Scholar] [CrossRef] [PubMed]
- Uhlén, M.; Fagerberg, L.; Hallström, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, Å.; Kampf, C.; Sjöstedt, E.; Asplund, A.; et al. Tissue-based map of the human proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef] [PubMed]
- Mercadante, A.A.; Dimri, M.; Mohiuddin, S.S. Biochemistry, Replication and Transcription; [Updated 2023]; StatPearls Publishing: Treasure Island, FL, USA, 2025. Available online: https://www.ncbi.nlm.nih.gov/books/NBK540152/ (accessed on 15 May 2025).
- Casamassimi, A.; Ciccodicola, A.; Rienzo, M. Transcriptional Regulation and Its Misregulation in Human Diseases. Int. J. Mol. Sci. 2023, 24, 8640. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.L.; Lin, Z.J.; Li, C.C.; Lin, X.; Shan, S.K.; Guo, B.; Zheng, M.H.; Li, F.; Yuan, L.Q.; Li, Z.H. Epigenetic regulation in metabolic diseases: Mechanisms and advances in clinical study. Signal Transduct. Target. Ther. 2023, 8, 98. [Google Scholar] [CrossRef]
- Torii, T.; Kawarai, T.; Nakamura, S.; Kawakami, H. Organization of the human orphan nuclear receptor Nurr1 gene. Gene 1999, 230, 225–232. [Google Scholar] [CrossRef]
- Mages, H.W.; Rilke, O.; Bravo, R.; Senger, G.; Kroczek, R.A. NOT, a human immediate-early response gene closely related to the steroid/thyroid hormone receptor NAK1/TR3. Mol. Endocrinol. 1994, 8, 583–1591. [Google Scholar] [CrossRef][Green Version]
- Saucedo-Cardenas, O.; Kardon, R.; Ediger, T.R.; Lydon, J.P.; Conneely, O.M. Cloning and structural organization of the gene encoding the murine nuclear receptor transcription factor, NURR1. Gene 1997, 187, 135–139. [Google Scholar] [CrossRef]
- Larsen, K.; Momeni, J.; Farajzadeh, L.; Callesen, H.; Bendixen, C. Molecular characterization and analysis of the porcine NURR1 gene. Biochim. Open 2016, 3, 26–39. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ichinose, H.; Ohye, T.; Suzuki, T.; Sumi-Ichinose, C.; Nomura, T.; Hagino, Y.; Nagatsu, T. Molecular cloning of the human Nurr1 gene: Characterization of the human gene and cDNAs. Gene 1999, 230, 233–239. [Google Scholar] [CrossRef] [PubMed]
- Siegel, D.A.; Le Tonqueze, O.; Biton, A.; Zaitlen, N.; Erle, D.J. Massively parallel analysis of human 3′ UTRs reveals that AU-rich element length and registration predict mRNA destabilization. G3 2022, 12, jkab404. [Google Scholar] [CrossRef] [PubMed]
- ENSEMBL. Available online: https://www.ensembl.org (accessed on 15 May 2025).
- UCSC Genome Browser on Human (GRCh38/hg38). Available online: http://genome.ucsc.edu (accessed on 15 May 2025).
- García-Campayo, J.; Puebla-Guedea, M.; Labarga, A.; Urdánoz, A.; Roldán, M.; Pulido, L.; Martínez de Morentin, X.; Perdones-Montero, A.; Montero-Marín, J.; Mendioroz, M. Epigenetic Response to Mindfulness in Peripheral Blood Leukocytes Involves Genes Linked to Common Human Diseases. Mindfulness 2018, 9, 1146–1159. [Google Scholar] [CrossRef]
- Lee, R.S.; Tamashiro, K.L.; Aryee, M.J.; Murakami, P.; Seifuddin, F.; Herb, B.; Huo, Y.; Rongione, M.; Feinberg, A.P.; Moran, T.H.; et al. Adaptation of the CHARM DNA methylation platform for the rat genome reveals novel brain region-specific differences. Epigenetics 2011, 6, 1378–1390. [Google Scholar] [CrossRef][Green Version]
- Volakakis, N.; Kadkhodaei, B.; Joodmardi, E.; Wallis, K.; Panman, L.; Silvaggi, J.; Spiegelman, B.M.; Perlmann, T. NR4A orphan nuclear receptors as mediators of CREB-dependent neuroprotection. Proc. Natl. Acad. Sci. USA 2010, 107, 12317–12322. [Google Scholar] [CrossRef]
- Parra-Damas, A.; Rubió-Ferrarons, L.; Shen, J.; Saura, C.A. CRTC1 mediates preferential transcription at neuronal activity-regulated CRE/TATA promoters. Sci. Rep. 2017, 7, 18004. [Google Scholar] [CrossRef]
- McEvoy, A.N.; Murphy, E.A.; Ponnio, T.; Conneely, O.M.; Bresnihan, B.; FitzGerald, O.; Murphy, E.P. Activation of nuclear orphan receptor NURR1 transcription by NF-kappa B and cyclic adenosine 5′-monophosphate response element-binding protein in rheumatoid arthritis synovial tissue. J. Immunol. 2002, 168, 2979–2987. [Google Scholar] [CrossRef]
- Eichler, E.E. Genetic Variation, Comparative Genomics, and the Diagnosis of Disease. N. Engl. J. Med. 2019, 381, 64–74. [Google Scholar] [CrossRef]
- Marian, A.J. Clinical Interpretation and Management of Genetic Variants. JACC Basic. Transl. Sci. 2020, 5, 1029–1042. [Google Scholar] [CrossRef]
- Ellingford, J.M.; Ahn, J.W.; Bagnall, R.D.; Baralle, D.; Barton, S.; Campbell, C.; Downes, K.; Ellard, S.; Duff-Farrier, C.; FitzPatrick, D.R.; et al. Recommendations for clinical interpretation of variants found in non-coding regions of the genome. Genome Med. 2022, 14, 73. [Google Scholar] [CrossRef] [PubMed]
- Vihinen, M. Functional effects of protein variants. Biochimie 2021, 180, 104–120. [Google Scholar] [CrossRef] [PubMed]
- Sinnott-Armstrong, N.; Fields, S.; Roth, F.; Starita, L.M.; Trapnell, C.; Villen, J.; Fowler, D.M.; Queitsch, C. Understanding genetic variants in context. eLife 2024, 13, e88231. [Google Scholar] [CrossRef]
- Prokop, J.W.; Jdanov, V.; Savage, L.; Morris, M.; Lamb, N.; VanSickle, E.; Stenger, C.L.; Rajasekaran, S.; Bupp, C.P. Computational and Experimental Analysis of Genetic Variants. Compr. Physiol. 2022, 12, 3303–3336. [Google Scholar] [CrossRef]
- Hernandez-Pacheco, N.; Kere, M.; Melén, E. Gene-environment interactions in childhood asthma revisited; expanding the interaction concept. Pediatr. Allergy Immunol. 2022, 33, e13780. [Google Scholar] [CrossRef]
- Gabaldon-Albero, A.; Mayo, S.; Martinez, F. NR4A2 as a Novel Target Gene for Developmental and Epileptic Encephalopathy: A Systematic Review of Related Disorders and Therapeutic Strategies. Int. J. Mol. Sci. 2024, 25, 5198. [Google Scholar] [CrossRef] [PubMed]
- Ramos, L.L.P.; Monteiro, F.P.; Sampaio, L.P.B.; Costa, L.A.; Ribeiro, M.D.O.; Freitas, E.L.; Kitajima, J.P.; Kok, F. Heterozygous loss of function of NR4A2 is associated with intellectual deficiency, rolandic epilepsy, and language impairment. Clin. Case Rep. 2019, 7, 1582–1584. [Google Scholar] [CrossRef]
- Le, W.D.; Xu, P.; Jankovic, J.; Jiang, H.; Appel, S.H.; Smith, R.G.; Vassilatis, D.K. Mutations in NR4A2 associated with familial Parkinson disease. Nat. Genet. 2003, 33, 85–89, Erratum in Nat. Genet. 2003, 33, 214. [Google Scholar] [CrossRef]
- Liu, H.; Liu, H.; Li, T.; Cui, J.; Fu, Y.; Ren, J.; Sun, X.; Jiang, P.; Yu, S.; Li, C. NR4A2 genetic variation and Parkinson’s disease: Evidence from a systematic review and meta-analysis. Neurosci. Lett. 2017, 650, 25–32. [Google Scholar] [CrossRef]
- Song, X.; Sun, N.; Zhang, A.; Lei, L.; Li, X.; Liu, Z.; Wang, Y.; Yang, C.; Zhang, K. Association Between NR4A2 Gene Polymorphism and Depressive Symptoms and Antidepressant Effect. Neuropsychiatr. Dis. Treat. 2021, 17, 2613–2623. [Google Scholar] [CrossRef] [PubMed]
- Ancín, I.; Cabranes, J.A.; Vázquez-Álvarez, B.; Santos, J.L.; Sánchez-Morla, E.; Alaerts, M.; Del-Favero, J.; Barabash, A. NR4A2: Effects of an “orphan” receptor on sustained attention in a schizophrenic population. Schizophr. Bull. 2013, 39, 555–563. [Google Scholar] [CrossRef]
- Wei, Y.M.; Du, Y.L.; Nie, Y.Q.; Li, Y.Y.; Wan, Y.J. Nur-related receptor 1 gene polymorphisms and alcohol dependence in Mexican Americans. World J. Gastroenterol. 2012, 18, 5276–5282. [Google Scholar]
- Pardo, L.; Valor, L.M.; Eraso-Pichot, A.; Barco, A.; Golbano, A.; Hardingham, G.E.; Masgrau, R.; Galea, E. CREB Regulates Distinct Adaptive Transcriptional Programs in Astrocytes and Neurons. Sci. Rep. 2017, 7, 6390. [Google Scholar] [CrossRef] [PubMed]
- Català-Solsona, J.; Lituma, P.J.; Lutzu, S.; Siedlecki-Wullich, D.; Fábregas-Ordoñez, C.; Miñano-Molina, A.J.; Saura, C.A.; Castillo, P.E.; Rodriguez-Álvarez, J. Activity-Dependent Nr4a2 Induction Modulates Synaptic Expression of AMPA Receptors and Plasticity via a Ca2+/CRTC1/CREB Pathway. J. Neurosci. 2023, 43, 3028–3041. [Google Scholar] [CrossRef]
- España, J.; Valero, J.; Miñano-Molina, A.J.; Masgrau, R.; Martín, E.; Guardia-Laguarta, C.; Lleó, A.; Giménez-Llort, L.; Rodríguez-Alvarez, J.; Saura, C.A. beta-Amyloid disrupts activity-dependent gene transcription required for memory through the CREB coactivator CRTC1. J. Neurosci. 2010, 30, 9402–9410. [Google Scholar] [CrossRef] [PubMed]
- Ji, R.; Sanchez, C.M.; Chou, C.L.; Chen, X.B.; Woodward, D.F.; Regan, J.W. Prostanoid EP1 receptors mediate up-regulation of the orphan nuclear receptor Nurr1 by cAMP-independent activation of protein kinase A, CREB and NF-κB. Br. J. Pharmacol. 2012, 166, 1033–1046, Erratum in Br. J. Pharmacol. 2017, 174, 2156. https://doi.org/10.1111/bph.13858. [Google Scholar] [CrossRef]
- Jia, C.; Qi, H.; Cheng, C.; Wu, X.; Yang, Z.; Cai, H.; Chen, S.; Le, W. α-Synuclein Negatively Regulates Nurr1 Expression Through NF-κB-Related Mechanism. Front. Mol. Neurosci. 2020, 13, 64. [Google Scholar] [CrossRef]
- Yuan, Y.; Jin, J.; Yang, B.; Zhang, W.; Hu, J.; Zhang, Y.; Chen, N.H. Overexpressed alpha-synuclein regulated the nuclear factor-kappaB signal pathway. Cell Mol. Neurobiol. 2008, 28, 21–33. [Google Scholar] [CrossRef]
- García-Yagüe, Á.J.; Lastres-Becker, I.; Stefanis, L.; Vassilatis, D.K.; Cuadrado, A. α-Synuclein Induces the GSK-3-Mediated Phosphorylation and Degradation of NURR1 and Loss of Dopaminergic Hallmarks. Mol. Neurobiol. 2021, 58, 6697–6711. [Google Scholar] [CrossRef]
- Rossetti, C.; Cherix, A.; Guiraud, L.F.; Cardinaux, J.R. New Insights into the Pivotal Role of CREB-Regulated Transcription Coactivator 1 in Depression and Comorbid Obesity. Front. Mol. Neurosci. 2022, 15, 810641. [Google Scholar] [CrossRef]
- Dresselhaus, E.C.; Meffert, M.K. Cellular Specificity of NF-κB Function in the Nervous System. Front. Immunol. 2019, 10, 1043. [Google Scholar] [CrossRef]
- Hu, L.; Si, L.; Dai, X.; Dong, H.; Ma, Z.; Sun, Z.; Li, N.; Sha, H.; Chen, Y.; Qian, Y.; et al. Exosomal miR-409-3p secreted from activated mast cells promotes microglial migration, activation and neuroinflammation by targeting Nr4a2 to activate the NF-κB pathway. J. Neuroinflamm. 2021, 18, 68. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.; Wang, D.; Jiang, S.; Mao, J.; Yang, X. NF κB is negatively associated with Nurr1 to reduce the inflammatory response in Parkinson’s disease. Mol. Med. Rep. 2021, 23, 396. [Google Scholar] [CrossRef] [PubMed]
- Popichak, K.A.; Hammond, S.L.; Moreno, J.A.; Afzali, M.F.; Backos, D.S.; Slayden, R.D.; Safe, S.; Tjalkens, R.B. Compensatory Expression of Nur77 and Nurr1 Regulates NF-κB-Dependent Inflammatory Signaling in Astrocytes. Mol. Pharmacol. 2018, 94, 1174–1186. [Google Scholar] [CrossRef] [PubMed]
- Saijo, K.; Winner, B.; Carson, C.T.; Collier, J.G.; Boyer, L.; Rosenfeld, M.G.; Gage, F.H.; Glass, C.K. A Nurr1/CoREST pathway in microglia and astrocytes protects dopaminergic neurons from inflammation-induced death. Cell 2009, 137, 47–59. [Google Scholar] [CrossRef]
- Crean, D.; Cummins, E.P.; Bahar, B.; Mohan, H.; McMorrow, J.P.; Murphy, E.P. Adenosine Modulates NR4A Orphan Nuclear Receptors To Attenuate Hyperinflammatory Responses in Monocytic Cells. J. Immunol. 2015, 195, 1436–1448. [Google Scholar] [CrossRef]
- De Miranda, B.R.; Popichak, K.A.; Hammond, S.L.; Jorgensen, B.A.; Phillips, A.T.; Safe, S.; Tjalkens, R.B. The Nurr1 Activator 1,1-Bis(3′-Indolyl)-1-(p-Chlorophenyl)Methane Blocks Inflammatory Gene Expression in BV-2 Microglial Cells by Inhibiting Nuclear Factor κB. Mol. Pharmacol. 2015, 87, 1021–1034. [Google Scholar] [CrossRef]
- McEvoy, C.; de Gaetano, M.; Giffney, H.E.; Bahar, B.; Cummins, E.P.; Brennan, E.P.; Barry, M.; Belton, O.; Godson, C.G.; Murphy, E.P.; et al. NR4A Receptors Differentially Regulate NF-κB Signaling in Myeloid Cells. Front. Immunol. 2017, 8, 7. [Google Scholar] [CrossRef]
- Shao, Q.H.; Yan, W.F.; Zhang, Z.; Ma, K.L.; Peng, S.Y.; Cao, Y.L.; Yuan, Y.H.; Chen, N.H. Nurr1: A vital participant in the TLR4-NF-κB signal pathway stimulated by α-synuclein in BV-2 cells. Neuropharmacology 2019, 144, 388–399, Erratum in Neuropharmacology 2024, 252, 109962. https://doi.org/10.1016/j.neuropharm.2024.109962. [Google Scholar] [CrossRef]
- Park, H.; Kam, T.I.; Dawson, V.L.; Dawson, T.M. α-Synuclein pathology as a target in neurodegenerative diseases. Nat. Rev. Neurol. 2025, 21, 32–47. [Google Scholar] [CrossRef] [PubMed]
- Al-Nusaif, M.; Lin, Y.; Li, T.; Cheng, C.; Le, W. Advances in NURR1-Regulated Neuroinflammation Associated with Parkinson’s Disease. Int. J. Mol. Sci. 2022, 23, 16184. [Google Scholar] [CrossRef] [PubMed]
- Chu, Y.; Kordower, J.H. Age-associated increases of alpha-synuclein in monkeys and humans are associated with nigrostriatal dopamine depletion: Is this the target for Parkinson’s disease? Neurobiol. Dis. 2007, 25, 134–149. [Google Scholar] [CrossRef]
- Chu, Y.; Kompoliti, K.; Cochran, E.J.; Mufson, E.J.; Kordower, J.H. Age-related decreases in Nurr1 immunoreactivity in the human substantia nigra. J. Comp. Neurol. 2002, 450, 203–214. [Google Scholar] [CrossRef]
- Guo, Q.; Jin, Y.; Chen, X.; Ye, X.; Shen, X.; Lin, M.; Zeng, C.; Zhou, T.; Zhang, J. NF-κB in biology and targeted therapy: New insights and translational implications. Signal Transduct. Target. Ther. 2024, 9, 53. [Google Scholar] [CrossRef]
- Yang, Y.X.; Latchman, D.S. Nurr1 transcriptionally regulates the expression of alpha-synuclein. Neuroreport 2008, 19, 867–871. [Google Scholar] [CrossRef]
- Volakakis, N.; Tiklova, K.; Decressac, M.; Papathanou, M.; Mattsson, B.; Gillberg, L.; Nobre, A.; Björklund, A.; Perlmann, T. Nurr1 and Retinoid X Receptor Ligands Stimulate Ret Signaling in Dopamine Neurons and Can Alleviate α-Synuclein Disrupted Gene Expression. J. Neurosci. 2015, 35, 14370–14385. [Google Scholar] [CrossRef] [PubMed]
- Abdelaziz, A.M. Alpha-Synuclein drives NURR1 and NLRP3 Inflammasome dysregulation in Parkinson’s disease: From pathogenesis to potential therapeutic strategies. Int. Immunopharmacol. 2025, 156, 114692. [Google Scholar] [CrossRef]
- Hawk, J.D.; Bookout, A.L.; Poplawski, S.G.; Bridi, M.; Rao, A.J.; Sulewski, M.E.; Kroener, B.T.; Manglesdorf, D.J.; Abel, T. NR4A nuclear receptors support memory enhancement by histone deacetylase inhibitors. J. Clin. Investig. 2012, 122, 3593–3602. [Google Scholar] [CrossRef]
- McNulty, S.E.; Barrett, R.M.; Vogel-Ciernia, A.; Malvaez, M.; Hernandez, N.; Davatolhagh, M.F.; Matheos, D.P.; Schiffman, A.; Wood, M.A. Differential roles for Nr4a1 and Nr4a2 in object location vs. object recognition long-term memory. Learn. Mem. 2012, 19, 588–592. [Google Scholar] [CrossRef]
- Kwapis, J.L.; Alaghband, Y.; López, A.J.; White, A.O.; Campbell, R.R.; Dang, R.T.; Rhee, D.; Tran, A.V.; Carl, A.E.; Matheos, D.P.; et al. Context and Auditory Fear are Differentially Regulated by HDAC3 Activity in the Lateral and Basal Subnuclei of the Amygdala. Neuropsychopharmacology 2017, 42, 1284–1294. [Google Scholar] [CrossRef]
- Campbell, R.R.; Kramár, E.A.; Pham, L.; Beardwood, J.H.; Augustynski, A.S.; López, A.J.; Chitnis, O.S.; Delima, G.; Banihani, J.; Matheos, D.P.; et al. HDAC3 Activity within the Nucleus Accumbens Regulates Cocaine-Induced Plasticity and Behavior in a Cell-Type-Specific Manner. J. Neurosci. 2021, 41, 2814–2827. [Google Scholar] [CrossRef] [PubMed]
- Kwapis, J.L.; Alaghband, Y.; López, A.J.; Long, J.M.; Li, X.; Shu, G.; Bodinayake, K.K.; Matheos, D.P.; Rapp, P.R.; Wood, M.A. HDAC3-Mediated Repression of the Nr4a Family Contributes to Age-Related Impairments in Long-Term Memory. J. Neurosci. 2019, 39, 4999–5009. [Google Scholar] [CrossRef]
- Rogge, G.A.; Singh, H.; Dang, R.; Wood, M.A. HDAC3 is a negative regulator of cocaine-context-associated memory formation. J. Neurosci. 2013, 33, 6623–6632. [Google Scholar] [CrossRef] [PubMed]
- McQuown, S.C.; Barrett, R.M.; Matheos, D.P.; Post, R.J.; Rogge, G.A.; Alenghat, T.; Mullican, S.E.; Jones, S.; Rusche, J.R.; Lazar, M.A.; et al. HDAC3 is a critical negative regulator of long-term memory formation. J. Neurosci. 2011, 31, 764–774. [Google Scholar] [CrossRef] [PubMed]
- Wei, W.; Zhao, Q.; Wang, Z.; Liau, W.S.; Basic, D.; Ren, H.; Marshall, P.R.; Zajaczkowski, E.L.; Leighton, L.J.; Madugalle, S.U.; et al. ADRAM is an experience-dependent long noncoding RNA that drives fear extinction through a direct interaction with the chaperone protein 14-3-3. Cell Rep. 2022, 38, 110546. [Google Scholar] [CrossRef]
- Amelio, A.L.; Caputi, M.; Conkright, M.D. Bipartite functions of the CREB co-activators selectively direct alternative splicing or transcriptional activation. EMBO J. 2009, 28, 2733–2747. [Google Scholar] [CrossRef]
- Pereira, L.A.; Munita, R.; González, M.P.; Andrés, M.E. Long 3′UTR of Nurr1 mRNAs is targeted by miRNAs in mesencephalic dopamine neurons. PLoS ONE 2017, 12, e0188177. [Google Scholar] [CrossRef]
- Yang, D.; Li, T.; Wang, Y.; Tang, Y.; Cui, H.; Tang, Y.; Zhang, X.; Chen, D.; Shen, N.; Le, W. miR-132 regulates the differentiation of dopamine neurons by directly targeting Nurr1 expression. J. Cell Sci. 2012, 125, 1673–1682. [Google Scholar] [CrossRef] [PubMed]
- Tsai, Z.Y.; Singh, S.; Yu, S.L.; Kao, L.P.; Chen, B.Z.; Ho, B.C.; Yang, P.C.; Li, S.S. Identification of microRNAs regulated by activin A in human embryonic stem cells. J. Cell Biochem. 2010, 109, 93–102. [Google Scholar] [CrossRef] [PubMed]
- Beard, J.A.; Tenga, A.; Hills, J.; Hoyer, J.D.; Cherian, M.T.; Wang, Y.D.; Chen, T. The orphan nuclear receptor NR4A2 is part of a p53-microRNA-34 network. Sci. Rep. 2016, 6, 25108. [Google Scholar] [CrossRef]
- Yao, H.; Yang, L.; Tian, L.; Guo, Y.; Li, Y. LncRNA MSC-AS1 aggravates nasopharyngeal carcinoma progression by targeting miR-524-5p/nuclear receptor subfamily 4 group A member 2 (NR4A2). Cancer Cell Int. 2020, 20, 138. [Google Scholar] [CrossRef]
- Xie, J.; Xie, L.; Wei, H.; Li, X.J.; Lin, L. Dynamic Regulation of DNA Methylation and Brain Functions. Biology 2023, 12, 152. [Google Scholar] [CrossRef]
- Park, J.; Farris, S. Spatiotemporal Regulation of Transcript Isoform Expression in the Hippocampus. Front. Mol. Neurosci. 2021, 14, 694234. [Google Scholar] [CrossRef]
- Naftelberg, S.; Schor, I.E.; Ast, G.; Kornblihtt, A.R. Regulation of alternative splicing through coupling with transcription and chromatin structure. Annu. Rev. Biochem. 2015, 84, 165–198. [Google Scholar] [CrossRef] [PubMed]
- Kochmanski, J.; VanOeveren, S.E.; Patterson, J.R.; Bernstein, A.I. Developmental Dieldrin Exposure Alters DNA Methylation at Genes Related to Dopaminergic Neuron Development and Parkinson’s Disease in Mouse Midbrain. Toxicol. Sci. 2019, 169, 593–607. [Google Scholar] [CrossRef]
- Park, J.; Lee, K.; Kim, K.; Yi, S.J. The role of histone modifications: From neurodevelopment to neurodiseases. Signal Transduct. Target. Ther. 2022, 7, 217. [Google Scholar] [CrossRef]
- Seto, E.; Yoshida, M. Erasers of histone acetylation: The histone deacetylase enzymes. Cold Spring Harb. Perspect. Biol. 2014, 6, a018713. [Google Scholar] [CrossRef]
- Alberini, C.M.; Kandel, E.R. The regulation of transcription in memory consolidation. Cold Spring Harb. Perspect. Biol. 2014, 7, a021741. [Google Scholar] [CrossRef]
- Johnston, M.V.; Alemi, L.; Harum, K.H. Learning, memory, and transcription factors. Pediatr. Res. 2003, 53, 369–374. [Google Scholar] [CrossRef]
- Hawk, J.D.; Abel, T. The role of NR4A transcription factors in memory formation. Brain Res. Bull. 2011, 85, 21–29. [Google Scholar] [CrossRef] [PubMed]
- Malvaez, M.; McQuown, S.C.; Rogge, G.A.; Astarabadi, M.; Jacques, V.; Carreiro, S.; Rusche, J.R.; Wood, M.A. HDAC3-selective inhibitor enhances extinction of cocaine-seeking behavior in a persistent manner. Proc. Natl. Acad. Sci. USA 2013, 110, 2647–2652. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.Y.; Zhang, S.Y.; Wen, R.; Zhang, T.N.; Yang, N. Role of histone deacetylases and their inhibitors in neurological diseases. Pharmacol. Res. 2024, 208, 107410. [Google Scholar] [CrossRef]
- Alaghband, Y.; Kwapis, J.L.; López, A.J.; White, A.O.; Aimiuwu, O.V.; Al-Kachak, A.; Bodinayake, K.K.; Oparaugo, N.C.; Dang, R.; Astarabadi, M.; et al. Distinct roles for the deacetylase domain of HDAC3 in the hippocampus and medial prefrontal cortex in the formation and extinction of memory. Neurobiol. Learn. Mem. 2017, 145, 94–104. [Google Scholar] [CrossRef] [PubMed]
- Das, T.; Das, T.K.; Khodarkovskaya, A.; Dash, S. Non-coding RNAs and their bioengineering applications for neurological diseases. Bioengineered 2021, 12, 11675–11698. [Google Scholar] [CrossRef]
- Mangiavacchi, A.; Morelli, G.; Orlando, V. Behind the scenes: How RNA orchestrates the epigenetic regulation of gene expression. Front. Cell Dev. Biol. 2023, 11, 1123975. [Google Scholar] [CrossRef]
- Gaggi, G.; Hausman, C.; Cho, S.; Badalamenti, B.C.; Trinh, B.Q.; Di Ruscio, A.; Ummarino, S. LncRNAs Ride the Storm of Epigenetic Marks. Genes 2025, 16, 313. [Google Scholar] [CrossRef]
- Weinreb, A.; Varol, E.; Barrett, A.; McWhirter, R.M.; Taylor, S.R.; Courtney, I.; Basavaraju, M.; Poff, A.; Tipps, J.A.; Collings, B.; et al. Alternative splicing across the C. elegans nervous system. Nat. Commun. 2025, 16, 4508. [Google Scholar] [CrossRef] [PubMed]
- Tasoulas, J.; Rodon, L.; Kaye, F.J.; Montminy, M.; Amelio, A.L. Adaptive Transcriptional Responses by CRTC Coactivators in Cancer. Trends Cancer 2019, 5, 111–127. [Google Scholar] [CrossRef]
- Michelhaugh, S.K.; Vaitkevicius, H.; Wang, J.; Bouhamdan, M.; Krieg, A.R.; Walker, J.L.; Mendiratta, V.; Bannon, M.J. Dopamine neurons express multiple isoforms of the nuclear receptor nurr1 with diminished transcriptional activity. J. Neurochem. 2005, 95, 1342–1350. [Google Scholar] [CrossRef]
- Saura, C.A.; Cardinaux, J.R. Emerging Roles of CREB-Regulated Transcription Coactivators in Brain Physiology and Pathology. Trends Neurosci. 2017, 40, 720–733. [Google Scholar] [CrossRef]
- Van Meter, E.N.; Onyango, J.A.; Teske, K.A. A review of currently identified small molecule modulators of microRNA function. Eur. J. Med. Chem. 2020, 188, 112008. [Google Scholar] [CrossRef]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef]
- Lungu, G.; Stoica, G.; Ambrus, A. MicroRNA profiling and the role of microRNA-132 in neurodegeneration using a rat model. Neurosci. Lett. 2013, 553, 153–158. [Google Scholar] [CrossRef]
- Yang, Z.; Li, T.; Li, S.; Wei, M.; Qi, H.; Shen, B.; Chang, R.C.; Le, W.; Piao, F. Altered Expression Levels of MicroRNA-132 and Nurr1 in Peripheral Blood of Parkinson’s Disease: Potential Disease Biomarkers. ACS Chem. Neurosci. 2019, 10, 2243–2249. [Google Scholar] [CrossRef] [PubMed]
- Xie, X.; Peng, L.; Zhu, J.; Zhou, Y.; Li, L.; Chen, Y.; Yu, S.; Zhao, Y. miR-145-5p/Nurr1/TNF-α Signaling-Induced Microglia Activation Regulates Neuron Injury of Acute Cerebral Ischemic/Reperfusion in Rats. Front. Mol. Neurosci. 2017, 10, 383. [Google Scholar] [CrossRef] [PubMed]
- Moon, M.; Jung, E.S.; Jeon, S.G.; Cha, M.Y.; Jang, Y.; Kim, W.; Lopes, C.; Mook-Jung, I.; Kim, K.S. Nurr1 (NR4A2) regulates Alzheimer’s disease-related pathogenesis and cognitive function in the 5XFAD mouse model. Aging Cell 2019, 18, e12866. [Google Scholar] [CrossRef] [PubMed]
- Montarolo, F.; Martire, S.; Perga, S.; Spadaro, M.; Brescia, I.; Allegra, S.; De Francia, S.; Bertolotto, A. NURR1 deficiency is associated to ADHD-like phenotypes in mice. Transl. Psychiatry 2019, 9, 207. [Google Scholar] [CrossRef]
- Vuillermot, S.; Joodmardi, E.; Perlmann, T.; Ove Ögren, S.; Feldon, J.; Meyer, U. Schizophrenia-relevant behaviors in a genetic mouse model of constitutive Nurr1 deficiency. Genes. Brain Behav. 2011, 10, 589–603. [Google Scholar] [CrossRef] [PubMed]
- Rojas, P.; Joodmardi, E.; Hong, Y.; Perlmann, T.; Ogren, S.O. Adult mice with reduced Nurr1 expression: An animal model for schizophrenia. Mol. Psychiatry 2007, 12, 756–766. [Google Scholar] [CrossRef]
- Corley, S.M.; Tsai, S.Y.; Wilkins, M.R.; Shannon Weickert, C. Transcriptomic Analysis Shows Decreased Cortical Expression of NR4A1, NR4A2 and RXRB in Schizophrenia and Provides Evidence for Nuclear Receptor Dysregulation. PLoS ONE 2016, 11, e0166944. [Google Scholar] [CrossRef] [PubMed]
- Poewe, W.; Seppi, K.; Tanner, C.M.; Halliday, G.M.; Brundin, P.; Volkmann, J.; Schrag, A.E.; Lang, A.E. Parkinson disease. Nat. Rev. Dis. Primers 2017, 3, 17013. [Google Scholar] [CrossRef]
- Prajjwal, P.; Flores Sanga, H.S.; Acharya, K.; Tango, T.; John, J.; Rodriguez, R.S.C.; Dheyaa Marsool Marsool, M.; Sulaimanov, M.; Ahmed, A.; Hussin, O.A. Parkinson’s disease updates: Addressing the pathophysiology, risk factors, genetics, diagnosis, along with the medical and surgical treatment. Ann. Med. Surg. 2023, 85, 4887–4902. [Google Scholar] [CrossRef] [PubMed]
- Kummari, E.; Guo-Ross, S.X.; Partington, H.S.; Nutter, J.M.; Eells, J.B. Quantitative Immunohistochemistry to Measure Regional Expression of Nurr1 in the Brain and the Effect of the Nurr1 Heterozygous Genotype. Front. Neuroanat. 2021, 15, 563854. [Google Scholar] [CrossRef]
- Jiang, C.; Wan, X.; He, Y.; Pan, T.; Jankovic, J.; Le, W. Age-dependent dopaminergic dysfunction in Nurr1 knockout mice. Exp. Neurol. 2005, 191, 154–162. [Google Scholar] [CrossRef]
- Le, W.; Conneely, O.M.; He, Y.; Jankovic, J.; Appel, S.H. Reduced Nurr1 expression increases the vulnerability of mesencephalic dopamine neurons to MPTP-induced injury. J. Neurochem. 1999, 73, 2218–2221. [Google Scholar]
- Glaab, E.; Schneider, R. Comparative pathway and network analysis of brain transcriptome changes during adult aging and in Parkinson’s disease. Neurobiol. Dis. 2015, 74, 1–13. [Google Scholar] [CrossRef]
- Chu, Y.; Le, W.; Kompoliti, K.; Jankovic, J.; Mufson, E.J.; Kordower, J.H. Nurr1 in Parkinson’s disease and related disorders. J. Comp. Neurol. 2006, 494, 495–514. [Google Scholar] [CrossRef] [PubMed]
- Montarolo, F.; Perga, S.; Martire, S.; Navone, D.N.; Marchet, A.; Leotta, D.; Bertolotto, A. Altered NR4A Subfamily Gene Expression Level in Peripheral Blood of Parkinson’s and Alzheimer’s Disease Patients. Neurotox. Res. 2016, 30, 338–344, Erratum in Neurotox. Res. 2017, 31, 317. https://doi.org/10.1007/s12640-016-9685-6. [Google Scholar] [CrossRef] [PubMed]
- Le, W.; Pan, T.; Huang, M.; Xu, P.; Xie, W.; Zhu, W.; Zhang, X.; Deng, H.; Jankovic, J. Decreased NURR1 gene expression in patients with Parkinson’s disease. J. Neurol. Sci. 2008, 273, 29–33. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Wei, L.; Tao, Q.; Deng, H.; Ming, M.; Xu, P.; Le, W. Decreased NURR1 and PITX3 gene expression in Chinese patients with Parkinson’s disease. Eur. J. Neurol. 2012, 19, 870–875. [Google Scholar] [CrossRef]
- Li, T.; Yang, Z.; Li, S.; Cheng, C.; Shen, B.; Le, W. Alterations of NURR1 and Cytokines in the Peripheral Blood Mononuclear Cells: Combined Biomarkers for Parkinson’s Disease. Front. Aging Neurosci. 2018, 10, 392. [Google Scholar] [CrossRef]
- Ruiz-Sánchez, E.; Yescas, P.; Rodríguez-Violante, M.; Martínez-Rodríguez, N.; Díaz-López, J.N.; Ochoa, A.; Valdes-Rojas, S.S.; Magos-Rodríguez, D.; Rojas-Castañeda, J.C.; Cervantes-Arriaga, A.; et al. Association of polymorphisms and reduced expression levels of the NR4A2 gene with Parkinson’s disease in a Mexican population. J. Neurol. Sci. 2017, 379, 58–63. [Google Scholar] [CrossRef]
- Kochmanski, J.; Kuhn, N.C.; Bernstein, A.I. Parkinson’s disease-associated, sex-specific changes in DNA methylation at PARK7 (DJ-1), SLC17A6 (VGLUT2), PTPRN2 (IA-2β), and NR4A2 (NURR1) in cortical neurons. NPJ Parkinsons Dis. 2022, 8, 120. [Google Scholar] [CrossRef]
- Smith, G.A.; Rocha, E.M.; Rooney, T.; Barneoud, P.; McLean, J.R.; Beagan, J.; Osborn, T.; Coimbra, M.; Luo, Y.; Hallett, P.J.; et al. A Nurr1 agonist causes neuroprotection in a Parkinson’s disease lesion model primed with the toll-like receptor 3 dsRNA inflammatory stimulant poly(I:C). PLoS ONE 2015, 10, e0121072. [Google Scholar] [CrossRef]
- Rojas, P.; Ruiz-Sánchez, E.; Rojas, C.; Ogren, S.O. Ginkgo biloba extract (EGb 761) modulates the expression of dopamine-related genes in 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine-induced Parkinsonism in mice. Neuroscience 2012, 223, 246–257. [Google Scholar] [CrossRef]
- Breijyeh, Z.; Karaman, R. Comprehensive Review on Alzheimer’s Disease: Causes and Treatment. Molecules 2020, 25, 5789. [Google Scholar] [CrossRef]
- Wang, R.; Reddy, P.H. Role of Glutamate and NMDA Receptors in Alzheimer’s Disease. J. Alzheimers Dis. 2017, 57, 1041–1048. [Google Scholar] [CrossRef]
- Moon, M.; Jeong, I.; Kim, C.H.; Kim, J.; Lee, P.K.; Mook-Jung, I.; Leblanc, P.; Kim, K.S. Correlation between orphan nuclear receptor Nurr1 expression and amyloid deposition in 5XFAD mice, an animal model of Alzheimer’s disease. J. Neurochem. 2015, 132, 254–262. [Google Scholar] [CrossRef] [PubMed]
- Terzioglu-Usak, S.; Negis, Y.; Karabulut, D.S.; Zaim, M.; Isik, S. Cellular Model of Alzheimer’s Disease: Aβ1-42 Peptide Induces Amyloid Deposition and a Decrease in Topo Isomerase IIβ and Nurr1 Expression. Curr. Alzheimer Res. 2017, 14, 636–644. [Google Scholar] [CrossRef] [PubMed]
- Parra-Damas, A.; Valero, J.; Chen, M.; España, J.; Martín, E.; Ferrer, I.; Rodríguez-Alvarez, J.; Saura, C.A. Crtc1 activates a transcriptional program deregulated at early Alzheimer’s disease-related stages. J. Neurosci. 2014, 34, 5776–5787. [Google Scholar] [CrossRef]
- Chatterjee, S.; Walsh, E.N.; Yan, A.L.; Giese, K.P.; Safe, S.; Abel, T. Pharmacological activation of Nr4a rescues age-associated memory decline. Neurobiol. Aging 2020, 85, 140–144. [Google Scholar] [CrossRef] [PubMed]
- Morris-Rosendahl, D.J.; Crocq, M.A. Neurodevelopmental disorders-the history and future of a diagnostic concept. Dialogues Clin. Neurosci. 2020, 22, 65–72. [Google Scholar] [CrossRef]
- Palmieri, R.; Albano, V.; Guerriero, S.; Craig, F.; La Torre, F.; Filoni, S.; Sardella, D.; Petruzzelli, M.G.; Lecce, P.; De Giacomo, A. Beyond Diagnosis: Preliminary Study of Impact on Children and Parents in Neurodevelopmental Disorders and Juvenile Idiopathic Arthritis-Associated Uveitis. Diagnostics 2024, 14, 275. [Google Scholar] [CrossRef]
- Du, Y.; Chen, L.; Yan, M.C.; Wang, Y.L.; Zhong, X.L.; Xv, C.X.; Li, Y.B.; Cheng, Y. Neurometabolite levels in the brains of patients with autism spectrum disorders: A meta-analysis of proton magnetic resonance spectroscopy studies (N = 1501). Mol. Psychiatry 2023, 28, 3092–3103. [Google Scholar] [CrossRef]
- Bordoni, L.; Petracci, I.; Calleja-Agius, J.; Lalor, J.G.; Gabbianelli, R. NURR1 Alterations in Perinatal Stress: A First Step towards Late-Onset Diseases? A Narrative Review. Biomedicines 2020, 8, 584. [Google Scholar] [CrossRef]
- Montes, P.; Ruiz-Sánchez, E.; Calvillo, M.; Rojas, P. Active coping of prenatally stressed rats in the forced swimming test: Involvement of the Nurr1 gene. Stress 2016, 19, 506–515. [Google Scholar] [CrossRef]
- Song, X.; Xu, W.; Xiao, M.; Lu, Y.; Lan, X.; Tang, X.; Xu, N.; Yu, G.; Zhang, H.; Wu, S. Two novel heterozygous truncating variants in NR4A2 identified in patients with neurodevelopmental disorder and brief literature review. Front. Neurosci. 2022, 16, 956429. [Google Scholar] [CrossRef]
- Orsolini, L.; Pompili, S.; Volpe, U. Schizophrenia: A Narrative Review of Etiopathogenetic, Diagnostic and Treatment Aspects. J. Clin. Med. 2022, 11, 5040. [Google Scholar] [CrossRef]
- McCutcheon, R.A.; Keefe, R.S.E.; McGuire, P.K. Cognitive impairment in schizophrenia: Aetiology, pathophysiology, and treatment. Mol. Psychiatry 2023, 28, 1902–1918, Erratum in Mol. Psychiatry 2023, 28, 1919. https://doi.org/10.1038/s41380-023-01984-6. [Google Scholar] [CrossRef]
- Fabbri, C.; Leggio, G.M.; Drago, F.; Serretti, A. Imputed expression of schizophrenia-associated genes and cognitive measures in patients with schizophrenia. Mol. Genet. Genomic. Med. 2022, 10, e1942. [Google Scholar] [CrossRef]
- Liang, J.; Chen, L.; Li, Y.; Chen, Y.; Yuan, L.; Qiu, Y.; Ma, S.; Fan, F.; Cheng, Y. Unraveling the Prefrontal Cortex-Basolateral Amygdala Pathway’s Role on Schizophrenia’s Cognitive Impairments: A Multimodal Study in Patients and Mouse Models. Schizophr. Bull. 2024, 50, 913–923. [Google Scholar] [CrossRef]
- San-Martin, R.; Castro, L.A.; Menezes, P.R.; Fraga, F.J.; Simões, P.W.; Salum, C. Meta-Analysis of Sensorimotor Gating Deficits in Patients with Schizophrenia Evaluated by Prepulse Inhibition Test. Schizophr. Bull. 2020, 46, 1482–1497. [Google Scholar] [CrossRef]
- Guillozet-Bongaarts, A.L.; Hyde, T.M.; Dalley, R.A.; Hawrylycz, M.J.; Henry, A.; Hof, P.R.; Hohmann, J.; Jones, A.R.; Kuan, C.L.; Royall, J.; et al. Altered gene expression in the dorsolateral prefrontal cortex of individuals with schizophrenia. Mol. Psychiatry 2014, 19, 478–485. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Sánchez, E.; Jiménez-Genchi, J.; Alcántara-Flores, Y.M.; Castañeda-González, C.J.; Aviña-Cervantes, C.L.; Yescas, P.; Del Socorro González-Valadez, M.; Martínez-Rodríguez, N.; Ríos-Ortiz, A.; González-González, M.; et al. Working memory deficits in schizophrenia are associated with the rs34884856 variant and expression levels of the NR4A2 gene in a sample Mexican population: A case control study. BMC Psychiatry 2021, 21, 86. [Google Scholar] [CrossRef] [PubMed]
- Marx, W.; Penninx, B.W.J.H.; Solmi, M.; Furukawa, T.A.; Firth, J.; Carvalho, A.F.; Berk, M. Major depressive disorder. Nat. Rev. Dis. Primers 2023, 9, 44. [Google Scholar] [CrossRef] [PubMed]
- Malhi, G.S.; Mann, J.J. Depression. Lancet 2018, 392, 2299–2312. [Google Scholar] [CrossRef]
- Bains, N.; Abdijadid, S. Major Depressive Disorder; [Updated 10 April 2023]; StatPearls Publishing: Treasure Island, FL, USA, 2025. Available online: https://www.ncbi.nlm.nih.gov/books/NBK559078/ (accessed on 15 May 2025).
- Cui, L.; Li, S.; Wang, S.; Wu, X.; Liu, Y.; Yu, W.; Wang, Y.; Tang, Y.; Xia, M.; Li, B. Major depressive disorder: Hypothesis, mechanism, prevention and treatment. Signal Transduct. Target. Ther. 2024, 9, 30. [Google Scholar] [CrossRef]
- Huang, K.W.; Ochandarena, N.E.; Philson, A.C.; Hyun, M.; Birnbaum, J.E.; Cicconet, M.; Sabatini, B.L. Molecular and anatomical organization of the dorsal raphe nucleus. eLife 2019, 8, e46464. [Google Scholar] [CrossRef]
- Kerman, I.A.; Bernard, R.; Bunney, W.E.; Jones, E.G.; Schatzberg, A.F.; Myers, R.M.; Barchas, J.D.; Akil, H.; Watson, S.J.; Thompson, R.C. Evidence for transcriptional factor dysregulation in the dorsal raphe nucleus of patients with major depressive disorder. Front Neurosci 2012, 6, 135. [Google Scholar] [CrossRef]
- Rojas, P.; Joodmardi, E.; Perlmann, T.; Ogren, S.O. Rapid increase of Nurr1 mRNA expression in limbic and cortical brain structures related to coping with depression-like behavior in mice. J. Neurosci. Res. 2010, 88, 2284–2293. [Google Scholar] [CrossRef]
- Ruiz-Sánchez, E.; López-Ramírez, A.M.; Ruiz-Chow, Á.; Calvillo, M.; Reséndiz-Albor, A.A.; Anguiano, B.; Rojas, P. Variability in Behavioral Phenotypes after Forced Swimming-Induced Stress in Rats Is Associated with Expression of the Glucocorticoid Receptor, Nurr1, and IL-1β in the Hippocampus. Int. J. Mol. Sci. 2021, 22, 12700. [Google Scholar] [CrossRef] [PubMed]
- Volkow, N.D.; Blanco, C. Substance use disorders: A comprehensive update of classification, epidemiology, neurobiology, clinical aspects, treatment and prevention. World Psychiatry 2023, 22, 203–229. [Google Scholar] [CrossRef] [PubMed]
- Swimmer, K.R.; Sandelich, S. Substance Use Disorder. Emerg. Med. Clin. N. Am. 2024, 42, 53–67. [Google Scholar] [CrossRef] [PubMed]
- American Psychiatric Association, DSM-5 Task Force. Diagnostic and Statistical Manual of Mental Disorders: DSM-5™, 5th ed.; American Psychiatric Publishing, Inc.: Washington, DC, USA, 2013. [Google Scholar] [CrossRef]
- Polak, K.; Haug, N.A.; Dillon, P.; Svikis, D.S. Substance Use Disorders in Women. Psychiatr. Clin. N. Am. 2023, 46, 487–503. [Google Scholar] [CrossRef]
- Danpanichkul, P.; Duangsonk, K.; Díaz, L.A.; Chen, V.L.; Rangan, P.; Sukphutanan, B.; Dutta, P.; Wanichthanaolan, O.; Ramadoss, V.; Sim, B.; et al. The burden of alcohol and substance use disorders in adolescents and young adults. Drug Alcohol. Depend. 2025, 266, 112495. [Google Scholar] [CrossRef]
- Caffino, L.; Mottarlini, F.; Zita, G.; Gawliński, D.; Gawlińska, K.; Wydra, K.; Przegaliński, E.; Fumagalli, F. The effects of cocaine exposure in adolescence: Behavioural effects and neuroplastic mechanisms in experimental models. Br. J. Pharmacol. 2022, 179, 4233–4253. [Google Scholar] [CrossRef]
- Walker, D.M.; Cates, H.M.; Loh, Y.E.; Purushothaman, I.; Ramakrishnan, A.; Cahill, K.M.; Lardner, C.K.; Godino, A.; Kronman, H.G.; Rabkin, J.; et al. Cocaine Self-administration Alters Transcriptome-wide Responses in the Brain’s Reward Circuitry. Biol. Psychiatry 2018, 84, 867–880. [Google Scholar] [CrossRef]
- Leo, D.; di Porzio, U.; Racagni, G.; Riva, M.A.; Fumagalli, F.; Perrone-Capano, C. Chronic cocaine administration modulates the expression of transcription factors involved in midbrain dopaminergic neuron function. Exp. Neurol. 2007, 203, 472–480. [Google Scholar] [CrossRef]
- Bannon, M.J.; Pruetz, B.; Manning-Bog, A.B.; Whitty, C.J.; Michelhaugh, S.K.; Sacchetti, P.; Granneman, J.G.; Mash, D.C.; Schmidt, C.J. Decreased expression of the transcription factor NURR1 in dopamine neurons of cocaine abusers. Proc. Natl. Acad. Sci. USA 2002, 99, 6382–6385. [Google Scholar] [CrossRef]
- Bannon, M.J.; Pruetz, B.; Barfield, E.; Schmidt, C.J. Transcription factors specifying dopamine phenotype are decreased in cocaine users. Neuroreport 2004, 15, 401–404. [Google Scholar] [CrossRef]
- Nestler, E.J.; Lüscher, C. The Molecular Basis of Drug Addiction: Linking Epigenetic to Synaptic and Circuit Mechanisms. Neuron 2019, 102, 48–59. [Google Scholar] [CrossRef] [PubMed]
- Potvin, S.; Stavro, K.; Rizkallah, E.; Pelletier, J. Cocaine and cognition: A systematic quantitative review. J. Addict. Med. 2014, 8, 368–376. [Google Scholar] [CrossRef] [PubMed]
- Mañas-Padilla, M.C.; Ávila-Gámiz, F.; Gil-Rodríguez, S.; Ladrón de Guevara-Miranda, D.; Rodríguez de Fonseca, F.; Santín, L.J.; Castilla-Ortega, E. Persistent changes in exploration and hyperactivity coexist with cognitive impairment in mice withdrawn from chronic cocaine. Physiol. Behav. 2021, 240, 113542. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Xu, P.; Xu, Y.; Teng, H.; Tian, W.; Du, Q.; Zhao, M. Dynamic Expression Changes in the Transcriptome of the Prefrontal Cortex after Repeated Exposure to Cocaine in Mice. Front. Pharmacol. 2017, 8, 142. [Google Scholar] [CrossRef]
- Lu, H.; Cheng, P.L.; Lim, B.K.; Khoshnevisrad, N.; Poo, M.M. Elevated BDNF after cocaine withdrawal facilitates LTP in medial prefrontal cortex by suppressing GABA inhibition. Neuron 2010, 67, 821–833. [Google Scholar] [CrossRef]
- Teague, C.D.; Nestler, E.J. Key transcription factors mediating cocaine-induced plasticity in the nucleus accumbens. Mol. Psychiatry 2022, 27, 687–709. [Google Scholar] [CrossRef]
- Anderson, E.M.; Taniguchi, M. Epigenetic Effects of Addictive Drugs in the Nucleus Accumbens. Front Mol Neurosci 2022, 15, 828055. [Google Scholar] [CrossRef]
- He, R.; Liu, B.; Geng, B.; Li, N.; Geng, Q. The role of HDAC3 and its inhibitors in regulation of oxidative stress and chronic diseases. Cell Death Discov. 2023, 9, 131. [Google Scholar] [CrossRef] [PubMed]
- López, A.J.; Jia, Y.; White, A.O.; Kwapis, J.L.; Espinoza, M.; Hwang, P.; Campbell, R.; Alaghband, Y.; Chitnis, O.; Matheos, D.P.; et al. Medial habenula cholinergic signaling regulates cocaine-associated relapse-like behavior. Addict. Biol. 2019, 24, 403–413. [Google Scholar] [CrossRef]
- Childs, J.E.; Morabito, S.; Das, S.; Santelli, C.; Pham, V.; Kusche, K.; Vera, V.A.; Reese, F.; Campbell, R.R.; Matheos, D.P.; et al. Relapse to cocaine seeking is regulated by medial habenula NR4A2/NURR1 in mice. Cell Rep. 2024, 43, 113956. [Google Scholar] [CrossRef] [PubMed]
- López, A.J.; Hemstedt, T.J.; Jia, Y.; Hwang, P.H.; Campbell, R.R.; Kwapis, J.L.; White, A.O.; Chitnis, O.; Scarfone, V.M.; Matheos, D.P.; et al. Epigenetic regulation of immediate-early gene Nr4a2/Nurr1 in the medial habenula during reinstatement of cocaine-associated behavior. Neuropharmacology 2019, 153, 13–19. [Google Scholar] [CrossRef]
- Kim, W.; Tripathi, M.; Kim, C.; Vardhineni, S.; Cha, Y.; Kandi, S.K.; Feitosa, M.; Kholiya, R.; Sah, E.; Thakur, A.; et al. An optimized Nurr1 agonist provides disease-modifying effects in Parkinson’s disease models. Nat. Commun. 2023, 14, 4283. [Google Scholar] [CrossRef]
- Jang, Y.; Kim, W.; Leblanc, P.; Kim, C.H.; Kim, K.S. Potent synthetic and endogenous ligands for the adopted orphan nuclear receptor Nurr1. Exp. Mol. Med. 2021, 53, 19–29. [Google Scholar] [CrossRef]
- Mill, J.; Tang, T.; Kaminsky, Z.; Khare, T.; Yazdanpanah, S.; Bouchard, L.; Jia, P.; Assadzadeh, A.; Flanagan, J.; Schumacher, A.; et al. Epigenomic profiling reveals DNA-methylation changes associated with major psychosis. Am. J. Hum. Genet. 2008, 82, 696–711. [Google Scholar] [CrossRef] [PubMed]
- Bhatia, S.; Pooja; Yadav, S.K. CRISPR-Cas for genome editing: Classification, mechanism, designing and applications. Int. J. Biol. Macromol. 2023, 238, 124054. [Google Scholar] [CrossRef]
- Kovalev, M.A.; Mamaeva, N.Y.; Kristovskiy, N.V.; Feskin, P.G.; Vinnikov, R.S.; Oleinikov, P.D.; Sosnovtseva, A.O.; Yakovlev, V.A.; Glukhov, G.S.; Shaytan, A.K. Epigenome Engineering Using dCas Systems for Biomedical Applications and Biotechnology: Current Achievements, Opportunities and Challenges. Int. J. Mol. Sci. 2025, 26, 6371. [Google Scholar] [CrossRef]
- Li, C.; Samulski, R.J. Engineering adeno-associated virus vectors for gene therapy. Nat. Rev. Genet. 2020, 21, 255–272. [Google Scholar] [CrossRef]
- Zhou, X.; Liu, J.; Xiao, S.; Liang, X.; Li, Y.; Mo, F.; Xin, X.; Yang, Y.; Gao, C. Adeno-Associated Virus Engineering and Load Strategy for Tropism Modification, Immune Evasion and Enhanced Transgene Expression. Int. J. Nanomed. 2024, 19, 7691–7708. [Google Scholar] [CrossRef]
- Yim, Y.Y.; Teague, C.D.; Nestler, E.J. In vivo locus-specific editing of the neuroepigenome. Nat. Rev. Neurosci. 2020, 21, 471–484. [Google Scholar] [CrossRef]
- Sun, H.; Fu, S.; Cui, S.; Yin, X.; Sun, X.; Qi, X.; Cui, K.; Wang, J.; Ma, L.; Liu, F.Y.; et al. Development of a CRISPR-SaCas9 system for projection- and function-specific gene editing in the rat brain. Sci. Adv. 2020, 6, eaay6687. [Google Scholar] [CrossRef]
- Asmamaw, M.; Zawdie, B. Mechanism and Applications of CRISPR/Cas-9-Mediated Genome Editing. Biologics 2021, 15, 353–361. [Google Scholar] [CrossRef]
- Yang, Y.; Seok, M.J.; Kim, Y.E.; Choi, Y.; Song, J.J.; Sulistio, Y.A.; Kim, S.H.; Chang, M.Y.; Oh, S.J.; Nam, M.H.; et al. Adeno-associated virus (AAV) 9-mediated gene delivery of Nurr1 and Foxa2 ameliorates symptoms and pathologies of Alzheimer disease model mice by suppressing neuro-inflammation and glial pathology. Mol. Psychiatry 2023, 28, 5359–5374, Erratum in Mol. Psychiatry 2023, 28, 5378–5379. https://doi.org/10.1038/s41380-023-02054-7; Erratum in Mol. Psychiatry 2023, 28, 5375–5377. https://doi.org/10.1038/s41380-023-02169-x. [Google Scholar] [CrossRef]
- Qian, Y.; Chen, X.X.; Wang, W.; Li, J.J.; Wang, X.P.; Tang, Z.W.; Xu, J.T.; Lin, H.; Yang, Z.Y.; Li, L.Y.; et al. Transplantation of Nurr1-overexpressing neural stem cells and microglia for treating parkinsonian rats. CNS Neurosci. Ther. 2020, 26, 55–65. [Google Scholar] [CrossRef]
- Giehrl-Schwab, J.; Giesert, F.; Rauser, B.; Lao, C.L.; Hembach, S.; Lefort, S.; Ibarra, I.L.; Koupourtidou, C.; Luecken, M.D.; Truong, D.J.; et al. Parkinson’s disease motor symptoms rescue by CRISPRa-reprogramming astrocytes into GABAergic neurons. EMBO Mol. Med. 2022, 14, e14797. [Google Scholar] [CrossRef]
- Zhang, G.; Song, C.; Yin, M.; Liu, L.; Zhang, Y.; Li, Y.; Zhang, J.; Guo, M.; Li, C. TRAPT: A multi-stage fused deep learning framework for predicting transcriptional regulators based on large-scale epigenomic data. Nat. Commun. 2025, 16, 3611. [Google Scholar] [CrossRef]
- Hu, E.; Li, Z.; Li, T.; Yang, X.; Ding, R.; Jiang, H.; Su, H.; Cheng, M.; Yu, Z.; Li, H.; et al. A novel microbial and hepatic biotransformation-integrated network pharmacology strategy explores the therapeutic mechanisms of bioactive herbal products in neurological diseases: The effects of Astragaloside IV on intracerebral hemorrhage as an example. Chin. Med. 2023, 18, 40. [Google Scholar] [CrossRef]


| Neurodegenerative Disease/ Psychiatric Disorder | NR4A2 Expression in Patients | Relevant Studies in Animal Models | Potential Mechanisms Implicated in Gene Regulation |
|---|---|---|---|
| Parkinson’s disease (PD) | In patients with PD:
|
|
|
| Alzheimer’s disease (AD) | In patients with AD: |
| The precise mechanisms underlying altered NR4A2 expression in AD remain unclear. However, in animal models of cognition, particularly memory and learning, histone deacetylases have been shown to be part of the mechanism involved in changes in the NR4A2 gene expression levels [100]. |
| Neurodevelopmental Disorders (NDD) |
|
|
|
| Schizophrenia (SZ) | In SZ patients:
| It is unclear how DNA methylation impacts NR4A2 expression or if they play a part in disease progression. No research has been conducted on this subject. However, in animal models of cognition, particularly memory and learning, have shown that histone deacetylases are part of the mechanism involved in changes in the NR4A2 gene expression levels [100]. | |
| Major depressive disorder (MDD) | In MDD patients: |
|
|
| Substance use disorders | Postmortem human individuals with history of cocaine and heroin abuse: |
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ruiz-Sánchez, E.; Rojas, C.; Yescas Gómez, P.; Martínez-Rodríguez, N.; Ruiz-Chow, Á.A.; Nava-Ruiz, C.; Ibáñéz-Cervantes, G.; Arciniega-Martínez, I.M.; Reséndiz-Albor, A.A.; Rojas, P. Regulation of NR4A2 Gene Expression and Its Importance in Neurodegenerative and Psychiatric Diseases. Int. J. Mol. Sci. 2025, 26, 9162. https://doi.org/10.3390/ijms26189162
Ruiz-Sánchez E, Rojas C, Yescas Gómez P, Martínez-Rodríguez N, Ruiz-Chow ÁA, Nava-Ruiz C, Ibáñéz-Cervantes G, Arciniega-Martínez IM, Reséndiz-Albor AA, Rojas P. Regulation of NR4A2 Gene Expression and Its Importance in Neurodegenerative and Psychiatric Diseases. International Journal of Molecular Sciences. 2025; 26(18):9162. https://doi.org/10.3390/ijms26189162
Chicago/Turabian StyleRuiz-Sánchez, Elizabeth, Carolina Rojas, Petra Yescas Gómez, Nancy Martínez-Rodríguez, Ángel Alberto Ruiz-Chow, Concepción Nava-Ruiz, Gabriela Ibáñéz-Cervantes, Ivonne Maciel Arciniega-Martínez, Aldo Arturo Reséndiz-Albor, and Patricia Rojas. 2025. "Regulation of NR4A2 Gene Expression and Its Importance in Neurodegenerative and Psychiatric Diseases" International Journal of Molecular Sciences 26, no. 18: 9162. https://doi.org/10.3390/ijms26189162
APA StyleRuiz-Sánchez, E., Rojas, C., Yescas Gómez, P., Martínez-Rodríguez, N., Ruiz-Chow, Á. A., Nava-Ruiz, C., Ibáñéz-Cervantes, G., Arciniega-Martínez, I. M., Reséndiz-Albor, A. A., & Rojas, P. (2025). Regulation of NR4A2 Gene Expression and Its Importance in Neurodegenerative and Psychiatric Diseases. International Journal of Molecular Sciences, 26(18), 9162. https://doi.org/10.3390/ijms26189162







