The Role of the Beclin1 Complex in Rab9-Dependent Alternative Autophagy
Abstract
1. Introduction
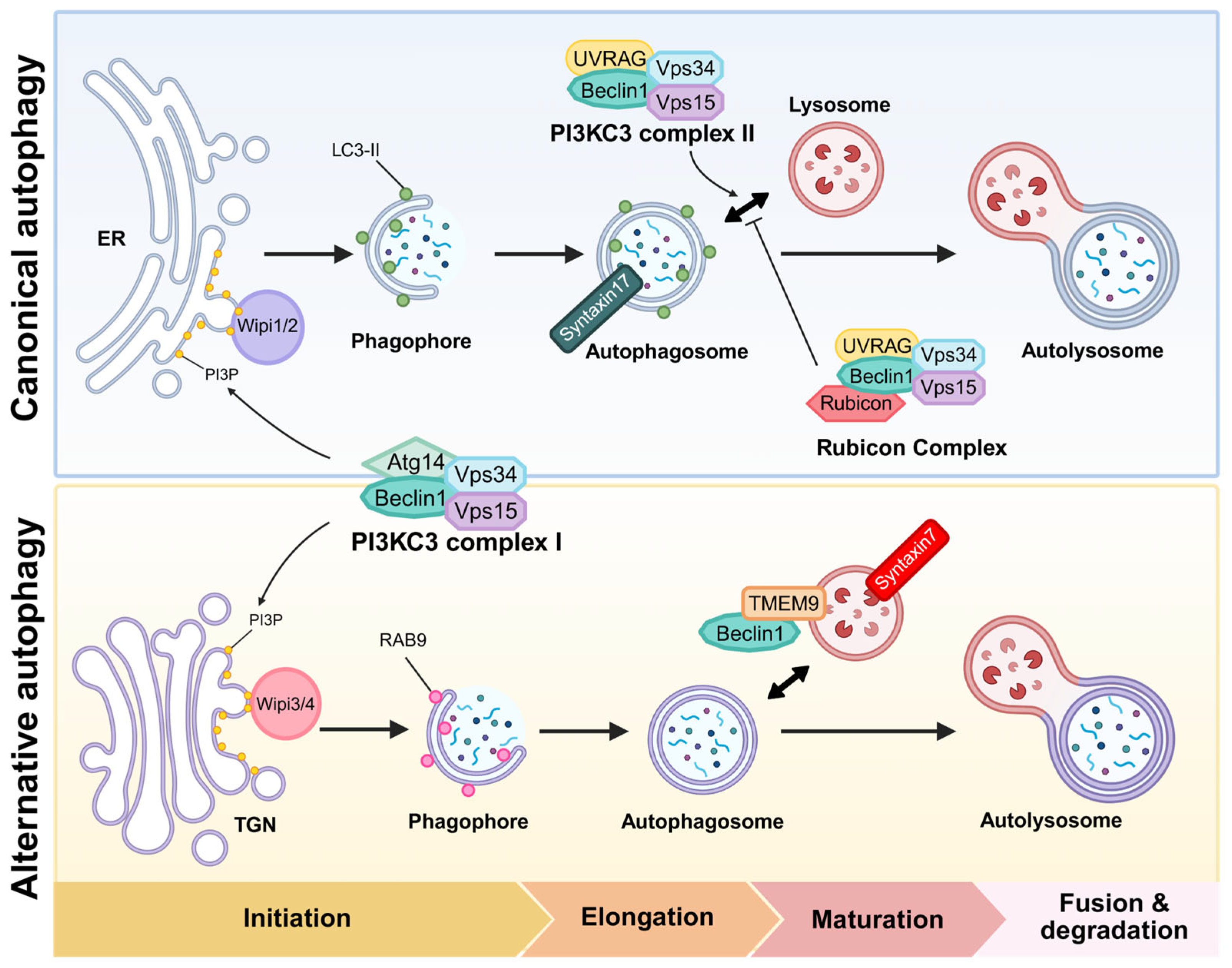
2. The Beclin1 Complex in Canonical Autophagy
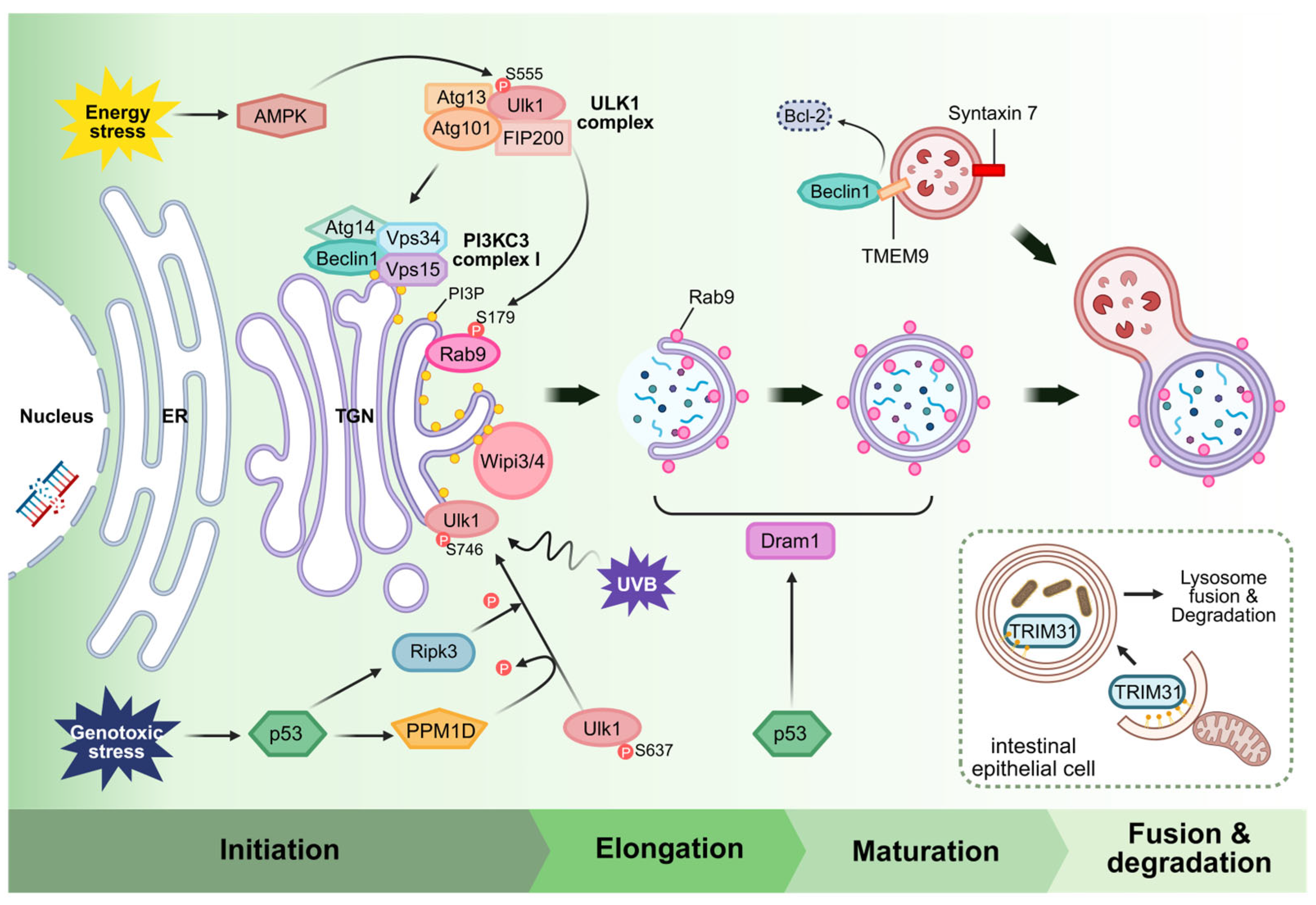
3. Rab9-Dependent Alternative Autophagy: Overview and Detailed Mechanism
3.1. Wipi3/4 in Alternative Autophagy
3.2. Isolation Membrane Closure via Dram1
3.3. Syntaxin7 in Membrane Closure
4. The Beclin1 Complex in Alternative Autophagy
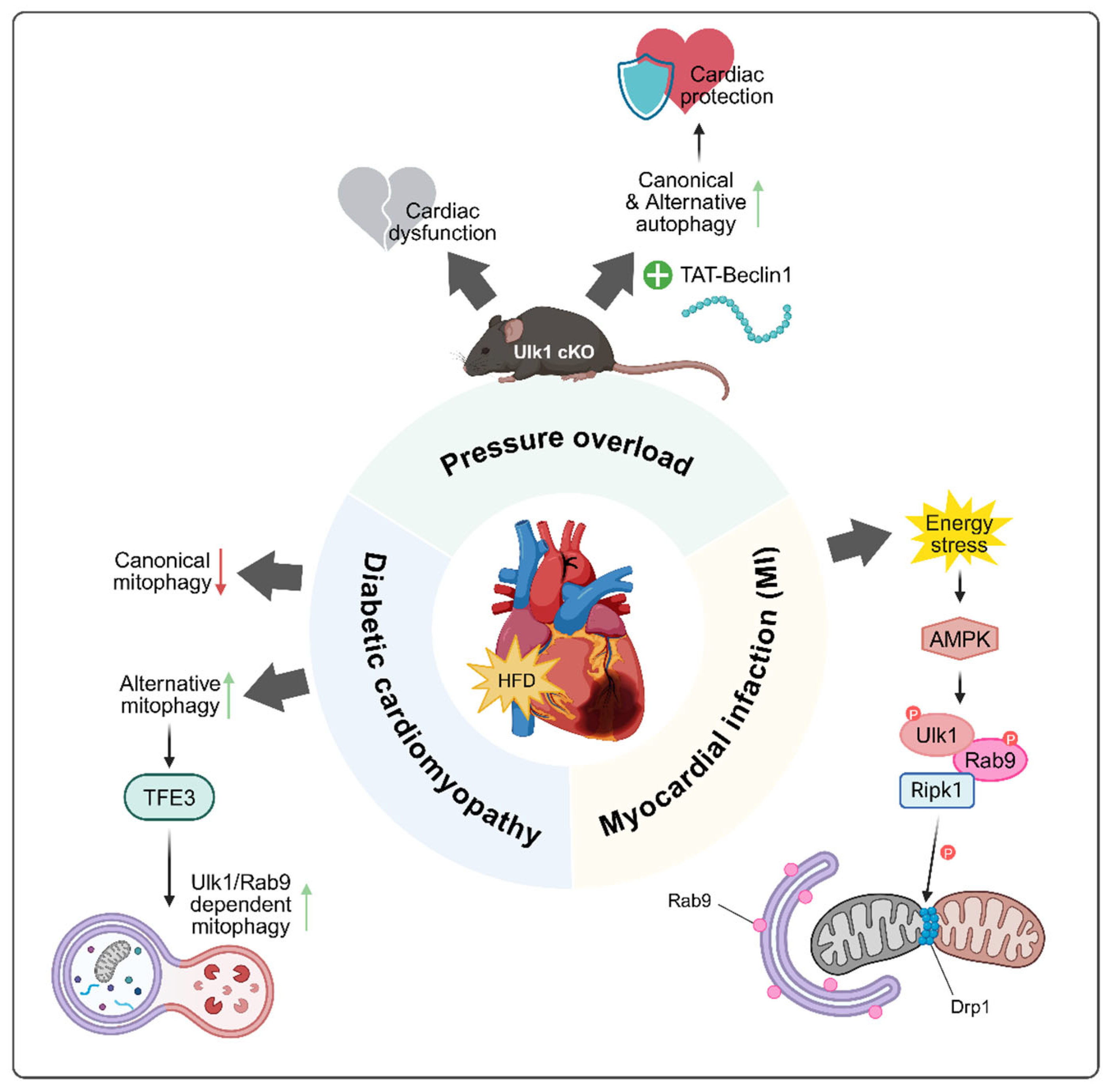
5. Physiological and Pathophysiological Roles of Alternative Autophagy
5.1. Myocardial Infarction
5.2. Pressure Overload Condition
5.3. Diabetic Cardiomyopathy
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Correction Statement
Abbreviations
| AMBRA1 | Autophagy and Beclin1 regulator 1 |
| AMPK | AMP-activated catalytic subunit alpha 1 |
| Atg | Autophagy-related |
| BARA | β-α repeated autophagic-specific domain |
| BH3 | Bcl-2 homology 3 |
| CCD | Coiled-coil domain |
| DAPK | Death-associated protein kinase 1 |
| DFCP1 | Double FYVE domain-containing protein 1 |
| DHHC5 | DHHC-type palmitoyltransferase 5 |
| DLC1 | Dynein Light Chain 1 |
| Dram1 | Damage-regulated autophagy modulator 1 |
| Drp1 | Dynamin-related protein 1 |
| ECD | Evolutionarily conserved domain |
| EPG5 | Ectopic P-granules autophagy protein 5 |
| ER | Endoplasmic reticulum |
| GABARAP | Gamma-aminobutyric acid receptor-associated protein |
| GAPR-1 | Golgi-associated plant pathogenesis-related protein 1 |
| HEK293A | Human embryonic kidney 293A |
| HFD | High-fat diet |
| HOPS | Homotypic fusion and protein sorting |
| IL-1β | Interleukin-1β |
| JNK | C-Jun N-terminal kinase |
| KO | Knockout |
| LC3 | Microtubule-associated protein 1 light chain 3 |
| LIR | LC3-interacting region |
| MEF | Mouse embryonic fibroblast |
| MI | Myocardial infarction |
| Mst1 | Mammalian Ste20-like kinases 1 |
| mTOR1 | Mammalian target of rapamycin complex 1 |
| NLRP3 | NLR family pyrin domain containing 3 |
| PE | Phosphatidylethanolamine |
| PI | Phosphatidylinositol |
| PI3K | Class III phosphatidylinositol 3-kinase |
| PI3P | Phosphatidylinositol-3-phosphate |
| PLEKHM1 | Pleckstrin homology domain-containing family M member 1 |
| PTM | Post-translational modification |
| Rab | Member RAS oncogene family |
| Ripk | Receptor-interacting serine–threonine kinase |
| Rubicon | Run domain Beclin1-interacting and cysteine-rich domain-containing protein |
| SNAP29 | Synaptosomal-associated protein 29 |
| SNARE | Soluble N-ethylmaleimide-sensitive factor attachment protein receptor |
| STX17 | Syntaxin 17 |
| TAC | Transverse aortic constriction |
| TEM | Transmission electron microscopy |
| TFE3 | Transcription factor binding to IGHM enhancer 3 |
| TFEB | Transcription factor EB |
| TGN | Trans-Golgi network |
| TMEM9 | Transmembrane protein 9 |
| TRIM | Tripartite motif |
| TSC | Tuberous Sclerosis Complex |
| Ulk1 | Unc-51 like autophagy-activating kinase 1 |
| UVB | Ultraviolet B |
| UVRAG | UV radiation resistance-associated gene protein |
| VAMP8 | Vesicle-associated membrane protein 8 |
| Wipi | WD-repeat protein, phosphoinositide-interacting |
References
- Kabeya, Y.; Mizushima, N.; Ueno, T.; Yamamoto, A.; Kirisako, T.; Noda, T.; Kominami, E.; Ohsumi, Y.; Yoshimori, T. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000, 19, 5720–5728. [Google Scholar] [CrossRef] [PubMed]
- Mizushima, N.; Yamamoto, A.; Hatano, M.; Kobayashi, Y.; Kabeya, Y.; Suzuki, K.; Tokuhisa, T.; Ohsumi, Y.; Yoshimori, T. Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. J. Cell Biol. 2001, 152, 657–668. [Google Scholar] [CrossRef] [PubMed]
- Komatsu, M.; Waguri, S.; Ueno, T.; Iwata, J.; Murata, S.; Tanida, I.; Ezaki, J.; Mizushima, N.; Ohsumi, Y.; Uchiyama, Y.; et al. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J. Cell Biol. 2005, 169, 425–434. [Google Scholar] [CrossRef]
- Hosokawa, N.; Hara, T.; Kaizuka, T.; Kishi, C.; Takamura, A.; Miura, Y.; Iemura, S.-I.; Natsume, T.; Takehana, K.; Yamada, N.; et al. Nutrient-dependent mTORC1 Association with the ULK1–Atg13–FIP200 complex required for autophagy. Mol. Biol. Cell 2009, 20, 1981–1991. [Google Scholar] [CrossRef]
- Nishida, Y.; Arakawa, S.; Fujitani, K.; Yamaguchi, H.; Mizuta, T.; Kanaseki, T.; Komatsu, M.; Otsu, K.; Tsujimoto, Y.; Shimizu, S. Discovery of Atg5/Atg7-independent alternative macroautophagy. Nature 2009, 461, 654–658. [Google Scholar] [CrossRef]
- Shang, L.; Chen, S.; Du, F.; Li, S.; Zhao, L.; Wang, X. Nutrient starvation elicits an acute autophagic response mediated by Ulk1 dephosphorylation and its subsequent dissociation from AMPK. Proc. Natl. Acad. Sci. USA 2011, 108, 4788–4793. [Google Scholar] [CrossRef]
- Kim, J.; Kundu, M.; Viollet, B.; Guan, K.-L. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat. Cell Biol. 2011, 13, 132–141. [Google Scholar] [CrossRef]
- Hara, T.; Takamura, A.; Kishi, C.; Iemura, S.-I.; Natsume, T.; Guan, J.-L.; Mizushima, N. FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. J. Cell Biol. 2008, 181, 497–510. [Google Scholar] [CrossRef]
- Park, J.-M.; Jung, C.H.; Seo, M.; Otto, N.M.; Grunwald, D.; Kim, K.H.; Moriarity, B.; Kim, Y.-M.; Starker, C.; Nho, R.S.; et al. The ULK1 complex mediates MTORC1 signaling to the autophagy initiation machinery via binding and phosphorylating ATG14. Autophagy 2016, 12, 547–564. [Google Scholar] [CrossRef]
- Axe, E.L.; Walker, S.A.; Manifava, M.; Chandra, P.; Roderick, H.L.; Habermann, A.; Griffiths, G.; Ktistakis, N.T. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J. Cell Biol. 2008, 182, 685–701. [Google Scholar] [CrossRef] [PubMed]
- Mizushima, N.; Noda, T.; Ohsumi, Y. Apg16p is required for the function of the Apg12p–Apg5p conjugate in the yeast autophagy pathway. EMBO J. 1999, 18, 3888–3896. [Google Scholar] [CrossRef]
- Ichimura, Y.; Kirisako, T.; Takao, T.; Satomi, Y.; Shimonishi, Y.; Ishihara, N.; Mizushima, N.; Tanida, I.; Kominami, E.; Ohsumi, M.; et al. A ubiquitin-like system mediates protein lipidation. Nature 2000, 408, 488–492. [Google Scholar] [CrossRef] [PubMed]
- Saito, T.; Nah, J.; Oka, S.-I.; Mukai, R.; Monden, Y.; Maejima, Y.; Ikeda, Y.; Sciarretta, S.; Liu, T.; Li, H.; et al. An alternative mitophagy pathway mediated by Rab9 protects the heart against ischemia. J. Clin. Investig. 2019, 129, 802–819. [Google Scholar] [CrossRef]
- Nah, J.; Shirakabe, A.; Mukai, R.; Zhai, P.; Sung, E.A.; Ivessa, A.; Mizushima, W.; Nakada, Y.; Saito, T.; Hu, C.; et al. Ulk1-dependent alternative mitophagy plays a protective role during pressure overload in the heart. Cardiovasc. Res. 2022, 118, 2638–2651. [Google Scholar] [CrossRef]
- Ra, E.A.; Lee, T.A.; Kim, S.W.; Park, A.; Choi, H.J.; Jang, I.; Kang, S.; Cheon, J.H.; Cho, J.W.; Lee, J.E.; et al. TRIM31 promotes Atg5/Atg7-independent autophagy in intestinal cells. Nat. Commun. 2016, 7, 11726. [Google Scholar] [CrossRef]
- Urbańska, K.; Orzechowski, A. The Secrets of Alternative Autophagy. Cells 2021, 10, 3241. [Google Scholar] [CrossRef]
- Baek, S.; Chang, J.-W.; Yoo, S.-M.; Choo, J.; Jung, S.; Nah, J.; Jung, Y.-K. TMEM9 activates Rab9-dependent alternative autophagy through interaction with Beclin1. Cell. Mol. Life Sci. 2024, 81, 322. [Google Scholar] [CrossRef]
- Liang, X.H.; Kleeman, L.K.; Jiang, H.H.; Gordon, G.; Goldman, J.E.; Berry, G.; Herman, B.; Levine, B. Protection against fatal Sindbis virus encephalitis by beclin, a novel Bcl-2-interacting protein. J. Virol. 1998, 72, 8586–8596. [Google Scholar] [CrossRef]
- He, C.; Levine, B. The Beclin 1 interactome. Curr. Opin. Cell Biol. 2010, 22, 140–149. [Google Scholar] [CrossRef] [PubMed]
- Volinia, S.; Dhand, R.; Vanhaesebroeck, B.; MacDougall, L.K.; Stein, R.; Zvelebil, M.J.; Domin, J.; Panaretou, C.; Waterfield, M.D. A human phosphatidylinositol 3-kinase complex related to the yeast Vps34p-Vps15p protein sorting system. EMBO J. 1995, 14, 3339–3348. [Google Scholar] [CrossRef]
- Matsunaga, K.; Saitoh, T.; Tabata, K.; Omori, H.; Satoh, T.; Kurotori, N.; Maejima, I.; Shirahama-Noda, K.; Ichimura, T.; Isobe, T.; et al. Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nat. Cell Biol. 2009, 11, 385–396. [Google Scholar] [CrossRef] [PubMed]
- Itakura, E.; Kishi, C.; Inoue, K.; Mizushima, N.; Subramani, S. Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol. Biol. Cell 2008, 19, 5360–5372. [Google Scholar] [CrossRef]
- Maiuri, M.C.; Le Toumelin, G.; Criollo, A.; Rain, J.-C.; Gautier, F.; Juin, P.; Tasdemir, E.; Pierron, G.; Troulinaki, K.; Tavernarakis, N.; et al. Functional and physical interaction between Bcl-X(L) and a BH3-like domain in Beclin-1. EMBO J. 2007, 26, 2527–2539. [Google Scholar] [CrossRef]
- Pattingre, S.; Tassa, A.; Qu, X.; Garuti, R.; Liang, X.H.; Mizushima, N.; Packer, M.; Schneider, M.D.; Levine, B. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell 2005, 122, 927–939. [Google Scholar] [CrossRef]
- Di Bartolomeo, S.; Corazzari, M.; Nazio, F.; Oliverio, S.; Lisi, G.; Antonioli, M.; Pagliarini, V.; Matteoni, S.; Fuoco, C.; Giunta, L.; et al. The dynamic interaction of AMBRA1 with the dynein motor complex regulates mammalian autophagy. J. Cell Biol. 2010, 191, 155–168. [Google Scholar] [CrossRef]
- Zhong, Y.; Wang, Q.J.; Li, X.; Yan, Y.; Backer, J.M.; Chait, B.T.; Heintz, N.; Yue, Z. Distinct regulation of autophagic activity by Atg14L and Rubicon associated with Beclin 1–phosphatidylinositol-3-kinase complex. Nat. Cell Biol. 2009, 11, 468–476. [Google Scholar] [CrossRef]
- Shoji-Kawata, S.; Sumpter, R.; Leveno, M.; Campbell, G.R.; Zou, Z.; Kinch, L.; Wilkins, A.D.; Sun, Q.; Pallauf, K.; MacDuff, D.; et al. Identification of a candidate therapeutic autophagy-inducing peptide. Nature 2013, 494, 201–206. [Google Scholar] [CrossRef]
- Takahashi, Y.; Coppola, D.; Matsushita, N.; Cualing, H.D.; Sun, M.; Sato, Y.; Liang, C.; Jung, J.U.; Cheng, J.Q.; Mul, J.J.; et al. Bif-1 interacts with Beclin 1 through UVRAG and regulates autophagy and tumorigenesis. Nat. Cell Biol. 2007, 9, 1142–1151. [Google Scholar] [CrossRef]
- Guo, R.; Liu, J.; Min, X.; Zeng, W.; Shan, B.; Zhang, M.; He, Z.; Zhang, Y.; He, K.; Yuan, J.; et al. Reduction of DHHC5-mediated beclin 1 S-palmitoylation underlies autophagy decline in aging. Nat. Struct. Mol. Biol. 2024, 31, 232–245. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Pattingre, S.; Sinha, S.; Bassik, M.; Levine, B. JNK1-mediated phosphorylation of Bcl-2 regulates starvation-induced autophagy. Mol. Cell 2008, 30, 678–688. [Google Scholar] [CrossRef] [PubMed]
- Berger, S.B.; Romero, X.; Ma, C.; Wang, G.; AFaubion, W.; Liao, G.; Compeer, E.; Keszei, M.; Rameh, L.; Wang, N.; et al. SLAM is a microbial sensor that regulates bacterial phagosome functions in macrophages. Nat. Immunol. 2010, 11, 920–927. [Google Scholar] [CrossRef]
- Strappazzon, F.; Vietri-Rudan, M.; Campello, S.; Nazio, F.; Florenzano, F.; Fimia, G.M.; Piacentini, M.; Levine, B.; Cecconi, F. Mitochondrial BCL-2 inhibits AMBRA1-induced autophagy. EMBO J. 2011, 30, 1195–1208. [Google Scholar] [CrossRef]
- Zalckvar, E.; Berissi, H.; Mizrachy, L.; Idelchuk, Y.; Koren, I.; Eisenstein, M.; Sabanay, H.; Pinkas-Kramarski, R.; Kimchi, A. DAP-kinase-mediated phosphorylation on the BH3 domain of beclin 1 promotes dissociation of beclin 1 from Bcl-XL and induction of autophagy. EMBO Rep. 2009, 10, 285–292. [Google Scholar] [CrossRef]
- Kim, J.; Kim, Y.C.; Fang, C.; Russell, R.C.; Kim, J.H.; Fan, W.; Liu, R.; Zhong, Q.; Guan, K.-L. Differential regulation of distinct Vps34 complexes by AMPK in nutrient stress and autophagy. Cell 2013, 152, 290–303. [Google Scholar] [CrossRef]
- Russell, R.C.; Tian, Y.; Yuan, H.; Park, H.W.; Chang, Y.-Y.; Kim, J.; Kim, H.; Neufeld, T.P.; Dillin, A.; Guan, K.-L. ULK1 induces autophagy by phosphorylating Beclin-1 and activating VPS34 lipid kinase. Nat. Cell Biol. 2013, 15, 741–750. [Google Scholar] [CrossRef]
- Wang, R.C.; Wei, Y.; An, Z.; Zou, Z.; Xiao, G.; Bhagat, G.; White, M.; Reichelt, J.; Levine, B. Akt-mediated regulation of autophagy and tumorigenesis through Beclin 1 phosphorylation. Science 2012, 338, 956–959. [Google Scholar] [CrossRef]
- Maejima, Y.; Kyoi, S.; Zhai, P.; Liu, T.; Li, H.; Ivessa, A.; Sciarretta, S.; Del Re, D.P.; Zablocki, D.K.; Hsu, C.-P.; et al. Mst1 inhibits autophagy by promoting the interaction between Beclin1 and Bcl-2. Nat. Med. 2013, 19, 1478–1488. [Google Scholar] [CrossRef] [PubMed]
- Liang, R.; Ren, J.; Zhang, Y.; Feng, W. Structural Conservation of the Two Phosphoinositide-Binding Sites in WIPI Proteins. J. Mol. Biol. 2019, 431, 1494–1505. [Google Scholar] [CrossRef] [PubMed]
- Dooley, H.C.; Razi, M.; Polson, H.E.; Girardin, S.E.; Wilson, M.I.; Tooze, S.A. WIPI2 links LC3 conjugation with PI3P, autophagosome formation, and pathogen clearance by recruiting Atg12–5-16L1. Mol. Cell 2014, 55, 238–252. [Google Scholar] [CrossRef] [PubMed]
- Bakula, D.; Müller, A.J.; Zuleger, T.; Takacs, Z.; Franz-Wachtel, M.; Thost, A.-K.; Brigger, D.; Tschan, M.P.; Frickey, T.; Robenek, H.; et al. WIPI3 and WIPI4 beta-propellers are scaffolds for LKB1-AMPK-TSC signalling circuits in the control of autophagy. Nat. Commun. 2017, 8, 15637. [Google Scholar] [CrossRef]
- Polson, H.E.; de Lartigue, J.; Rigden, D.J.; Reedijk, M.; Urbé, S.; Clague, M.J.; Tooze, S.A. Mammalian Atg18 (WIPI2) localizes to omegasome-anchored phagophores and positively regulates LC3 lipidation. Autophagy 2010, 6, 506–522. [Google Scholar] [CrossRef] [PubMed]
- Willinger, T.; Flavell, R.A. Canonical autophagy dependent on the class III phosphoinositide-3 kinase Vps34 is required for naive T-cell homeostasis. Proc. Natl. Acad. Sci. USA 2012, 109, 8670–8675. [Google Scholar] [CrossRef] [PubMed]
- Itakura, E.; Mizushima, N. Characterization of autophagosome formation site by a hierarchical analysis of mammalian Atg proteins. Autophagy 2010, 6, 764–776. [Google Scholar] [CrossRef]
- Torii, S.; Yamaguchi, H.; Nakanishi, A.; Arakawa, S.; Honda, S.; Moriwaki, K.; Nakano, H.; Shimizu, S. Identification of a phosphorylation site on Ulk1 required for genotoxic stress-induced alternative autophagy. Nat. Commun. 2020, 11, 1754. [Google Scholar] [CrossRef]
- Nagata, M.; Arakawa, S.; Yamaguchi, H.; Torii, S.; Endo, H.; Tsujioka, M.; Honda, S.; Nishida, Y.; Konishi, A.; Shimizu, S. Dram1 regulates DNA damage-induced alternative autophagy. Cell Stress 2018, 2, 55–65. [Google Scholar] [CrossRef]
- Tong, M.; Saito, T.; Zhai, P.; Oka, S.-I.; Mizushima, W.; Nakamura, M.; Ikeda, S.; Shirakabe, A.; Sadoshima, J. Alternative Mitophagy Protects the Heart Against Obesity-Associated Cardiomyopathy. Circ. Res. 2021, 129, 1105–1121. [Google Scholar] [CrossRef]
- Tong, M.; Mukai, R.; Mareedu, S.; Zhai, P.; Oka, S.-I.; Huang, C.-Y.; Hsu, C.-P.; Yousufzai, F.A.; Fritzky, L.; Mizushima, W.; et al. Distinct Roles of DRP1 in Conventional and Alternative Mitophagy in Obesity Cardiomyopathy. Circ. Res. 2023, 133, 6–21. [Google Scholar] [CrossRef]
- Jiang, P.; Nishimura, T.; Sakamaki, Y.; Itakura, E.; Hatta, T.; Natsume, T.; Mizushima, N.; Yoshimori, T. The HOPS complex mediates autophagosome–lysosome fusion through interaction with syntaxin 17. Mol. Biol. Cell 2014, 25, 1327–1337. [Google Scholar] [CrossRef]
- Lombardi, D.; Soldati, T.; Riederer, M.; Goda, Y.; Zerial, M.; Pfeffer, S. Rab9 functions in transport between late endosomes and the trans Golgi network. EMBO J. 1993, 12, 677–682. [Google Scholar] [CrossRef]
- Yamaguchi, H.; Honda, S.; Torii, S.; Shimizu, K.; Katoh, K.; Miyake, K.; Miyake, N.; Fujikake, N.; Sakurai, H.T.; Arakawa, S.; et al. Wipi3 is essential for alternative autophagy and its loss causes neurodegeneration. Nat. Commun. 2020, 11, 5311. [Google Scholar] [CrossRef]
- Proikas-Cezanne, T.; Takacs, Z.; Dönnes, P.; Kohlbacher, O. WIPI proteins: Essential PtdIns3P effectors at the nascent autophagosome. J. Cell Sci. 2015, 128, 207–217. [Google Scholar] [CrossRef]
- Crighton, D.; Wilkinson, S.; O’PRey, J.; Syed, N.; Smith, P.; Harrison, P.R.; Gasco, M.; Garrone, O.; Crook, T.; Ryan, K.M. DRAM, a p53-induced modulator of autophagy, is critical for apoptosis. Cell 2006, 126, 121–134. [Google Scholar] [CrossRef]
- Kihara, A.; Kabeya, Y.; Ohsumi, Y.; Yoshimori, T. Beclin-phosphatidylinositol 3-kinase complex functions at the trans-Golgi network. EMBO Rep. 2001, 2, 330–335. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Miao, G.; Xue, X.; Guo, X.; Yuan, C.; Wang, Z.; Zhang, G.; Chen, Y.; Feng, D.; Hu, J.; et al. The Vici Syndrome Protein EPG5 Is a Rab7 Effector that Determines the Fusion Specificity of Autophagosomes with Late Endosomes/Lysosomes. Mol. Cell 2016, 63, 781–795. [Google Scholar] [CrossRef] [PubMed]
- McEwan, D.G.; Popovic, D.; Gubas, A.; Terawaki, S.; Suzuki, H.; Stadel, D.; Coxon, F.P.; de Stegmann, D.M.; Bhogaraju, S.; Maddi, K.; et al. PLEKHM1 regulates autophagosome-Lysosome fusion through HOPS complex and LC3/GABARAP proteins. Mol. Cell 2015, 57, 39–54. [Google Scholar] [CrossRef] [PubMed]
- Narendra, D.; Tanaka, A.; Suen, D.-F.; Youle, R.J. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J. Cell Biol. 2008, 183, 795–803. [Google Scholar] [CrossRef]
- Honda, S.; Arakawa, S.; Nishida, Y.; Yamaguchi, H.; Ishii, E.; Shimizu, S. Ulk1-mediated Atg5-independent macroautophagy mediates elimination of mitochondria from embryonic reticulocytes. Nat. Commun. 2014, 5, 4004. [Google Scholar] [CrossRef]
- Ma, T.; Li, J.; Xu, Y.; Yu, C.; Xu, T.; Wang, H.; Liu, K.; Cao, N.; Nie, B.-M.; Zhu, S.-Y.; et al. Atg5-independent autophagy regulates mitochondrial clearance and is essential for iPSC reprogramming. Nat. Cell Biol. 2015, 17, 1379–1387. [Google Scholar] [CrossRef]
- Shirakabe, A.; Zhai, P.; Ikeda, Y.; Saito, T.; Maejima, Y.; Hsu, C.-P.; Nomura, M.; Egashira, K.; Levine, B.; Sadoshima, J. Drp1-Dependent Mitochondrial Autophagy Plays a Protective Role Against Pressure Overload–Induced Mitochondrial Dysfunction and Heart Failure. Circulation 2016, 133, 1249–1263. [Google Scholar] [CrossRef]
- Ikeda, Y.; Shirakabe, A.; Maejima, Y.; Zhai, P.; Sciarretta, S.; Toli, J.; Nomura, M.; Mihara, K.; Egashira, K.; Ohishi, M.; et al. Endogenous Drp1 mediates mitochondrial autophagy and protects the heart against energy stress. Circ. Res. 2015, 116, 264–278. [Google Scholar] [CrossRef]
- Wen, X.; Klionsky, D.J. Phosphorylation of ULK1 serine 746 dictates ATG5-independent autophagy. Autophagy 2020, 16, 1557–1558. [Google Scholar] [CrossRef]
- Dhingra, R.; Rabinovich-Nikitin, I.; Kirshenbaum, L.A. Ulk1/Rab9-mediated alternative mitophagy confers cardioprotection during energy stress. J. Clin. Investig. 2019, 129, 509–512. [Google Scholar] [CrossRef]
- Birgisdottir, Å.B.; Lamark, T.; Johansen, T. The LIR motif-crucial for selective autophagy. J. Cell Sci. 2013, 126, 3237–3247. [Google Scholar] [CrossRef]
- Valbuena, A.; Castro-Obregón, S.; Lazo, P.A.; Wu, G.S. Downregulation of VRK1 by p53 in response to DNA damage is mediated by the autophagic pathway. PLoS ONE 2011, 6, e17320. [Google Scholar] [CrossRef] [PubMed]
- Kveine, M.; Tenstad, E.; Døsen, G.; Funderud, S.; Rian, E. Characterization of the novel human transmembrane protein 9 (TMEM9) that localizes to lysosomes and late endosomes. Biochem. Biophys. Res. Commun. 2002, 297, 912–917. [Google Scholar] [CrossRef]
- Jung, Y.-S.; Jun, S.; Kim, M.J.; Lee, S.H.; Na Suh, H.; Lien, E.M.; Jung, H.-Y.; Lee, S.; Zhang, J.; Yang, J.-I.; et al. TMEM9 promotes intestinal tumorigenesis through vacuolar-ATPase-activated Wnt/β-catenin signalling. Nat. Cell Biol. 2018, 20, 1421–1433. [Google Scholar] [CrossRef]
- Wei, W.; Jiang, F.; Liu, X.-C.; Su, Q. TMEM9 mediates IL-6 and IL-1β secretion and is modulated by the Wnt pathway. Int. Immunopharmacol. 2018, 63, 253–260. [Google Scholar] [CrossRef] [PubMed]
- Hasegawa, T.; Noguchi, S.; Nakashima, M.; Miyai, M.; Goto, M.; Matsumoto, Y.; Torii, S.; Honda, S.; Shimizu, S. Alternative autophagy dampens UVB-induced NLRP3 inflammasome activation in human keratinocytes. J. Biol. Chem. 2024, 300, 107173. [Google Scholar] [CrossRef]
- Feng, H.; Wang, N.; Zhang, N.; Liao, H.-H. Alternative autophagy: Mechanisms and roles in different diseases. Cell Commun. Signal. 2022, 20, 43. [Google Scholar] [CrossRef]
- Mareninova, O.A.; Dillon, D.L.; Wightman, C.J.; Yakubov, I.; Takahashi, T.; Gaisano, H.Y.; Munson, K.; Ohmuraya, M.; Dawson, D.; Gukovsky, I.; et al. Rab9 Mediates Pancreatic Autophagy Switch From Canonical to Noncanonical, Aggravating Experimental Pancreatitis. Cell. Mol. Gastroenterol. Hepatol. 2022, 13, 599–622. [Google Scholar] [CrossRef]
- Thygesen, K.; Alpert, J.S.; White, H.D.; on behalf of the Joint ESC/ACCF/AHA/WHF Task Force for the Redefinition of Myocardial Infarction. Universal definition of myocardial infarction. J. Am. Coll. Cardiol. 2007, 50, 2173–2195. [Google Scholar] [CrossRef]
- Rockman, H.A.; Ross, R.S.; Harris, A.N.; Knowlton, K.U.; Steinhelper, M.E.; Field, L.J.; Ross, J.; Chien, K.R. Segregation of atrial-specific and inducible expression of an atrial natriuretic factor transgene in an in vivo murine model of cardiac hypertrophy. Proc. Natl. Acad. Sci. USA 1991, 88, 8277–8281, Erratum in Proc. Natl. Acad. Sci. USA 1991, 88, 9907. https://doi.org/10.1073/pnas.88.21.9907a. [Google Scholar] [CrossRef]
- Fang, Z.Y.; Prins, J.B.; Marwick, T.H. Diabetic cardiomyopathy: Evidence, mechanisms, and therapeutic implications. Endocr. Rev. 2004, 25, 543–567. [Google Scholar] [CrossRef] [PubMed]
- Evans, J.L.; Goldfine, I.D.; Maddux, B.A.; Grodsky, G.M. Oxidative stress and stress-activated signaling pathways: A unifying hypothesis of type 2 diabetes. Endocr. Rev. 2002, 23, 599–622. [Google Scholar] [CrossRef] [PubMed]
- Hirota, Y.; Yamashita, S.-I.; Kurihara, Y.; Jin, X.; Aihara, M.; Saigusa, T.; Kang, D.; Kanki, T. Mitophagy is primarily due to alternative autophagy and requires the MAPK1 and MAPK14 signaling pathways. Autophagy 2015, 11, 332–343. [Google Scholar] [CrossRef] [PubMed]
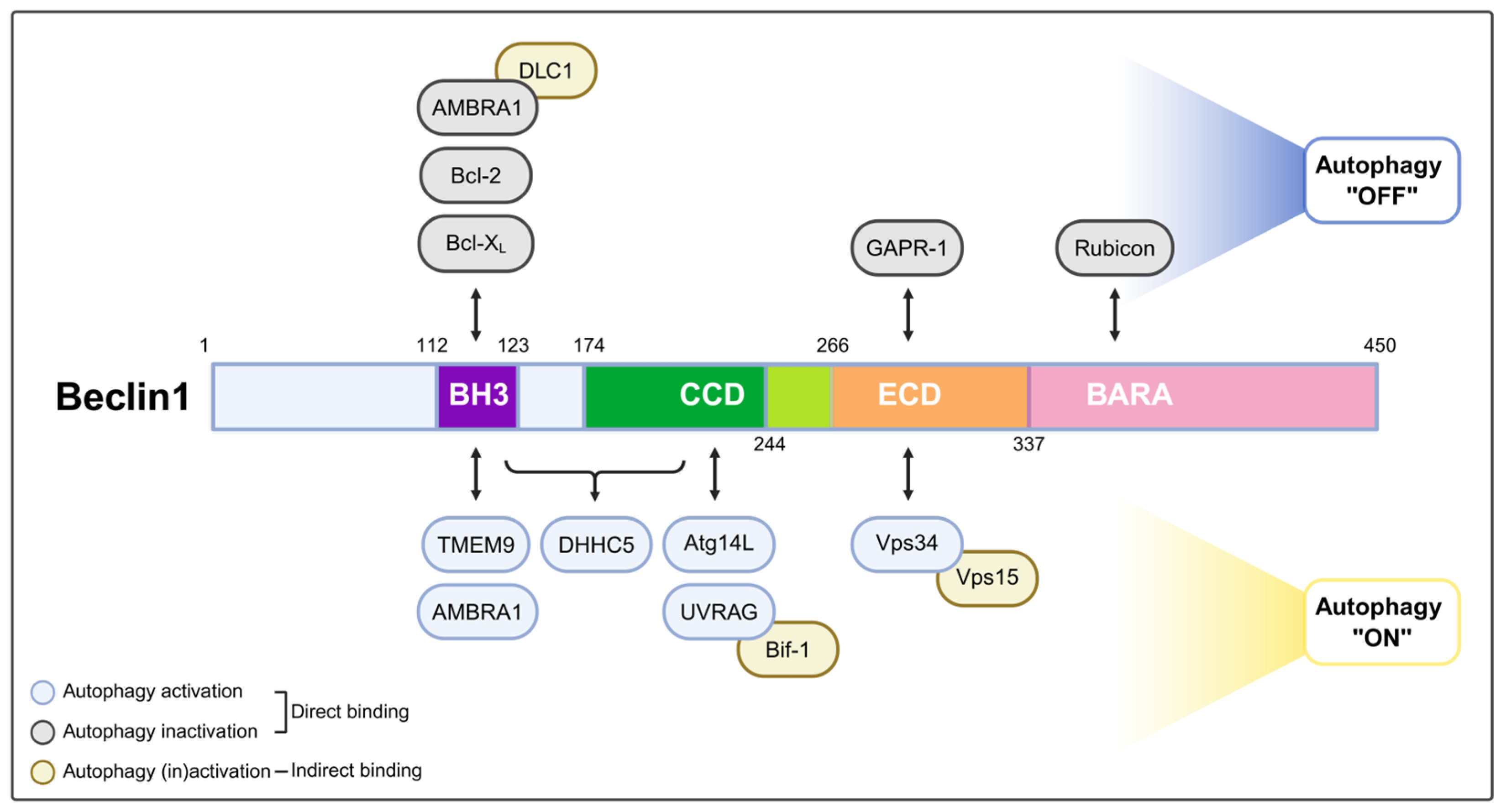
| Beclin1 Domain | Protein | Direct/Indirect | Function | Reference |
|---|---|---|---|---|
| BH3 | Bcl-2 or Bcl-XL(Bcl2L1) | Direct | Suppresses autophagy under normal conditions; stress or competition allows Beclin1-Vps34 complex formation | [18,23,24] |
| TMEM9 | Direct | Binds directly to Beclin1 and activates the Beclin1-Vps34 complex in alternative autophagy | [17] | |
| Autophagy and Beclin1 regulator 1 (AMBRA1) | Direct | Promotes Beclin1-Vps34 complex formation during starvation; enhances endoplasmic reticulum (ER) recruitment and autophagosome nucleation | [25] | |
| CCD | Atg14L | Direct | Recruits the Beclin1-Vps34-Vps15 complex to ER membranes, enabling PI3P production and initiating phagophore formation | [21,22,26] |
| UVRAG | Direct | Enhances endosomal trafficking, autophagosome maturation, and lysosome fusion through interaction with PI3KC3 complex II | [22] | |
| ECD and BARA | Vps34 | Direct | Produces PI3P, a crucial lipid signal for autophagy initiation | [20] |
| Rubicon | Direct | Inhibits autophagosome maturation by forming a complex with Beclin1-UVRAG-Vps34-Vps15 | [21,26] | |
| Golgi-associated plant pathogenesis-related protein 1 (GAPR1; GLIPR2) | Direct | Inhibits autophagy by preventing Beclin1 complex assembly at lysosomal or Golgi membranes | [27] | |
| Other interaction | Vps15 (p150) | Indirect (via Vps34) | Regulates Vps34 and promotes PI3P synthesis during autophagy initiation | [20] |
| Dynein light chain 1 (DLC1) | Indirect (via AMBRA1) | Anchors the AMBRA1-Beclin1-Vps34 complex to dynein under nutrient-rich conditions; releases upon Ulk1-mediated phosphorylation during starvation | [25] | |
| Bax-interacting factor 1 (Bif-1) | Indirect (via UVRAG) | Promotes autophagy by interacting with Beclin1 and UVRAG to activate the PI3KC3 complex, facilitating autophagosome formation | [28] | |
| DHHC-type palmitoyltransferase 5 (DHHC5) | BH3, CCD | Promotes the S-palmitoylation of Beclin1, enhancing the formation of the Atg14L-containing PI3KC3 complex critical for autophagy initiation | [29] |
| PTMs of Beclin1 | Site of PTMs | Kinases/Enzymes | Function | Reference |
|---|---|---|---|---|
| Phosphorylation | Ser14 | Ulk1 | Activation of the Vps34 complex kinase | [35] |
| Phosphorylation | Ser91 and Ser94 | AMPK | Promotion of autophagy | [34] |
| Phosphorylation | Thr108 | Mst1 | Promotion of the interaction between Beclin-1 and Bcl-2/Bcl-XL | [37] |
| Phosphorylation | Thr119 | DAPK | Disruption of the association with the anti-apoptotic protein Bcl-XL | [33] |
| Phosphorylation | Ser234 and Ser295 | Akt | Suppression of autophagy | [36] |
| S-palmitoylation | Cys137 and Cys159 | DHHC5 | Localization and autophagy initiation of Beclin1 | [29] |
| Characteristics | Canonical Autophagy | Alternative Autophagy | Reference |
|---|---|---|---|
| Primary stimuli | Nutrient deprivation, general stress | DNA damage, ischemia, prolonged pressure overload, prolonged metabolic stress | C—[4,6,7,8] |
| A—[13,14,44,45,46,47] | |||
| Phosphorylation sites of activated Ulk1 | At Ser317/Ser637/Ser777 by AMPK | At Ser746 by receptor-interacting serine-threonine kinase 3 (Ripk3) | C—[4,7] |
| A—[13,44] | |||
| Phosphorylation sites of inactivated Ulk1 | At Ser638/Ser758 by mTORC1 | - | C—[6] |
| Key regulators | Atg5, Atg7, LC3, Wipi1/2, PI3KC3 complex I/II, STX17 | Rab9, TMEM9, damage-regulated autophagy modulator 1 (Dram1), STX7, Wipi3/4 | C—[1,2,3,10,21,23,39,41,48] |
| A—[5,13,14,17,40,45,49,50,51,52] | |||
| Biomarker | Atg5, Atg7, LC3 | Rab9, TMEM9, Wipi3, Dram1 | C—[1,2,3] |
| A—[5,13,14,17,45,50] | |||
| Membrane source | ER-derived membranes | TGN-derived membranes | C—[10,43] |
| A—[5,13,49,53] | |||
| Components of Beclin1 complex | Beclin1-Vps34-Vps15 complex; regulated by Atg14L, UVRAG, Rubicon, AMBRA1 | Beclin1-TMEM9 | C—[19,20,21,22,25,26] |
| A—[17] | |||
| Membrane elongation/maturation | Atg12-Atg5-Atg16L1 complex, LC3 conjugation system | Rab9, Dram1, Wipi3 | C—[1,2,3,12] |
| A—[5,45,50] | |||
| Fusion machinery | STX17-synaptosomal-associated protein 29 (SNAP29)-vesicle-associated membrane protein 8 (VAMP8) with homotypic fusion and protein sorting (HOPS), pleckstrin homology domain-containing family M member 1 (PLEKHM1)/ectopic P-granules autophagy protein 5 (EPG5) | STX7 | C—[48,54,55] |
| A—[5,50] | |||
| Autophagosome marker | LC3 and gamma-aminobutyric acid receptor-associated protein (GABARAP) family | Rab9 | C—[1] |
| A—[5,13,14] | |||
| Cargo substrates | Bulk and selective substrates (mitochondria, etc.) | Mitochondria and intestinal bacteria | C—[1,2,3,56] |
| A—[13,14,15,46,57,58] | |||
| Function | Homeostasis, adaptation to starvation and disease | Cardioprotection in heart disease, neuronal proteostasis, mitochondrial clearance and antibacterial defense | C—[1,2,3,42,43] |
| A—[13,14,15,46,47,50,57,59,60] |
| Organ | Disease/Condition | Pathophysiological Role | Reference |
|---|---|---|---|
| Heart | Myocardial infarction (MI) | Protective | [13] |
| Pressure overload (TAC) | [14] | ||
| Diabetic cardiomyopathy | [47] | ||
| Brain | Neurodegenerative disorders | Protective | [50] |
| Intestine | Crohn’s disease | Protective | [15] |
| Pancreas | Acute pancreatitis | Detrimental | [70] |
| Skin | Sunburn | Protective | [68] |
| UVB-induced cutaneous inflammation |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Baek, S.; Jo, Y.; Nah, J. The Role of the Beclin1 Complex in Rab9-Dependent Alternative Autophagy. Int. J. Mol. Sci. 2025, 26, 9151. https://doi.org/10.3390/ijms26189151
Baek S, Jo Y, Nah J. The Role of the Beclin1 Complex in Rab9-Dependent Alternative Autophagy. International Journal of Molecular Sciences. 2025; 26(18):9151. https://doi.org/10.3390/ijms26189151
Chicago/Turabian StyleBaek, Sohyeon, Yunha Jo, and Jihoon Nah. 2025. "The Role of the Beclin1 Complex in Rab9-Dependent Alternative Autophagy" International Journal of Molecular Sciences 26, no. 18: 9151. https://doi.org/10.3390/ijms26189151
APA StyleBaek, S., Jo, Y., & Nah, J. (2025). The Role of the Beclin1 Complex in Rab9-Dependent Alternative Autophagy. International Journal of Molecular Sciences, 26(18), 9151. https://doi.org/10.3390/ijms26189151







