Exploring the Therapeutic Potential of Epigallocatechin-3-gallate (Green Tea) in Periodontitis Using Network Pharmacology and Molecular Modeling Approach
Abstract
1. Introduction
2. Results
2.1. Gene Targets of Epigallocatechin-3-gallate (EGCG)
2.2. Gene Targets of Periodontitis
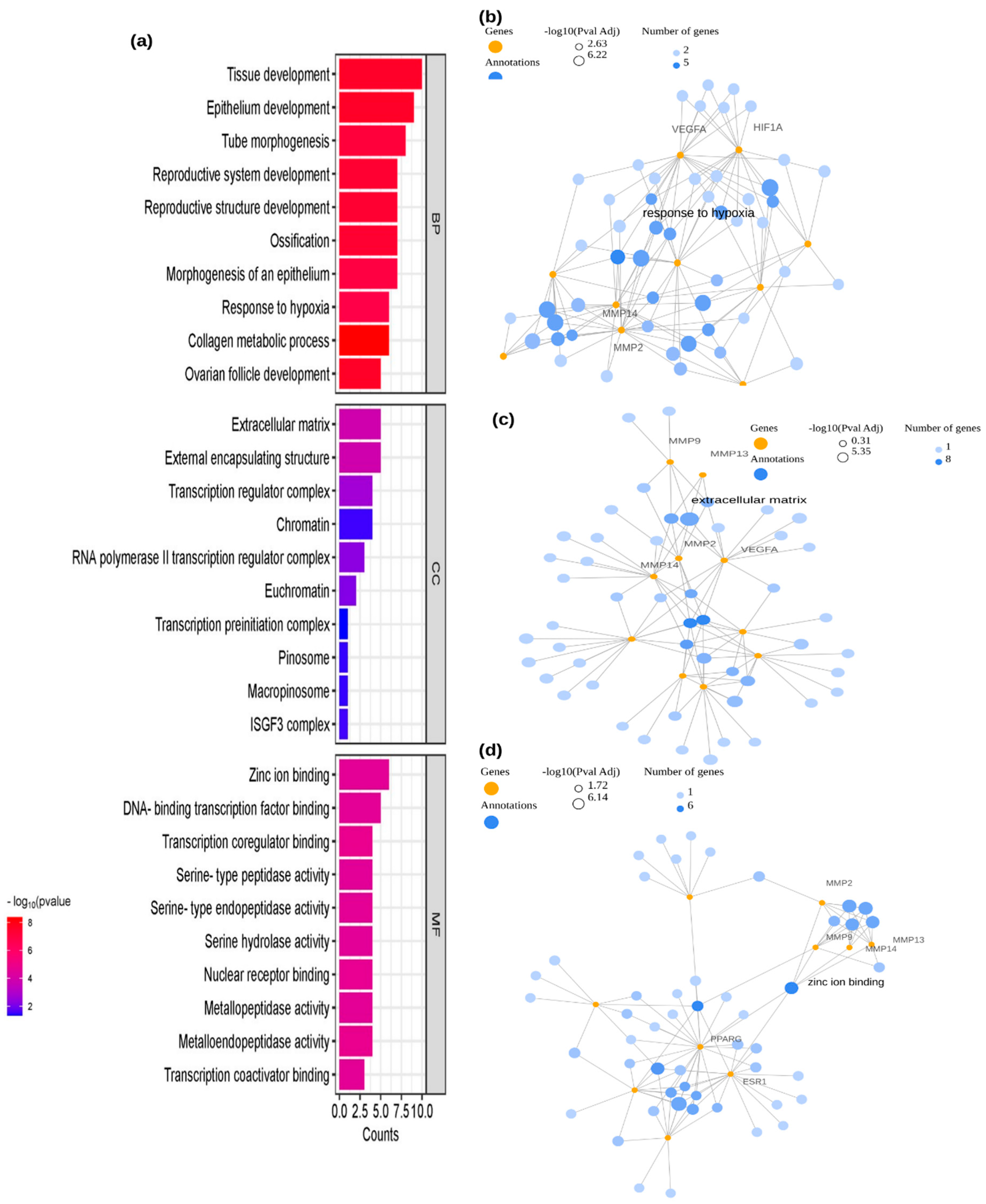
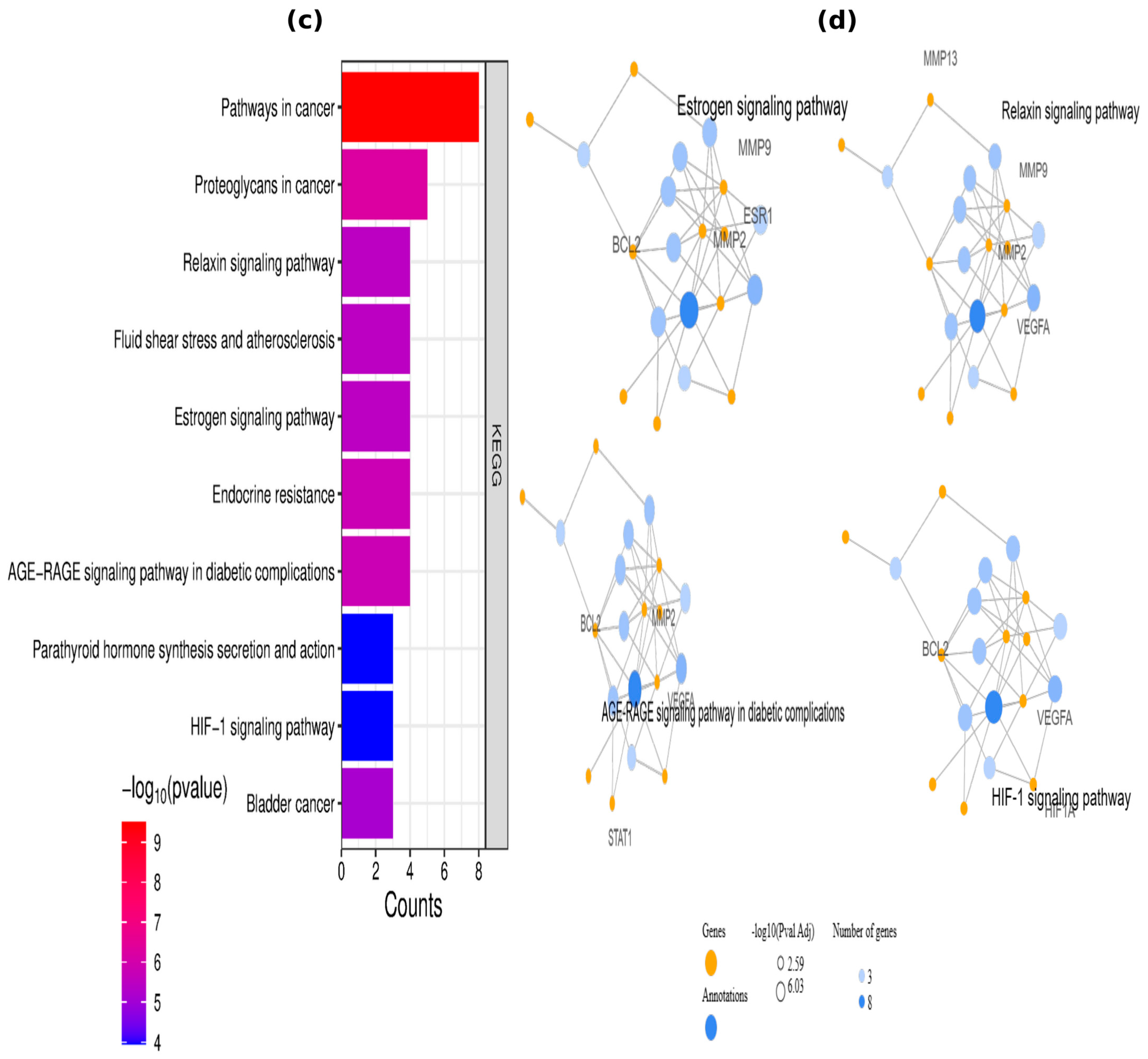
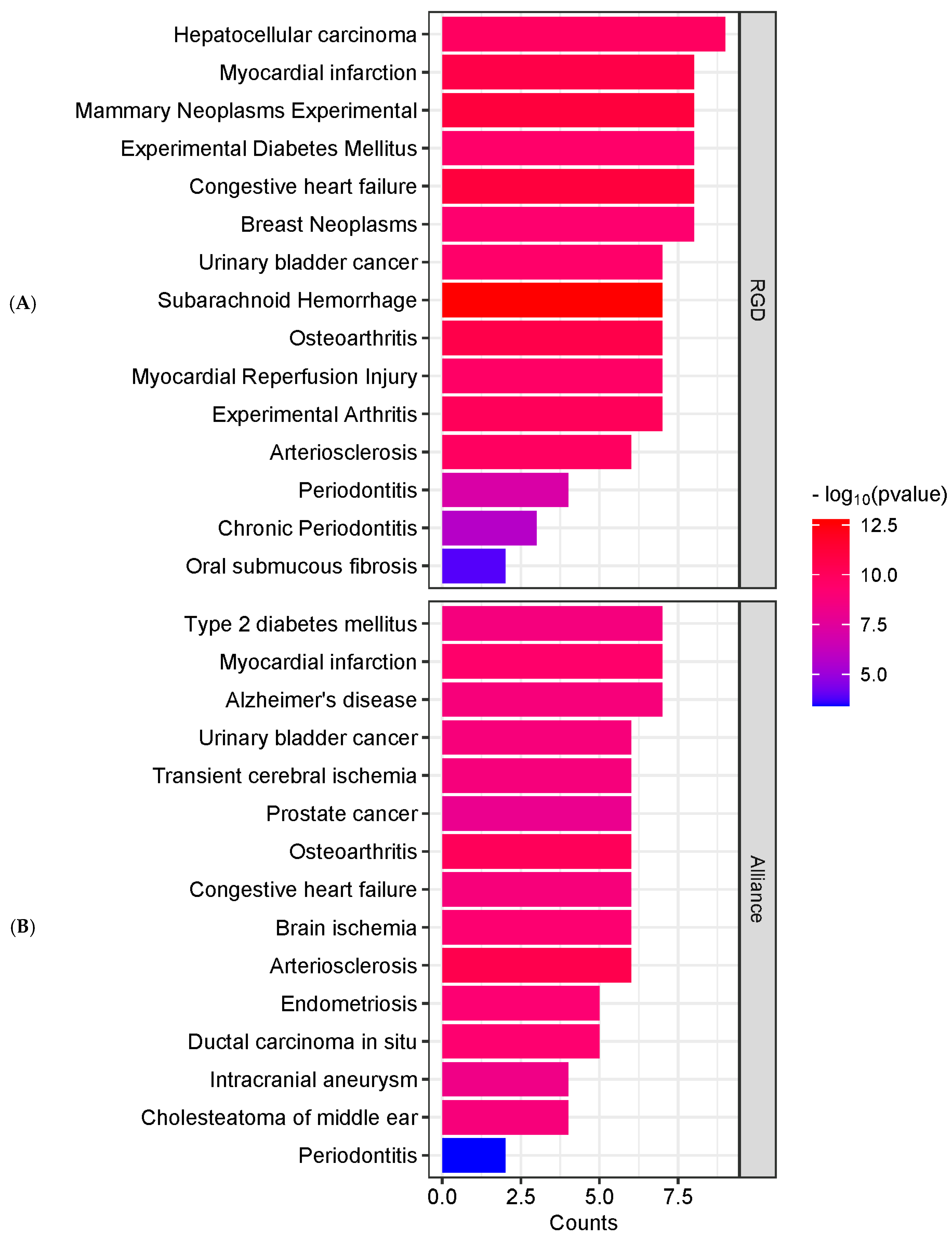
2.3. Enrichment Analysis
2.4. Construction and Analysis of PPI Networks
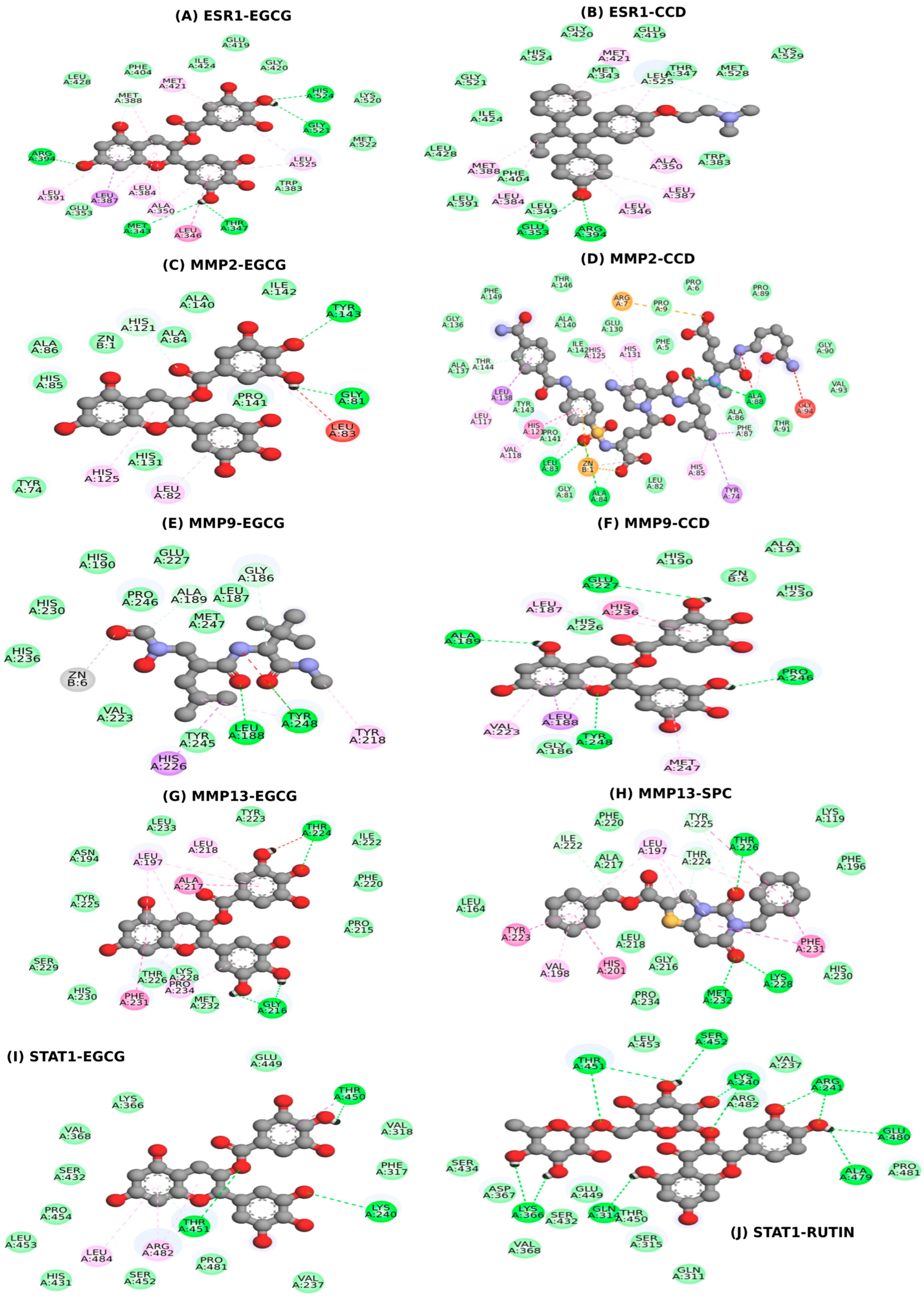
2.5. Molecular Docking
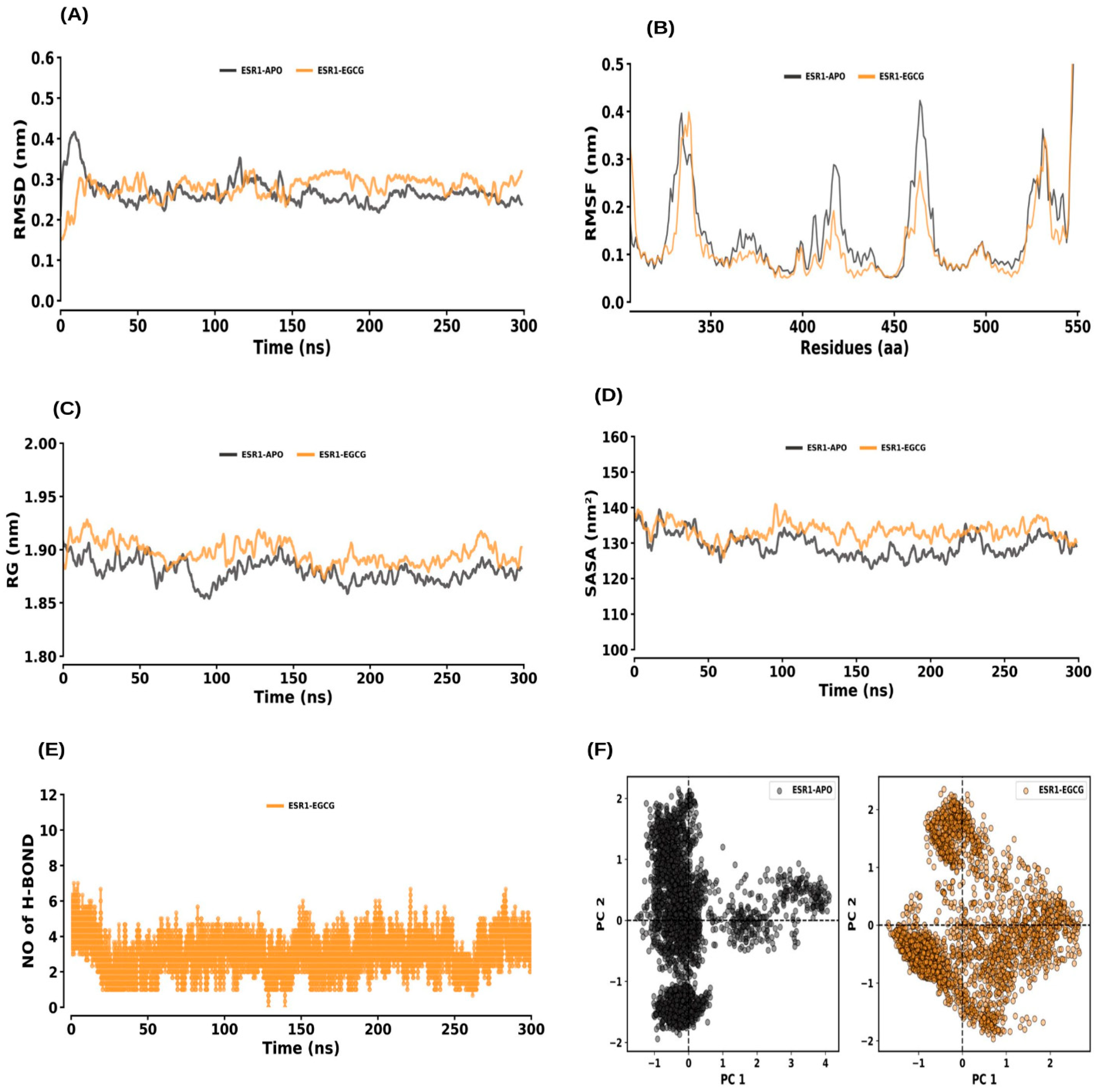
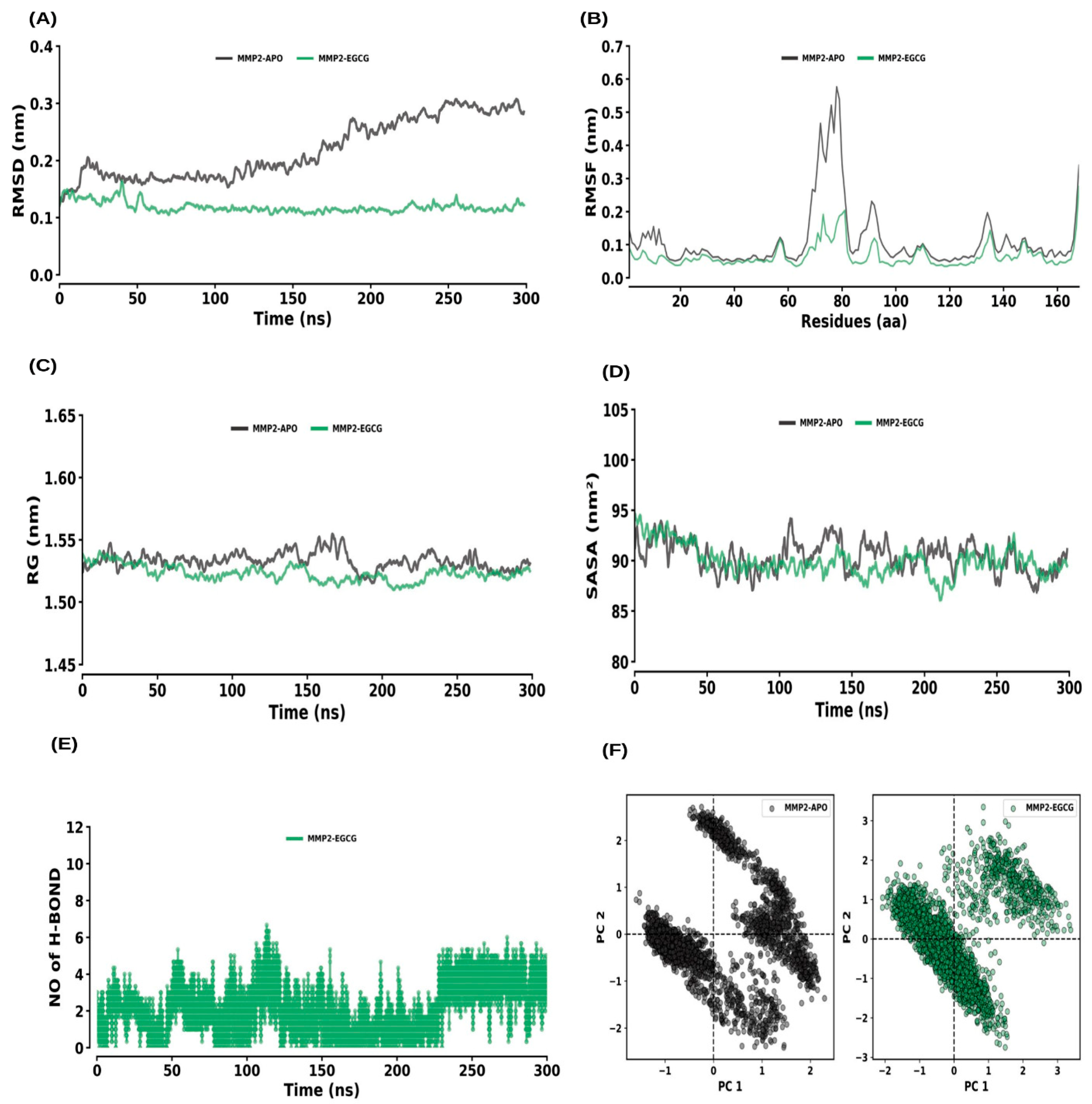
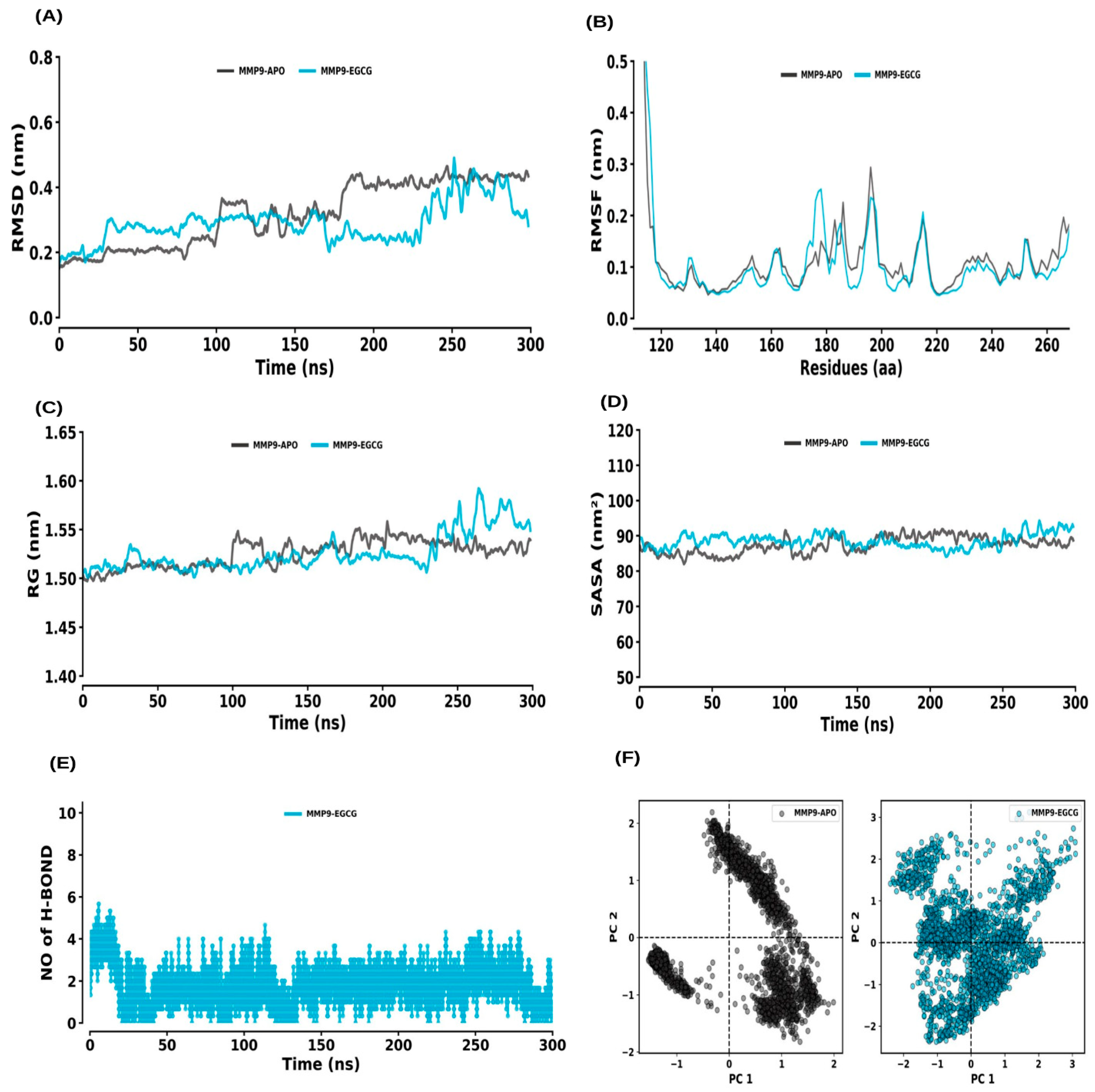
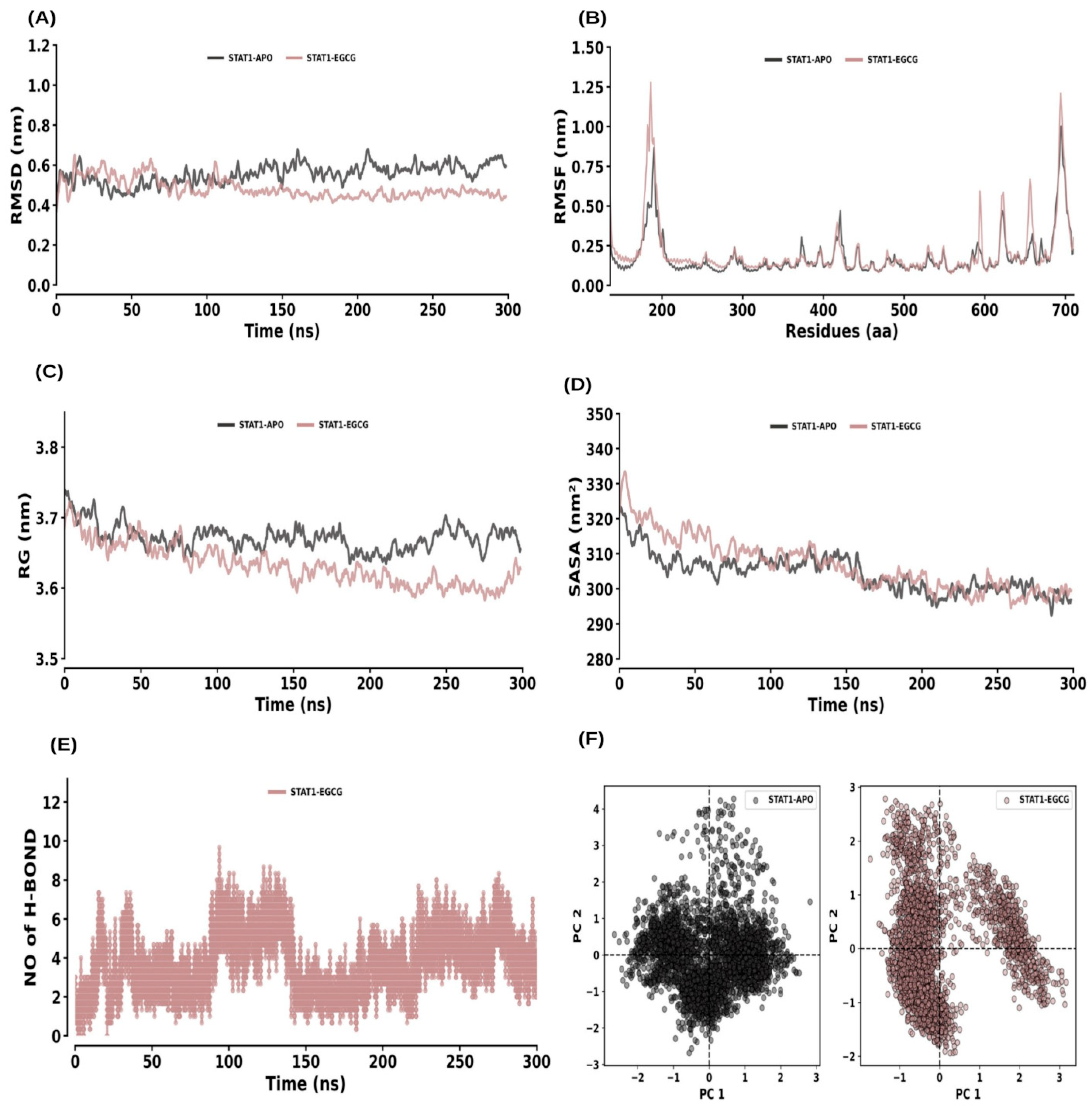
2.6. MD Simulations
2.6.1. ESR1 and ESR1–EGCG
2.6.2. MMP2 and MMP2–EGCG
2.6.3. MMP9 and MMP9–EGCG
2.6.4. MMP13 and MMP13–EGCG
2.6.5. STAT1 and STAT1–EGCG
2.7. MM–PBSA Calculation
3. Discussion
4. Materials and Methods
4.1. Epigallocatechin-3-gallate (EGCG) Compound Dataset and Target Prediction
4.2. Periodontitis Target Prediction
4.3. The Intersection of EGCG and Periodontitis Targets and Construction of PPI Networks
4.4. Enrichment Analysis
4.5. Docking
4.6. MD Simulation and MM_PBSA
5. Conclusions
Supplementary Materials
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Papapanou, P.N.; Sanz, M.; Buduneli, N.; Dietrich, T.; Feres, M.; Fine, D.H.; Flemmig, T.F.; Garcia, R.; Giannobile, W.V.; Tonetti, M.S.; et al. Periodontitis: Consensus report of workgroup 2 of the 2017 World Workshop on the Classification of Periodontal and Peri-Implant Diseases and Conditions. J. Periodontol. 2018, 89, S173–S182. [Google Scholar] [CrossRef]
- Muñoz-Carrillo, J.L.; Hernández-Reyes, V.E.; Díaz-Alfaro, L. Pathogenesis of Periodontal. In Periodontal Disease: Diagnostic and Adjunctive Non-Surgical Considerations; Springer: Berlin/Heidelberg, Germany, 2020. [Google Scholar]
- Könönen, E.; Gursoy, M.; Gursoy, U.K. Periodontitis: A multifaceted disease of tooth-supporting tissues. J. Clin. Med. 2019, 8, 1135. [Google Scholar] [CrossRef]
- Beck, J.D.; Papapanou, P.N.; Philips, K.H.; Offenbacher, S. Periodontal medicine: 100 years of progress. J. Dent. Res. 2019, 98, 1053–1062. [Google Scholar] [CrossRef] [PubMed]
- Dominy, S.S.; Lynch, C.; Ermini, F.; Benedyk, M.; Marczyk, A.; Konradi, A.; Nguyen, M.; Haditsch, U.; Raha, D.; Potempa, J.; et al. Porphyromonas gingivalis in Alzheimer’s disease brains: Evidence for disease causation and treatment with small-molecule inhibitors. Sci. Adv. 2019, 5, eaau3333. [Google Scholar] [CrossRef]
- Polak, D.; Shapira, L. An update on the evidence for pathogenic mechanisms that may link periodontitis and diabetes. J. Clin. Periodontol. 2018, 45, 150–166. [Google Scholar] [CrossRef] [PubMed]
- Herrera, D.; Meyle, J.; Renvert, S.; Jin, L. White Paper on Prevention and Management of Periodontal Diseases for Oral Health and General Health; FDI World Dental Federation: Geneva, Switzerland, 2018. [Google Scholar]
- Rajeshwari, H.R.; Dhamecha, D.; Jagwani, S.; Rao, M.; Jadhav, K.; Shaikh, S.; Puzhankara, L.; Jalalpure, S. Local drug delivery systems in the management of periodontitis: A scientific review. J. Control. Release 2019, 307, 393–409. [Google Scholar] [CrossRef] [PubMed]
- Isola, G. Current evidence of natural agents in oral and periodontal health. Nutrients 2020, 12, 585. [Google Scholar] [CrossRef]
- Song, P.; Hao, Y.; Lin, D.; Jin, Y.; Lin, J. Evaluation of the antibacterial effect of Epigallocatechin gallate on the major pathogens of canine periodontal disease and therapeutic effects on periodontal disease mice. Front. Microbiol. 2024, 14, 1329772. [Google Scholar] [CrossRef]
- Li, Y.; Cheng, L.; Li, M. Effects of Green Tea Extract Epigallocatechin-3-Gallate on oral diseases: A narrative review. Pathogens 2024, 13, 634. [Google Scholar] [CrossRef]
- Fan, Q.; Zhou, X.H.; Wang, T.F.; Zeng, F.J.; Liu, X.; Gu, Y.; Chen, B.; Yang, J.; Pang, Z.-Y.; Bai, G.H.; et al. Effects of epigallocatechin-3-gallate on oxidative stress, inflammation, and bone loss in a rat periodontitis model. J. Dent. Sci. 2023, 18, 1567–1575. [Google Scholar] [CrossRef]
- Chu, C.; Deng, J.; Man, Y.; Qu, Y. Green tea extracts epigallocatechin-3-gallate for different treatments. BioMed Res. Int. 2017, 2017, 5615647. [Google Scholar] [CrossRef]
- Garmash, O. Single-nucleotide polymorphisms of the esr1 [rs 2234693], il-1β [rs1143627], rankl [rs9594738] and [rs9594759] genes as possible risk markers for various variants of the course of periodontal disease in patients born macrosomic. Ukr. Dent. Alm. 2020, 5–18. [Google Scholar] [CrossRef]
- Shaffer, J.R.; Polk, D.E.; Wang, X.; Feingold, E.; Weeks, D.E.; Lee, M.K.; Cuenco, K.T.; Weyant, R.J.; Crout, R.J.; McNeil, D.W.; et al. Genome-wide association study of periodontal health measured by probing depth in adults ages 18–49 years. G3 Genes|Genomes|Genetics 2014, 4, 307–314. [Google Scholar] [CrossRef]
- Luchian, I.; Goriuc, A.; Sandu, D.; Covasa, M. The Role of Matrix Metalloproteinases (MMP-8, MMP-9, MMP-13) in Periodontal and Peri-Implant Pathological Processes. Int. J. Mol. Sci. 2022, 23, 1806. [Google Scholar] [CrossRef]
- Checchi, V.; Maravic, T.; Bellini, P.; Generali, L.; Consolo, U.; Breschi, L.; Mazzoni, A. The Role of Matrix Metalloproteinases in Periodontal Disease. Int. J. Environ. Res. Public Health 2020, 17, 4923. [Google Scholar] [CrossRef]
- Wei, W.; Xiao, X.; Li, J.; Ding, H.; Pan, W.; Deng, S.; Yin, W.; Xue, L.; Lu, Q.; Yue, Y.; et al. Activation of the STAT1 Pathway Accelerates Periodontitis in Nos3−/− Mice. J. Dent. Res. 2019, 98, 1027–1036. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Jiao, R.; Luo, S.; Liu, Z.; Song, J.; Chen, Z. Mechanism of action of Coptidis Rhizome in treating periodontitis based on network pharmacology and in vitro validation. BMC Oral Health 2024, 24, 530. [Google Scholar] [CrossRef]
- Shan, C.; Wu, Z.; Xia, Y.; Ji, X.; Zhang, W.; Peng, X.; Zhao, J. Network pharmacological study and in vitro studies validation-Molecular dynamics simulation of Cistanche deserticola in promoting periodontitis and bone remodeling. Int. Immunopharmacol. 2024, 135, 112299. [Google Scholar] [CrossRef] [PubMed]
- Tuerhong, K.; Liu, K.; Shen, D.; Zhang, Q.; Huang, Q.; Yang, M.; Huang, Z.; Wang, L.; Yang, S.; Li, Y. Integrating network pharmacology and experimental evaluation to explore the complementary therapeutic effect and mechanism of melatonin in periodontitis. Heliyon 2024, 10, e32494. [Google Scholar] [CrossRef] [PubMed]
- Shen, D.; Tuerhong, K.; Huang, Q.; Liu, K.; Li, Y.; Yang, S. Computational analysis of curcumin-mediated alleviation of inflammation in periodontitis patients with experimental validation in mice. J. Clin. Periodontol. 2024, 51, 787–799. [Google Scholar] [CrossRef]
- Zhao, Q.; Wang, K.; Hou, L.; Guo, L.; Liu, X. Based on network pharmacology and molecular docking to explore the potential mechanism of shikonin in periodontitis. BMC Oral Health 2024, 24, 839. [Google Scholar] [CrossRef]
- Liu, Y.; Cao, F.; Shi, M.; Deng, Z.; Guo, K.; Fan, T.; Meng, Y.; Bu, M.; Ma, Z. Investigation of the mechanism of baicalein in the treatment of periodontitis based on network pharmacology, molecular docking and experimental validation. BMC Oral Health 2024, 24, 987. [Google Scholar] [CrossRef]
- Wu, Z.; Ji, X.; Shan, C.; Song, J.; Zhao, J. Exploring the pharmacological components and effective mechanism of Mori Folium against periodontitis using network pharmacology and molecular docking. Arch. Oral Biol. 2022, 139, 105391. [Google Scholar] [CrossRef]
- Shah, A.B.; Lee, K.Y. Integrative metabolomics and system pharmacology reveal the antioxidant blueprint of Psoralea corylifolia. Sci. Rep. 2025, 15, 28632. [Google Scholar] [CrossRef]
- Taskan, M.M.; Karatas, O.; Yuce, H.B.; Kara, G.I.; Gevrek, F.; Yarkac, F.U. Hypoxia and collagen crosslinking in the healthy and affected sites of periodontitis patients. Acta Odontol. Scand. 2019, 77, 600–607. [Google Scholar] [CrossRef] [PubMed]
- Paula-Silva, F.W.; da Silva, L.A.; Kapila, Y.L. Matrix metalloproteinase expression in teeth with apical periodontitis is differentially modulated by the modality of root canal treatment. J. Endod. 2010, 36, 231–237. [Google Scholar] [CrossRef] [PubMed]
- Kong, C.; Zhang, H.; Li, L.; Liu, Z. Effects of green tea extract epigallocatechin-3-gallate (EGCG) on oral disease-associated microbes: A review. J. Oral Microbiol. 2022, 14, 2131117. [Google Scholar] [CrossRef]
- Miao, L.; Zhan, S.; Liu, J. Interleukin-12-mediated expression of matrix metalloproteinases in human periodontal ligament fibroblasts involves in NF-κB activation. Biosci. Rep. 2017, 37, BSR20170973. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Guan, M.; Wei, L.; Yan, H. IL-18 promotes the secretion of matrix metalloproteinases in human periodontal ligament fibroblasts by activating NF-κB signaling. Mol. Med. Rep. 2019, 19, 703–710. [Google Scholar] [CrossRef]
- Sorsa, T.; Tjäderhane, L.; Konttinen, Y.T.; Lauhio, A.; Salo, T.; Lee, H.; Golub, L.M.; Brown, D.L.; Mäntylä, P. Matrix metalloproteinases: Contribution to pathogenesis, diagnosis and treatment of periodontal inflammation. Ann. Med. 2006, 38, 306–321. [Google Scholar] [CrossRef]
- Robinson, J.L.; Johnson, P.M.; Kister, K.; Yin, M.T.; Chen, J.; Wadhwa, S. Estrogen signaling impacts temporomandibular joint and periodontal disease pathology. Odontology 2020, 108, 153–165. [Google Scholar] [CrossRef]
- Palanisamy, S. The impact of estrogen on periodontal tissue integrity and inflammation—A mini review. Front. Dent. Med. 2025, 6, 1455755. [Google Scholar] [CrossRef]
- Anton, D.-M.; Martu, M.-A.; Sufaru, I.-G.; Pasarin, L.; Luchian, I.; Maris, M.; Scutariu, M.; Martu, I. The role of AGE/RAGE in the pathophysiology of the diabetes-periodontitis relationship. Rom. J. Med. Dent. Educ. 2020, 9, 28–36. [Google Scholar]
- Celik, D.; Kantarci, A. Vascular changes and hypoxia in periodontal disease as a link to systemic complications. Pathogens 2021, 10, 1280. [Google Scholar] [CrossRef] [PubMed]
- Miguez, P.A.; Bash, E.; Musskopf, M.L.; Tuin, S.A.; Rivera-Concepcion, A.; Chapple, I.L.C.; Liu, J. Control of tissue homeostasis by the extracellular matrix: Synthetic heparan sulfate as a promising therapeutic for periodontal health and bone regeneration. Periodontology 2000 2024, 94, 510–531. [Google Scholar] [CrossRef] [PubMed]
- Jeon, H.H.; Huang, X.; Cortez, L.R.; Sripinun, P.; Lee, J.; Hong, J.J.; Graves, D.T. Inflammation and mechanical force-induced bone remodeling. Periodontology 2000 2024. online ahead of print. [Google Scholar] [CrossRef] [PubMed]
- Zazeri, G.; Povinelli, A.P.R.; Le Duff, C.S.; Tang, B.; Cornelio, M.L.; Jones, A.M. Synthesis and Spectroscopic Analysis of Piperine- and Piperlongumine-Inspired Natural Product Scaffolds and Their Molecular Docking with IL-1β and NF-κB Proteins. Molecules 2020, 25, 2841. [Google Scholar] [CrossRef]
- Zazeri, G.; Povinelli, A.P.R.; Pavan, N.M.; Jones, A.M.; Ximenes, V.F. Solvent-Induced Lag Phase during the Formation of Lysozyme Amyloid Fibrils Triggered by Sodium Dodecyl Sulfate: Biophysical Experimental and In Silico Study of Solvent Effects. Molecules 2023, 28, 6891. [Google Scholar] [CrossRef]
- Cai, Y.; Chen, Z.; Liu, H.; Xuan, Y.; Wang, X.; Luan, Q. Green tea epigallocatechin-3-gallate alleviates Porphyromonas gingivalis-induced periodontitis in mice. Int. Immunopharmacol. 2015, 29, 839–845. [Google Scholar] [CrossRef]
- Daina, A.; Michielin, O.; Zoete, V. SwissTargetPrediction: Updated data and new features for efficient prediction of protein targets of small molecules. Nucleic Acids Res. 2019, 47, W357–W364. [Google Scholar] [CrossRef]
- Ru, J.; Li, P.; Wang, J.; Zhou, W.; Li, B.; Huang, C.; Li, P.; Guo, Z.; Tao, W.; Yang, L.; et al. TCMSP: A database of systems pharmacology for drug discovery from herbal medicines. J. Cheminformatics 2014, 6, 13. [Google Scholar] [CrossRef]
- The UniProt Consortium. UniProt: The universal protein knowledgebase. Nucleic Acids Res. 2018, 46, 2699. [Google Scholar] [CrossRef]
- Safran, M.; Dalah, I.; Alexander, J.; Rosen, N.; Stein, T.I.; Shmoish, M.; Nativ, N.; Bahir, I.; Doniger, T.; Lancet, D.; et al. GeneCards Version 3: The human gene integrator. Database 2010, 2010, baq020. [Google Scholar] [CrossRef]
- Clough, E.; Barrett, T. The gene expression omnibus database. In Statistical Genomics: Methods and Protocols; Springer: Berlin/Heidelberg, Germany, 2016; pp. 93–110. [Google Scholar]
- Bardou, P.; Mariette, J.; Escudié, F.; Djemiel, C.; Klopp, C. jvenn: An interactive Venn diagram viewer. BMC Bioinform. 2014, 15, 293. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Mering, C.V.; et al. STRING v11: Protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Chin, C.H.; Chen, S.H.; Wu, H.H.; Ho, C.W.; Ko, M.T.; Lin, C.Y. cytoHubba: Identifying hub objects and sub-networks from complex interactome. BMC Syst. Biol. 2014, 8, S11. [Google Scholar] [CrossRef] [PubMed]
- Ge, S.X.; Jung, D.; Yao, R. ShinyGO: A graphical gene-set enrichment tool for animals and plants. Bioinformatics 2020, 36, 2628–2629. [Google Scholar] [CrossRef]
- Tang, D.; Chen, M.; Huang, X.; Zhang, G.; Zeng, L.; Zhang, G.; Wu, S.; Yin, Y.; Wang, Y. SRplot: A free online platform for data visualization and graphing. PLoS ONE 2023, 18, e0294236. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef]
- Kim, S. Getting the most out of PubChem for virtual screening. Expert Opin. Drug Discov. 2016, 11, 843–855. [Google Scholar] [CrossRef]
- O’Boyle, N.M.; Banck, M.; James, C.A.; Morley, C.; Vandermeersch, T.; Hutchison, G.R. Open Babel: An open chemical toolbox. J. Cheminform. 2011, 3, 33. [Google Scholar] [CrossRef]
- Hanwell, M.D.; Curtis, D.E.; Lonie, D.C.; Vandermeersch, T.; Zurek, E.; Hutchison, G.R. Avogadro: An advanced semantic chemical editor, visualization, and analysis platform. J. Cheminformatics 2012, 4, 17. [Google Scholar] [CrossRef] [PubMed]
- DeLano, W.L. The PyMOL Molecular Graphics System. 2002. Available online: http://www.pymol.org (accessed on 4 August 2025).
- Visualizer, D. Discovery Studio Visualizer, 2nd ed.; Accelrys Software Inc.: San Diego, CA, USA, 2005. [Google Scholar]
- Malde, A.K.; Zuo, L.; Breeze, M.; Stroet, M.; Poger, D.; Nair, P.C.; Oostenbrink, C.; Mark, A.E. An automated force field topology builder (ATB) and repository: Version 1.0. J. Chem. Theory Comput. 2011, 7, 4026–4037. [Google Scholar] [CrossRef] [PubMed]
- Abraham, M.J.; Van Der Spoel, D.; Lindahl, E.; Hess, B. GROMACS User Manual, Version 5.0.4; The GROMACS Development Team: Uppsala, Sweden, 2014. [Google Scholar]
- Gangadharappa, B.S.; Sharath, R.; Revanasiddappa, P.D.; Chandramohan, V.; Balasubramaniam, M.; Vardhineni, T.P. Structural insights of metallo-beta-lactamase revealed an effective way of inhibition of enzyme by natural inhibitors. J. Biomol. Struct. Dyn. 2020, 38, 3757–3771. [Google Scholar] [CrossRef] [PubMed]
- Kumari, R.; Kumar, R.; Open Source Drug Discovery Consortium; Lynn, A. g_mmpbsa—A GROMACS tool for high-throughput MM-PBSA calculations. J. Chem. Inf. Model. 2014, 54, 1951–1962. [Google Scholar] [CrossRef]




| Name | PubChem ID | MF | MW | Canonical Smiles | Structure |
|---|---|---|---|---|---|
| Epigallo catechin-3-gallate | 65064 | C22H18O11 | 458.4 | C1[C@H]([C@H](OC2=CC(=CC(=C21)O)O)C3=CC(=C(C(=C3)O)O)O)OC(=O)C4=CC (=C(C(=C4)O)O)O | 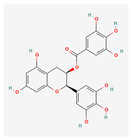 |
| Target Name | Protein–Ligand Complex | Docking Score | Interactions |
|---|---|---|---|
| ESR1 | ESR1–EGCG (Epigallocatechin gallate) | −8.2 | MET343, LEU346, THR347, ALA350, GLU353, TRP383, LEU384, LEU387, MET388, LEU391, ARG394, PHE404, GLU419, GLY420, MET421, LYS520, GLY521, MET522, HIS524, LEU525, |
| ESR1–CCD (Co-Crystal Ligand (Standard)) | −9.5 | MET343, LEU346, THR347, LEU349, ALA350, GLU353, TRP383, LEU384, LEU387, MET388, ARG394, PHE404, GLU419, GLY420, MET421, LEU428, GLY521, HIS524, LEU525, MET528, | |
| MMP2 | MMP2–EGCG (Epigallocatechin gallate) | −7.4 | ZN1, TYR74, GLY81, LEU82, LEU83, ALA84, HIS85, ALA86, HIS121, HIS125, ALA140, PRO141, ILE142, TYR143, |
| MMP2–CCD (Co-Crystal Ligand (Standard)) | −9.9 | ZN1, PHE5, PRO6, TYR74, GLY81, LEU82, LEU83, ALA84, HIS85, ALA86, PHE87, ALA88, PRO89, GLY90, THR91, VAL93, GLY94, LEU117, VAL118, HIS121, HIS125, GLU130, HIS131, GLY136, ALA137, LEU138, PRO141, ILE142, TYR143, THR144, PHE149, | |
| MMP9 | MMP9–EGCG (Epigallocatechin gallate) | −8.3 | ZN6, GLY186, LEU187, LEU188, ALA189, ALA191, VAL223, HIS226, GLU227, HIS230, HIS236, PRO246, MET247, TYR248, |
| MMP9–CCD (Co-Crystal Ligand (Standard)) | −6.7 | ZN6, GLY186, LEU187, LEU188, ALA189, TYR218, VAL223, HIS226, GLU227, HIS230, HIS236, TYR245, PRO246, MET247, TYR248, | |
| MMP13 | MMP13–EGCG (Epigallocatechin gallate) | −8.8 | LEU197, PRO215, GLY216, ALA217, LEU218, ILE222, TYR223, THR224, TYR225, THR226, LYS228, SER229, HIS230, PHE231, MET232, PRO234, |
| MMP13–SPC Co-Crystal Ligand (SPC) Standard | −12.3 | LYS119, PHE196, LEU197, VAL198, HIS201, GLY216, ALA217, LEU218, PHE220, ILE222, TYR223, THR224, TYR225, THR226, LYS228, HIS230, PHE231, MET232, PRO234, | |
| STAT1 | STAT1–EGCG (Epigallocatechin gallate) | −7.3 | LYS240, PHE317, LYS366, VAL368, HIS431, SER432, GLU449, THR450, THR451, SER452, LEU453, PRO454, ARG482, LEU484, |
| STAT1–Rutin (Standard) | −9.0 | VAL237, LYS240, ARG241, GLN314, SER315, LYS366, ASP367, SER432, SER434, GLU449, THR450, THR451, SER452, ALA479, GLU480, PRO481, ARG482, |
| Protein Type | RMSD (nm) | RMSF (nm) | Rg (nm) | SASA (nm2) | NH-Bond (Protein–Drug) | Covariance |
|---|---|---|---|---|---|---|
| ESR1–APO ESR1–EGCG | 0.27 ± 0.03 | 0.15 ± 0.12 | 1.88 ± 0.01 | 129.45 ± 3.66 | NA | 26.63 |
| 0.28 ± 0.03 | 0.13 ± 0.14 | 1.90 ± 0.01 | 133.32 ± 2.91 | 4.0 ± 1.0 | 26.77 | |
| MMP2–APO MMP2–EGCG | 0.22 ± 0.05 | 0.11 ± 0.10 | 1.53 ± 0.01 | 90.43 ± 2.01 | NA | 10.60 |
| 0.12 ± 0.01 | 0.07 ± 0.04 | 1.52 ± 0.01 | 89.96 ± 1.93 | 3.0 ± 1.0 | 02.73 | |
| MMP9–APO MMP9–EGCG | 0.32 ± 0.10 | 0.13 ± 0.17 | 1.53 ± 0.01 | 87.39 ± 2.70 | NA | 21.25 |
| 0.29 ± 0.06 | 0.12 ± 0.13 | 1.53 ± 0.02 | 88.58 ± 2.40 | 2.0 ± 1.0 | 26.12 | |
| MMP13–APO MMP13–EGCG | 0.44 ± 0.03 | 0.14 ± 0.09 | 1.57 ± 0.01 | 93.05 ± 3.32 | NA | 12.84 |
| 0.46 ± 0.07 | 0.16 ± 0.17 | 1.54 ± 0.02 | 98.05 ± 2.79 | 5.0 ± 1.4 | 14.02 | |
| STAT1–APO STAT1–EGCG | 0.55 ± 0.06 | 0.18 ± 0.13 | 3.67 ± 0.02 | 304.35 ± 5.97 | NA | 83.65 |
| 0.48 ± 0.06 | 0.21 ± 0.18 | 3.63 ± 0.03 | 306.82 ± 8.18 | 3.5 ± 1.7 | 125.44 |
| Complex Type | Binding Energy (K/Cal) | van der Waal Energy (K/Cal) | Electrostatic Energy (K/Cal) | Polar Solvation Energy (K/Cal) | SASA Energy (K/Cal) |
|---|---|---|---|---|---|
| ESR1–EGCG | −113.37 ± 10.75 | −197.13 ± 12.46 | −94.15 ± 17.39 | 199.88 ± 6.88 | −21.97 ± 0.84 |
| MMP2–EGCG | −31.63 ± 9.74 | −101.99 ± 9.42 | −14.67 ± 14.52 | 98.35 ± 22.39 | −13.33 ± 0.95 |
| MMP9–EGCG | −77.48 ± 21.45 | −138.86 ± 11.84 | −25.42 ± 37.36 | 103.03 ± 31.29 | −16.23 ± 0.82 |
| MMP13–EGCG | −101.94 ± 14.42 | −217.54 ± 18.83 | −147.19 ± 15.21 | 284.23 ± 13.69 | −21.45 ± 0.74 |
| STAT1–EGCG | −28.04 ± 13.04 | −135.28 ± 17.71 | −131.22 ± 34.87 | 256.03 ± 29.55 | −17.56 ± 0.98 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kamaraj, B. Exploring the Therapeutic Potential of Epigallocatechin-3-gallate (Green Tea) in Periodontitis Using Network Pharmacology and Molecular Modeling Approach. Int. J. Mol. Sci. 2025, 26, 9144. https://doi.org/10.3390/ijms26189144
Kamaraj B. Exploring the Therapeutic Potential of Epigallocatechin-3-gallate (Green Tea) in Periodontitis Using Network Pharmacology and Molecular Modeling Approach. International Journal of Molecular Sciences. 2025; 26(18):9144. https://doi.org/10.3390/ijms26189144
Chicago/Turabian StyleKamaraj, Balu. 2025. "Exploring the Therapeutic Potential of Epigallocatechin-3-gallate (Green Tea) in Periodontitis Using Network Pharmacology and Molecular Modeling Approach" International Journal of Molecular Sciences 26, no. 18: 9144. https://doi.org/10.3390/ijms26189144
APA StyleKamaraj, B. (2025). Exploring the Therapeutic Potential of Epigallocatechin-3-gallate (Green Tea) in Periodontitis Using Network Pharmacology and Molecular Modeling Approach. International Journal of Molecular Sciences, 26(18), 9144. https://doi.org/10.3390/ijms26189144






