Proteomic Screening for Cellular Targets of the Duck Enteritis Virus Protein VP26 Reveals That the Host Actin–Myosin II Network Regulates the Proliferation of the Virus
Abstract
1. Introduction
2. Results
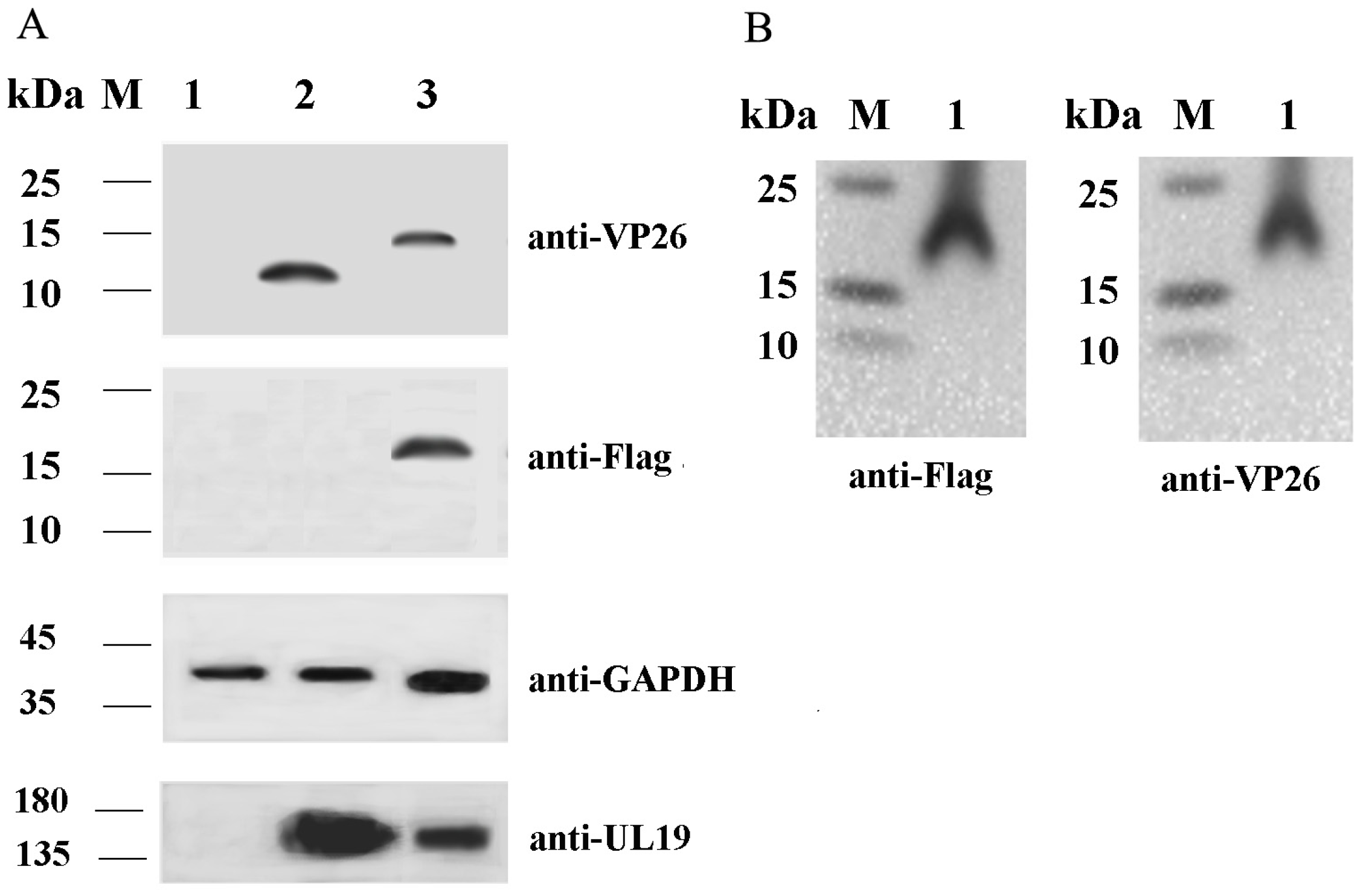
2.1. Validation of VP26 Expression in Recombinant Virus-Infected Cells Using Western Blot Analysis
2.2. Identification of the Interaction Between DEV VP26 and the Host Protein
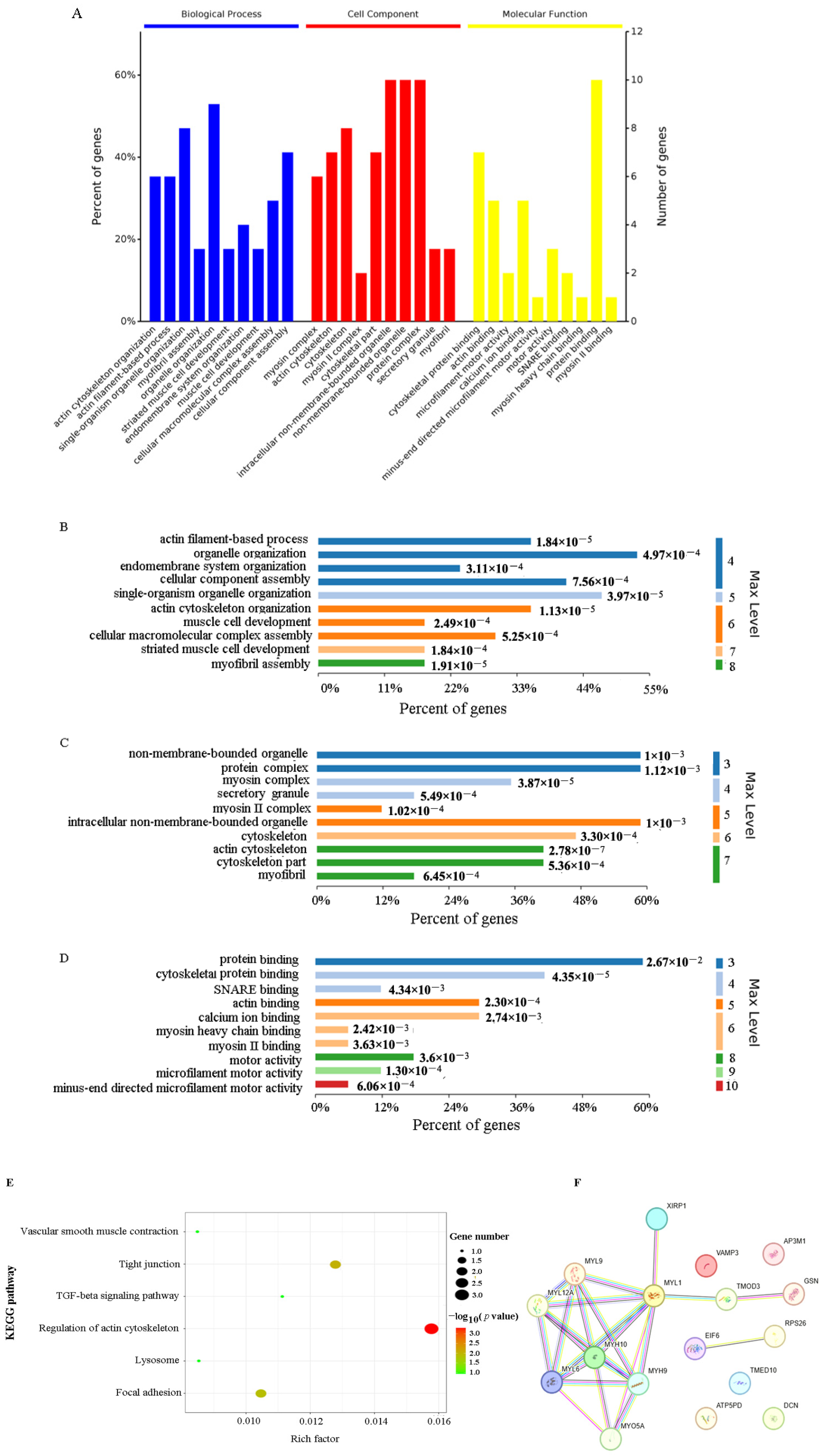
2.3. Functional Characterization of VP26-Associated Host Proteins
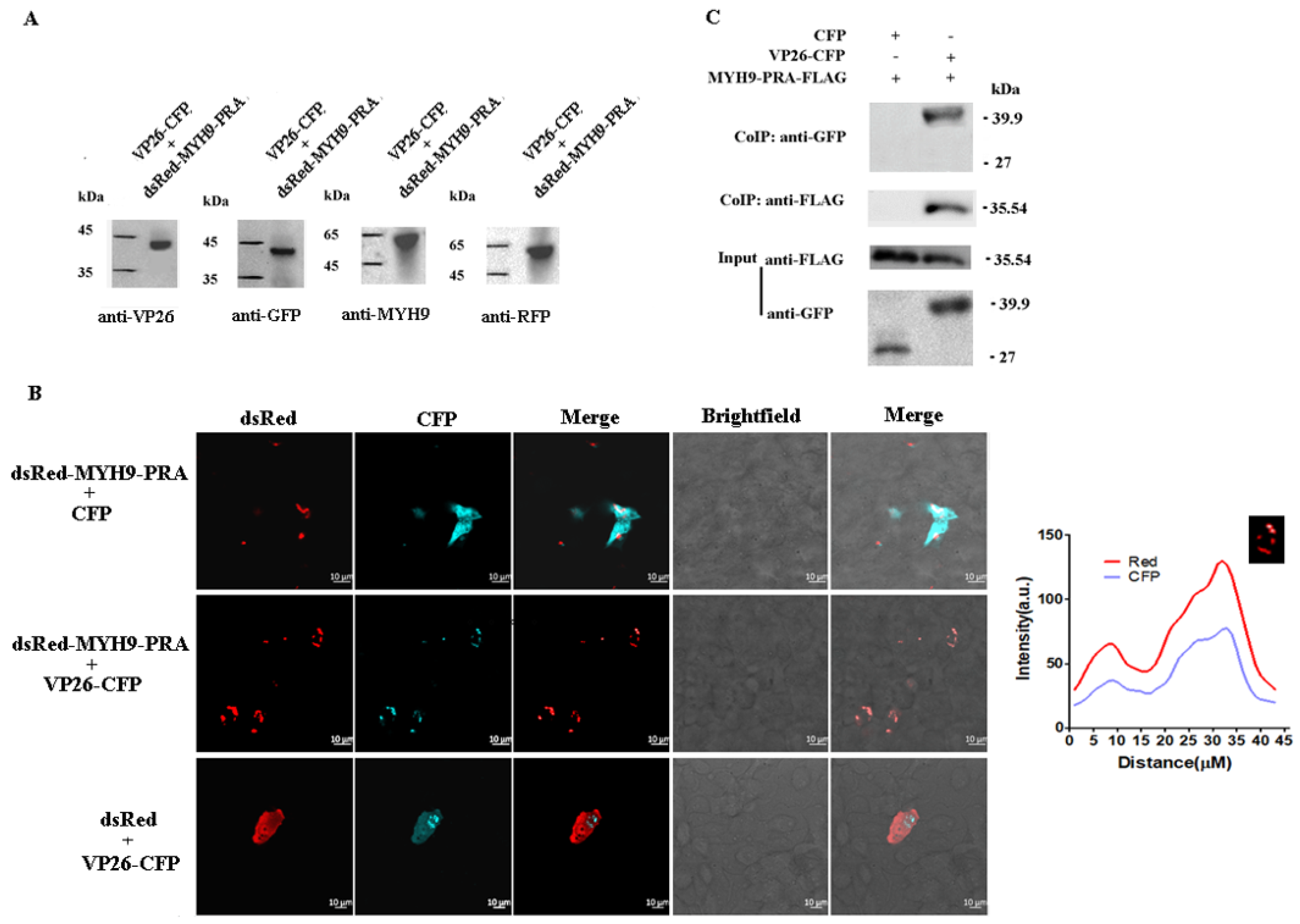
2.4. Verification of the Interaction Between DEV VP26 and MYH9 in CEF Cells
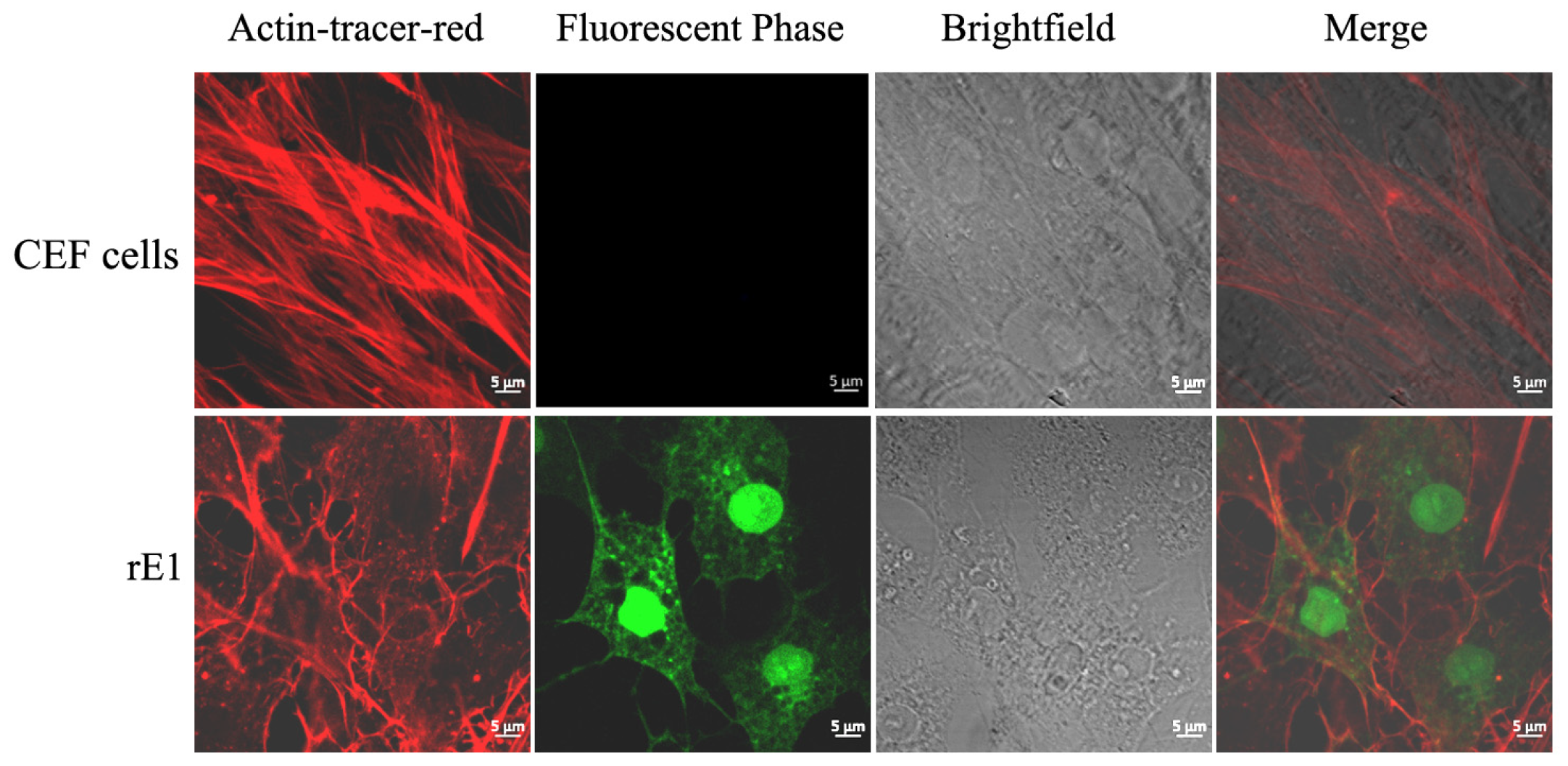
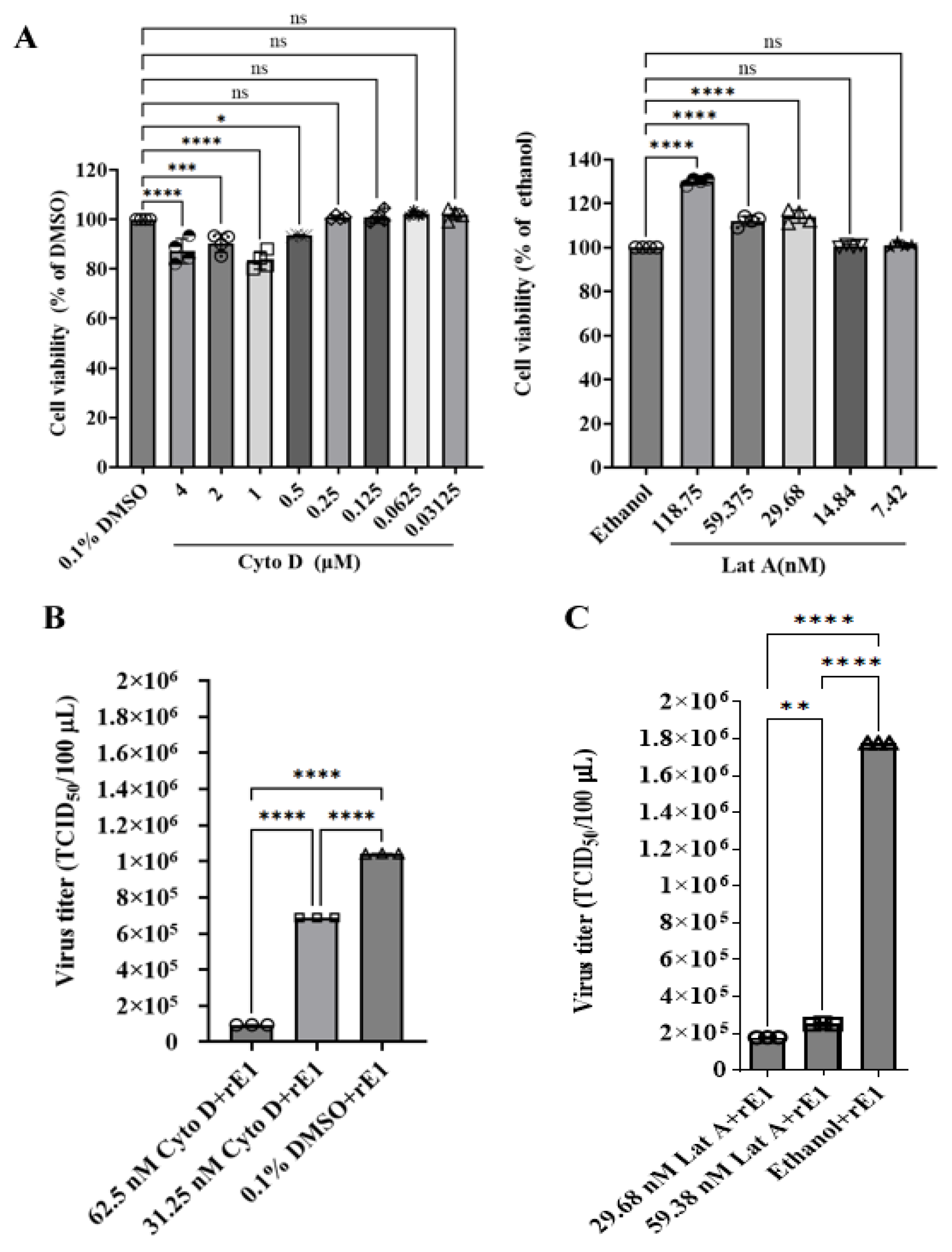
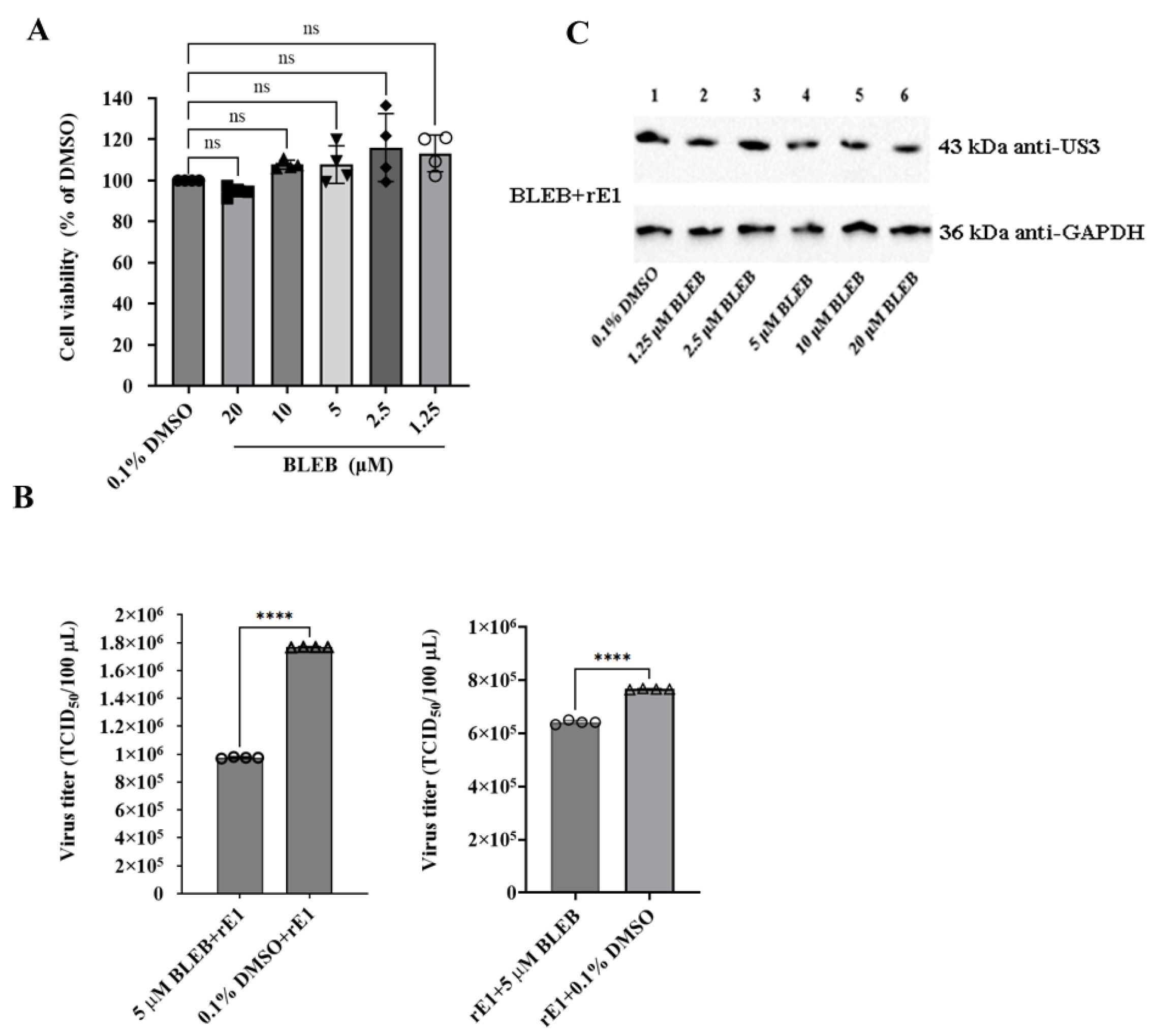
2.5. Effects of Drugs on Microfilament Cytoskeleton
2.6. Effect of Drugs on Viral Infection
2.7. The Deficiency of MYH9 Impairs Virus Replication in LMH Cells
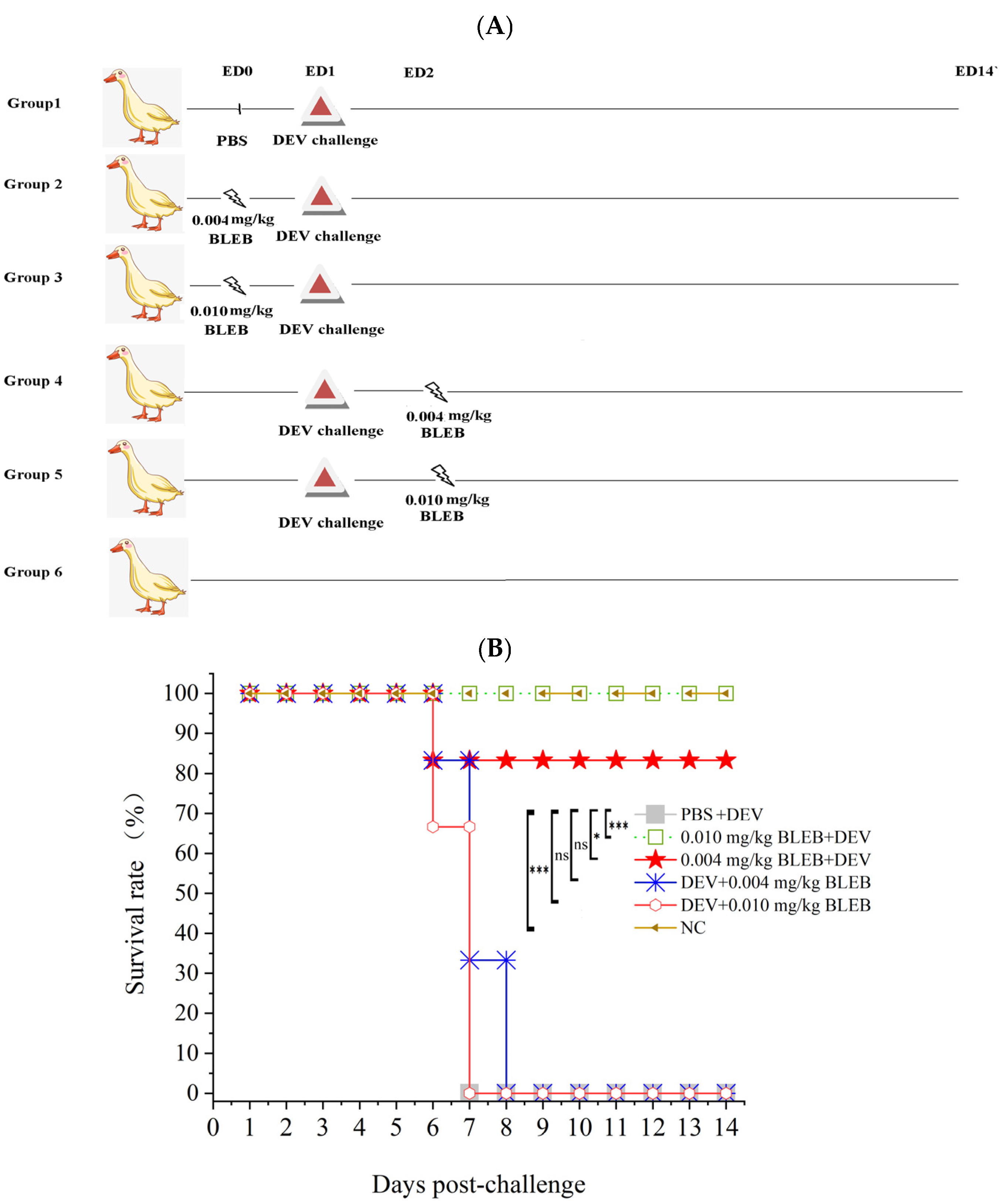
2.8. (-)-Blebbistatin Inhibited DEV Replication In Vitro and In Vivo
3. Discussion
4. Materials and Methods
4.1. Virus Strain, Cells, and Plasmids
4.2. Inhibitors
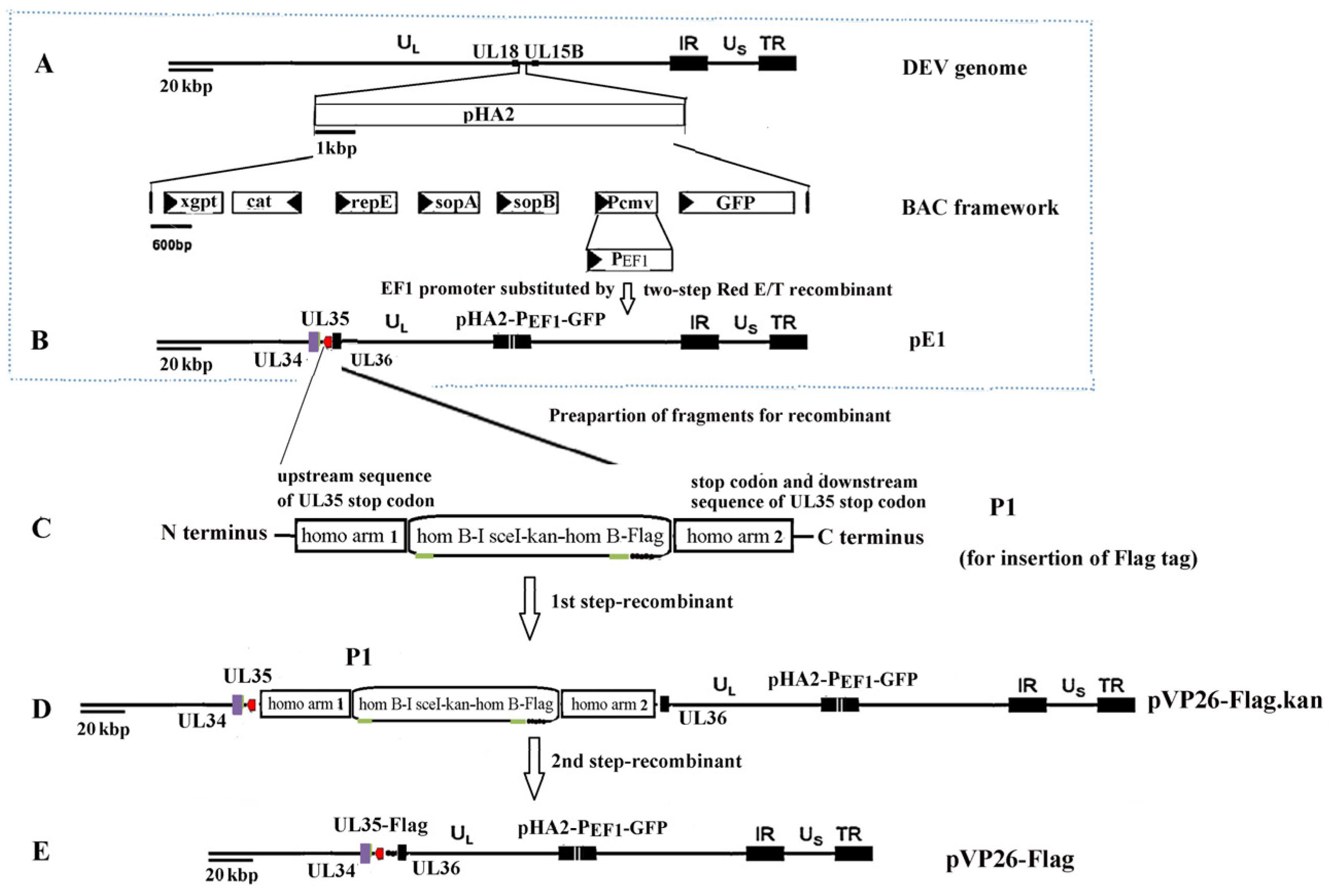
4.3. Reverse Genetics System
4.4. Virion Purification
4.5. Western Blot Analysis
4.6. Indirect Immunofluorescence (IF)
4.7. Co-Immunoprecipitation (Co-IP)
4.8. LC-MS/MS Analysis
4.9. Quantification and Bioinformatics Analysis
4.10. Immunofluorescence and Confocal Microscopy
4.11. Cell Viability Assay
4.12. Effect of Drugs on Microfilament Cytoskeleton
4.13. Inhibition Assay
4.14. siRNA-Mediated Gene Silencing
4.15. Overexpression Analysis
4.16. Animal Experiments
4.17. The Control Material and the Control Procedures
4.18. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| DEV | duck enteritis virus |
| HRP | horseradish peroxidase |
| MOI | multiplicity of infection |
| STRING | Search Tool for the Retrieval of Interacting Genes |
| GO | Gene Ontology |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| PPI | protein–protein interaction |
| Cyto D | cytochalasin D |
| Lat A | latrunculin A |
| BLEB | (-)-Blebbistatin |
| LMH | chicken liver hepatocellular carcinoma cell line |
| CEF | chicken embryo fibroblast |
References
- Dhama, K.; Kumar, N.; Saminathan, M.; Tiwari, R.; Karthik, K.; Kumar, M.A.; Palanivelu, M.; Shabbir, M.Z.; Malik, Y.S.; Singh, R.K. Duck virus enteritis (duck plague)—A comprehensive update. Vet. Q. 2017, 37, 57–80. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.F.; Huang, B.; Ma, X.L.; Wu, J.; Li, F.; Ai, W.; Song, M.X.; Yang, H.C. Molecular characterization of the genome of duck enteritis virus. Virology 2009, 391, 151–161. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Cheng, A.; Wang, M.; Yang, Q.; Zhu, D.; Jia, R.; Chen, S.; Zhou, Y.; Wang, X.; Chen, X. Complete genomic sequence of Chinese virulent duck enteritis virus. J. Virol. 2012, 86, 5965. [Google Scholar] [CrossRef] [PubMed]
- Cardone, G.; Heymann, J.B.; Cheng, N.; Trus, B.L.; Steven, A.C. Procapsid assembly, maturation, nuclear exit: Dynamic steps in the production of infectious herpesvirions. Adv. Exp. Med. Biol. 2012, 726, 423–439. [Google Scholar] [CrossRef]
- Desai, P.; DeLuca, N.A.; Person, S. Herpes simplex virus type 1 VP26 is not essential for replication in cell culture but influences production of infectious virus in the nervous system of infected mice. Virology 1998, 247, 115–124. [Google Scholar] [CrossRef]
- Krautwald, M.; Maresch, C.; Klupp, B.G.; Fuchs, W.; Mettenleiter, T.C. Deletion or green fluorescent protein tagging of the pUL35 capsid component of pseudorabies virus impairs virus replication in cell culture and neuroinvasion in mice. J. Gen. Virol. 2008, 89 Pt 6, 1346–1351. [Google Scholar] [CrossRef]
- Douglas, M.W.; Diefenbach, R.J.; Homa, F.L.; Miranda-Saksena, M.; Rixon, F.J.; Vittone, V.; Byth, K.; Cunningham, A.L. Herpes simplex virus type 1 capsid protein VP26 interacts with dynein light chains RP3 and Tctex1 and plays a role in retrograde cellular transport. J. Biol. Chem. 2004, 279, 28522–28530. [Google Scholar] [CrossRef]
- Kobayashi, R.; Kato, A.; Oda, S.; Koyanagi, N.; Oyama, M.; Kozuka-Hata, H.; Arii, J.; Kawaguchi, Y.Y. Function of the herpes simplex virus 1 small capsid protein VP26 is regulated by phosphorylation at a specific site. J. Virol. 2015, 89, 6141–6147. [Google Scholar] [CrossRef]
- Kobayashi, R.; Kato, A.; Sagara, H.; Watanabe, M.; Maruzuru, Y.; Koyanagi, N.; Arii, J.; Kawaguchi, Y. Herpes simplex virus 1 small capsomere-interacting protein VP26 regulates nucleocapsid maturation. J. Virol. 2017, 91, e01068-17. [Google Scholar] [CrossRef]
- Antinone, S.E.; Shubeita, G.T.; Coller, K.E.; Lee, J.I.; Haverlock-Moyns, S.; Gross, S.P.; Smith, G.A. The Herpesvirus capsid surface protein, VP26, and the majority of the tegument proteins are dispensable for capsid transport toward the nucleus. J. Virol. 2006, 80, 5494–5498. [Google Scholar] [CrossRef]
- Feierbach, B.; Piccinotti, S.; Bisher, M.; Denk, W.; Enquist, L.W. Alpha-herpesvirus infection induces the formation of nuclear actin filaments. PLoS Pathog. 2006, 2, e85, Erratum in PLoS Pathog. 2006, 2, e103. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Gao, J.M.; Xiao, S.Q.; Xiao, Y.H.; Wang, X.P.; Zhang, C.; Zhao, Q.; Nan, Y.C.; Huang, B.C.; Liu, H.L.; Liu, N.N.; et al. MYH9 is an essential factor for porcine reproductive and respiratory syndrome virus infection. Sci. Rep. 2016, 6, 25120. [Google Scholar] [CrossRef] [PubMed]
- Li, L.L.; Xue, B.Y.; Sun, W.Y.; Gu, G.Q.; Hou, G.P.; Zhang, L.; Wu, C.Y.; Zhao, Q.; Zhang, Y.J.; Zhang, G.P.; et al. Recombinant MYH9 protein C-terminal domain blocks porcine reproductive and respiratory syndrome virus internalization by direct interaction with viral glycoprotein 5. Antivir. Res. 2018, 156, 10–20. [Google Scholar] [CrossRef] [PubMed]
- Han, X.M.; Clark, J.J.; Sharma, P.; Bentley, E.G.; Kipar, A.; Alsayer, M.; Ren, X.L.; Robinson, A.; Alaidarous, S.; Mu, Y.; et al. Amino acids 1811-1960 of myosin heavy chain 9 is involved in murine gammaherpesvirus 68 infection. Virology 2023, 587, 109849. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Fan, J.; Chen, Z.L.; Zhang, M.M.; Peng, H.R.; Liu, J.; Ding, L.F.; Liu, M.B.; Zhao, C.; Zhao, P.; et al. Nonmuscle myosin heavy chain IIA facilitates SARS-CoV-2 infection in human pulmonary cells. Proc. Natl. Acad. Sci. USA 2021, 118, e2111011118. [Google Scholar] [CrossRef]
- Roman, B.I.; Verhasselt, S.; Stevens, C.V. Medicinal Chemistry and Use of Myosin II Inhibitor (S)-Blebbistatin and Its Derivatives. J. Med. Chem. 2018, 61, 9410–9428. [Google Scholar] [CrossRef]
- Zhang, J.; Pan, Z.; Zhou, J.; Zhang, L.; Tang, J.; Gong, S.; Li, F.; Yu, B.; Zhang, Y.; Kou, J. The myosin II inhibitor, blebbistatin, ameliorates pulmonary endothelial barrier dysfunction in acute lung injury induced by LPS via NMMHC IIA/Wnt5a/β-catenin pathway. Toxicol. Appl. Pharmacol. 2022, 450, 116132. [Google Scholar] [CrossRef]
- Huang, Y.X.; Ou, S.S.; Kuang, R.L.; Lin, W.Q. Study on duck plague virus. J. South China Agric. College 1980, 1, 1–16. (In Chinese) [Google Scholar]
- Huang, R.Y.; Xiao, J.F.; Chen, H.F.; Zhou, Q.Q.; Chen, Y. Experimental study on lethal effect of duck plague attenuated vaccine on chickens. Animal Husb. Vet. Med. 1981, 5, 14–16. (In Chinese) [Google Scholar]
- Chen, Y.; Huang, R.Y.; Xiao, J.F.; Chen, H.F. Lethal test of duck plague attenuated vaccine on chickens. Two, Cohabitation infection test (abstract). China Poul. 1982, 3, 45. (In Chinese) [Google Scholar]
- Li, L.G.; Li, B.D.; Guo, C.Y. Pathogenic test of duck plague virus on chicks. Animal Husb. Vet. Sci. Technol. 1981, 3, 18–24. (In Chinese) [Google Scholar]
- Kong, J.; Feng, K.; Zhao, Q.; Chen, Y.; Wang, J.; Chen, S.; Shao, G.; Liao, L.; Li, Y.; Xie, Z.; et al. Pathogenicity and transmissibility studies on live attenuated duck enteritis virus vaccine in non-target species. Front. Microbiol. 2022, 13, 979368. [Google Scholar] [CrossRef]
- Chen, L.; Yu, B.; Ni, Z.; Hua, J.G.; Ye, W.C.; Yun, T.; Zhang, C. Biological characteristics of DEV envelope glycoprotein gI gene deletion virus. Zhejiang J. Animal Sci. Vet. Med. 2014, 39, 9–13. (In Chinese) [Google Scholar]
- Chen, P.; Liu, J.; Jiang, Y.; Zhao, Y.; Li, Q.; Wu, L.; He, X.; Chen, H. The vaccine efficacy of recombinant duck enteritis virus expressing secreted E with or without PrM proteins of duck tembusu virus. Vaccine 2014, 32, 5271–5277. [Google Scholar] [CrossRef]
- Zou, Z.; Liu, Z.G.; Jin, M.L. Efficient strategy to generate a vectored Duck enteritis virus delivering envelope of Duck Tembusu virus. Viruses 2014, 6, 2428–2443. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.X.; Chen, P.C.; Jiang, Y.P.; Wu, L.; Zeng, X.Y.; Tian, G.B.; Ge, J.Y.; Kawaoka, Y.; Bu, Z.G.; Chen, H.L. A Duck enteritis virus-vectored bivalent live vaccine provides fast and complete protection against H5N1 avian influenza virus infection in Ducks. J. Virol. 2011, 85, 10989–10998. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.X.; Chen, P.C.; Jiang, Y.P.; Deng, G.H.; Shi, J.Z.; Wu, L.; Yuan, L.; Bu, Z.G.; Chen, H.L. Recombinant duck enteritis virus works as a single-dose vaccine in broilers providing rapid protection against H5N1 influenza infection. Antivir. Res. 2013, 97, 329–333. [Google Scholar] [CrossRef] [PubMed]
- Li, K.; Wang, C.; Yang, F.; Cao, W.; Zhu, Z.; Zheng, H. Virus-Host Interactions in Foot-and-Mouth Disease Virus Infection. Front. Immunol. 2021, 12, 571509. [Google Scholar] [CrossRef]
- Zhang, Y.; Xu, Z.; Cao, Y. Host-Virus Interaction: How Host Cells Defend against Influenza A Virus Infection. Viruses 2020, 12, 376. [Google Scholar] [CrossRef]
- Ma-Lauer, Y.; Lei, J.; Hilgenfeld, R.; von Brunn, A. Virus-host interactomes—Antiviral drug discovery. Curr. Opin. Virol. 2012, 2, 614–621. [Google Scholar] [CrossRef]
- Wong, K.Z.; Chu, J.J.H. The Interplay of Viral and Host Factors in Chikungunya Virus Infection: Targets for Antiviral Strategies. Viruses 2018, 10, 294. [Google Scholar] [CrossRef] [PubMed]
- Pollard, T.D.; Cooper, J.A. Actin, a central player in cell shape and movement. Science 2009, 326, 1208–1212. [Google Scholar] [CrossRef] [PubMed]
- Olson, M.F. Actin-Myosin Cytoskeleton Regulation and Function. Cells 2022, 12, 9. [Google Scholar] [CrossRef] [PubMed]
- Kumakura, M.; Kawaguchi, A.; Nagata, K. Actin-myosin network is required for proper assembly of influenza virus particles. Virology 2015, 476, 141–150. [Google Scholar] [CrossRef]
- Li, X.M.; Xu, K.; Wang, J.Y.; Guo, J.Y.; Wang, X.H.; Zeng, L.; Wan, B.; Wang, J.; Chu, B.B.; Yang, G.Y.; et al. The actin cytoskeleton is important for pseudorabies virus infection. Virology 2024, 600, 110233. [Google Scholar] [CrossRef]
- Shahriari, S.; Ghildyal, R. The actin-binding protein palladin associates with the respiratory syncytial virus matrix protein. J. Virol. 2024, 98, e0143524. [Google Scholar] [CrossRef]
- Kieser, Q.J.; Granoski, M.J.; McClelland, R.D.; Griffiths, C.; Bilawchuk, L.M.; Stojic, A.; Elawar, F.; Jamieson, K.; Proud, D.; Marchant, D.J. Actin cytoskeleton remodeling disrupts physical barriers to infection and presents entry receptors to respiratory syncytial virus. J. Gen. Virol. 2023, 104, 001923. [Google Scholar] [CrossRef]
- Trejo-Cerro, Ó.; Eichwald, C.; Schraner, E.M.; Silva-Ayala, D.; López, S.; Arias, C.F. Actin-Dependent Nonlytic Rotavirus Exit and Infectious Virus Morphogenetic Pathway in Nonpolarized Cells. J. Virol. 2018, 92, e02076-17. [Google Scholar] [CrossRef]
- Dietzel, E.; Kolesnikova, L.; Maisner, A. Actin filaments disruption and stabilization affect measles virus maturation by different mechanisms. Virol. J. 2013, 10, 249. [Google Scholar] [CrossRef]
- Horsington, J.; Lynn, H.; Turnbull, L.; Cheng, D.; Braet, F.; Diefenbach, R.J.; Whitchurch, C.B.; Karupiah, G.; Newsome, T.P. A36-dependent actin filament nucleation promotes release of vaccinia virus. PLoS Pathog. 2013, 9, e1003239. [Google Scholar] [CrossRef]
- Kim, M.J.; Kim, J.; Im, J.S.; Kang, I.; Ahn, J.K. Hepatitis B virus X protein enhances liver cancer cell migration by regulating calmodulin-associated actin polymerization. BMB Rep. 2021, 54, 614–619. [Google Scholar] [CrossRef]
- Wakimoto, H.; Shimodo, M.; Satoh, Y.; Kitagawa, Y.; Takeuchi, K.; Gotoh, B.; Itoh, M. F-actin modulates measles virus cell-cell fusion and assembly by altering the interaction between the matrix protein and the cytoplasmic tail of hemagglutinin. J. Virol. 2013, 87, 1974–1984. [Google Scholar] [CrossRef]
- Vetter, J.; Lee, M.; Eichwald, C. The Role of the Host Cytoskeleton in the Formation and Dynamics of Rotavirus Viroplasms. Viruses 2024, 16, 668. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Wang, L.; Zhang, X.; Wang, S.; Qin, Q. Nervous necrosis virus induced vacuolization is a Rab5- and actin-dependent process. Virulence 2024, 15, 2301244. [Google Scholar] [CrossRef] [PubMed]
- Stradal, T.E.B.; Schelhaas, M. Actin dynamics in host-pathogen interaction. FEBS Lett. 2018, 592, 3658–3669. [Google Scholar] [CrossRef] [PubMed]
- Denes, C.E.; Miranda-Saksena, M.; Cunningham, A.L.; Diefenbach, R.J. Cytoskeletons in the closet subversion in alphaherpes virus infections. Viruses 2018, 10, 79. [Google Scholar] [CrossRef]
- Roberts, K.L.; Baines, J.D. Actin in herpesvirus infection. Viruses 2011, 3, 336–346. [Google Scholar] [CrossRef]
- Arii, J.; Hirohata, Y.; Kato, A.; Kawaguchi, Y. Nonmuscle myosin heavy chain IIb mediates herpes simplex virus 1 entry. J. Virol. 2015, 89, 1879–1888. [Google Scholar] [CrossRef]
- van Leeuwen, H.; Elliott, G.; O’Hare, P. Evidence of a role for nonmuscle myosin II in herpes simplex virus type 1 egress. J. Virol. 2002, 76, 3471–3481. [Google Scholar] [CrossRef]
- Wang, C.; Liang, Q.; Sun, D.; He, Y.; Jiang, J.; Guo, R.; Malla, T.; Hamrah, P.; Liu, X.; Huang, Z.; et al. Nectin-1 and Non-muscle Myosin Heavy Chain-IIB: Major Mediators of Herpes Simplex Virus-1 Entry Into Corneal Nerves. Front. Microbiol. 2022, 13, 830699. [Google Scholar] [CrossRef]
- Concha, J.O.; Gutierrez, K.; Barbosa, N.; Rodrigues, R.L.; de Carvalho, A.N.; Tavares, L.A.; Rudd, J.S.; Costa, C.S.; Andrade, B.Y.G.; Espreafico, E.M.; et al. Rab27a GTPase and its effector Myosin Va are host factors required for efficient Oropouche virus cell egress. PLoS Pathog. 2024, 20, e1012504. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Liu, J.; Chen, Z.; Feng, D.; Zhu, C.; Fan, J.; Zhang, S.; Zhang, X.; Xu, J. Nonmuscle myosin IIA promotes the internalization of influenza A virus and regulates viral polymerase activity through interacting with nucleoprotein in human pulmonary cells. Virol. Sin. 2023, 38, 128–141. [Google Scholar] [CrossRef] [PubMed]
- Goddette, D.W.; Frieden, C. Actin polymerization. The mechanism of action of cytochalasin D. J. Biol. Chem. 1986, 261, 15974–15980. [Google Scholar] [CrossRef] [PubMed]
- Casella, J.F.; Flanagan, M.D.; Lin, S. Cytochalasin D inhibits actin polymerization and induces depolymerization of actin filaments formed during platelet shape change. Nature 1981, 293, 302–305. [Google Scholar] [CrossRef]
- Coué, M.; Brenner, S.L.; Spector, I.; Korn, E.D. Inhibition of actin polymerization by latrunculin A. FEBS Lett. 1987, 213, 316–318. [Google Scholar] [CrossRef]
- Fujiwara, I.; Zweifel, M.E.; Courtemanche, N.; Pollard, T.D. Latrunculin A Accelerates Actin Filament Depolymerization in Addition to Sequestering Actin Monomers. Curr. Biol. 2018, 28, 3183–3192.e2. [Google Scholar] [CrossRef]
- Sarmah, H.; Shah, M.; Pathak, M.; Barman, N.N.; Koul, M.; Gupta, A.; Sahariah, P.J.; Neher, S.; Das, S.K.; Gogoi, S.M.; et al. Pathodynamics of Circulating Strains of Duck Enteritis Virus: A Step Forward to Understand Its Pathogenesis. Avian Dis. 2020, 64, 166–173. [Google Scholar] [CrossRef]
- Qi, X.F.; Yang, X.Y.; Cheng, A.C.; Wang, M.S.; Zhu, D.K.; Jia, R.Y. The pathogenesis of duck virus enteritis in experimentally infected ducks: A quantitative time-course study using TaqMan polymerase chain reaction. Avian Pathol. 2008, 37, 307–310. [Google Scholar] [CrossRef]
- Qi, X.; Yang, X.; Cheng, A.; Wang, M.; Zhu, D.; Jia, R. Quantitative analysis of virulent duck enteritis virus loads in experimentally infected ducklings. Avian Dis. 2008, 52, 338–344. [Google Scholar] [CrossRef]
- Wen, J.l. Dynamic Analysis of Strong and Weak Strains of Duck Viral Fever in Duck Bodies. Master’s Thesis, China Veterinary Drug Administration, Beijing, China, 2014. (In Chinese). [Google Scholar]
- Chen, L.; Yu, B.; Hua, J.G.; Ye, W.C.; Ni, Z.; Yun, T.; Deng, X.H.; Zhang, C. Construction of a full-length infectious bacterial artificial chromosome clone of duck enteritis virus vaccine strain. Virol. J. 2013, 10, 328. [Google Scholar] [CrossRef]
- Chen, L.; Yu, B.; Ni, Z.; Hua, J.G.; Ye, W.C.; Yun, T.; Zhang, C. Construction and characterization of a recombinant duck enteritis virus expressing E protein of duck Tembusu virus. Acta Agric. Zhejiangensis 2015, 27, 1889–1895. (In Chinese) [Google Scholar]
- Karstischer, B.; von Einem, J.; Kaufer, B.; Osterrieder, N. Two-step Red-mediated recombination for versatile high-efficiency markerless DNA manipulation in Escherichia coli. BioTechniques 2006, 40, 191–197. [Google Scholar] [CrossRef]
- Tischer, B.K.; Smith, G.A.; Osterrieder, N. En Passant mutagenesis: A two step markerless red recombination system. In Vitro Mutagenesis Protocols; Braman, J., Ed.; Humana Press: Totowa, NJ, USA, 2010; pp. 421–430. [Google Scholar]
- Zhao, C.; He, T.; Xu, Y.; Wang, M.; Cheng, A.; Zhao, X.; Zhu, D.; Chen, S.; Liu, M.; Yang, Q.; et al. Molecular characterization and antiapoptotic function analysis of the duck plague virus Us5 gene. Sci. Rep. 2019, 9, 4851. [Google Scholar] [CrossRef]

| Protein ID | Name | Peptides | Sequence Coverage [%] | Average Intensity of Test | Average Intensity of Ctrl | FC (Test vs. Ctrl) | Amino Acid Sequence Homology Between Duck and Chicken |
|---|---|---|---|---|---|---|---|
| F1NA76 | TMED10 | 7 | 40.6 | 189,220,000 | 0 | Inf | 94.06% |
| F1P4I3 | VAMP3 | 4 | 53.8 | 47,205,000 | 0 | Inf | 89.42% |
| Q91957 | Xirp1 | 9 | 6.3 | 53,513,333.33 | 0 | Inf | 89.00% |
| Q5ZLY3 | TMOD3 | 7 | 31.8 | 141,886,666.7 | 0 | Inf | 97.72% |
| P28675 | DCN | 5 | 19.9 | 48,087,000 | 0 | Inf | 97.76% |
| E1C658 | ATP5PD | 6 | 37.3 | 81,129,333.33 | 0 | Inf | 74.19% |
| Q5ZMP7 | AP3M1 | 5 | 17 | 15,092,000 | 0 | Inf | 98.80% |
| P02604 | - | 8 | 56.2 | 407,123,333.3 | 0 | Inf | / |
| P02612 | - | 8 | 62.2 | 373,010,000 | 0 | Inf | / |
| Q02440 | MYO5A | 7 | 5.3 | 54,430,333.33 | 0 | Inf | / |
| Q789A6 | MYH10 | 147 | 66.8 | 19,682,333,333 | 197,000,000 | 99.91032149 | 99.00% |
| A0A1D5PM19 | MYH9 | 173 | 70 | 55,490,000,000 | 1,570,066,667 | 35.3424483 | 98.00% |
| A0A1L1RLN6 | - | 9 | 74 | 2,179,333,333 | 158,516,666.7 | 13.74829145 | / |
| P24032 | - | 10 | 62.8 | 2,327,600,000 | 174,780,000 | 13.31731319 | / |
| O93510 | GSN | 24 | 43.6 | 1,491,766,667 | 213,916,666.7 | 6.973587846 | 94.68% |
| Q5ZM66 | RPS26 | 3 | 29.6 | 274,823,333.3 | 41,238,333.33 | 6.664268682 | 100% |
| A0A1D5PNG3 | EIF6 | 2 | 13.1 | 35,480,333.33 | 5,989,000 | 5.924250014 | 98.37% |
| d.p.c | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | Survival Rate | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Group | ||||||||||||||||
| PBS+DEV | 6 | 0/6 | ||||||||||||||
| 0.010 mg/kg BLEB+DEV | 6/6 | |||||||||||||||
| 0.004 mg/kg BLEB+DEV | 1 | 5/6 | ||||||||||||||
| DEV+0.004 mg/kg BLEB | 1 | 3 | 2 | 0/6 | ||||||||||||
| DEV+0.010 mg/kg BLEB | 2 | 4 | 0/6 | |||||||||||||
| Name | Sequence (5′ to 3′) | Restriction Site Introduced |
|---|---|---|
| dsRed-myh9-PRA(SalI+) | gGTCGACatgcgggaactggaggacac | Sal I |
| dsRed-myh9-PRA(BamHI-) | cgGGATCCttattcagtagctttggcatcacc | Bam HI |
| UL35-CFP(BamHI+) | cgGGATCCatgtctaattctggaggttc | Bam HI |
| UL35-CFP(EcoRI-) | cgGAATTCtcgctgatcgtctggcgcg | Eco RI |
| siRNA-MYH9-1870-s | CUGGCAAGGUGGAUUAUAATT | |
| siRNA-MYH9-1870-as | UUAUAAUCCACCUUGCCAGTT | |
| siRNA-MYH9-5503-s | GCCAGAACAAGGAGCUUAATT | |
| siRNA-MYH9-5503-as | UUAAGCUCCUUGUUCUGGCTT | |
| siRNA-MYH9-2693-s | GGCCAAGGAAGAAGAACUATT | |
| siRNA-MYH9-2693-as | UAGUUCUUCUUCCUUGGCCTT | |
| siRNA-negative control (S) | UUCUCCGAACGUGUCACGUTT | |
| siRNA-negative control (AS) | ACGUGACACGUUCGGAGAATT |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, L.; Zhu, Y.-C.; Yun, T.; Ye, W.-C.; Ni, Z.; Hua, J.-G.; Zhang, C. Proteomic Screening for Cellular Targets of the Duck Enteritis Virus Protein VP26 Reveals That the Host Actin–Myosin II Network Regulates the Proliferation of the Virus. Int. J. Mol. Sci. 2025, 26, 9108. https://doi.org/10.3390/ijms26189108
Chen L, Zhu Y-C, Yun T, Ye W-C, Ni Z, Hua J-G, Zhang C. Proteomic Screening for Cellular Targets of the Duck Enteritis Virus Protein VP26 Reveals That the Host Actin–Myosin II Network Regulates the Proliferation of the Virus. International Journal of Molecular Sciences. 2025; 26(18):9108. https://doi.org/10.3390/ijms26189108
Chicago/Turabian StyleChen, Liu, Yin-Chu Zhu, Tao Yun, Wei-Cheng Ye, Zheng Ni, Jiong-Gang Hua, and Cun Zhang. 2025. "Proteomic Screening for Cellular Targets of the Duck Enteritis Virus Protein VP26 Reveals That the Host Actin–Myosin II Network Regulates the Proliferation of the Virus" International Journal of Molecular Sciences 26, no. 18: 9108. https://doi.org/10.3390/ijms26189108
APA StyleChen, L., Zhu, Y.-C., Yun, T., Ye, W.-C., Ni, Z., Hua, J.-G., & Zhang, C. (2025). Proteomic Screening for Cellular Targets of the Duck Enteritis Virus Protein VP26 Reveals That the Host Actin–Myosin II Network Regulates the Proliferation of the Virus. International Journal of Molecular Sciences, 26(18), 9108. https://doi.org/10.3390/ijms26189108





