Integrating MALDI-MSI-Based Spatial Proteomics and Machine Learning to Predict Chemoradiotherapy Outcomes in Head and Neck Cancer
Abstract
1. Introduction
2. Results
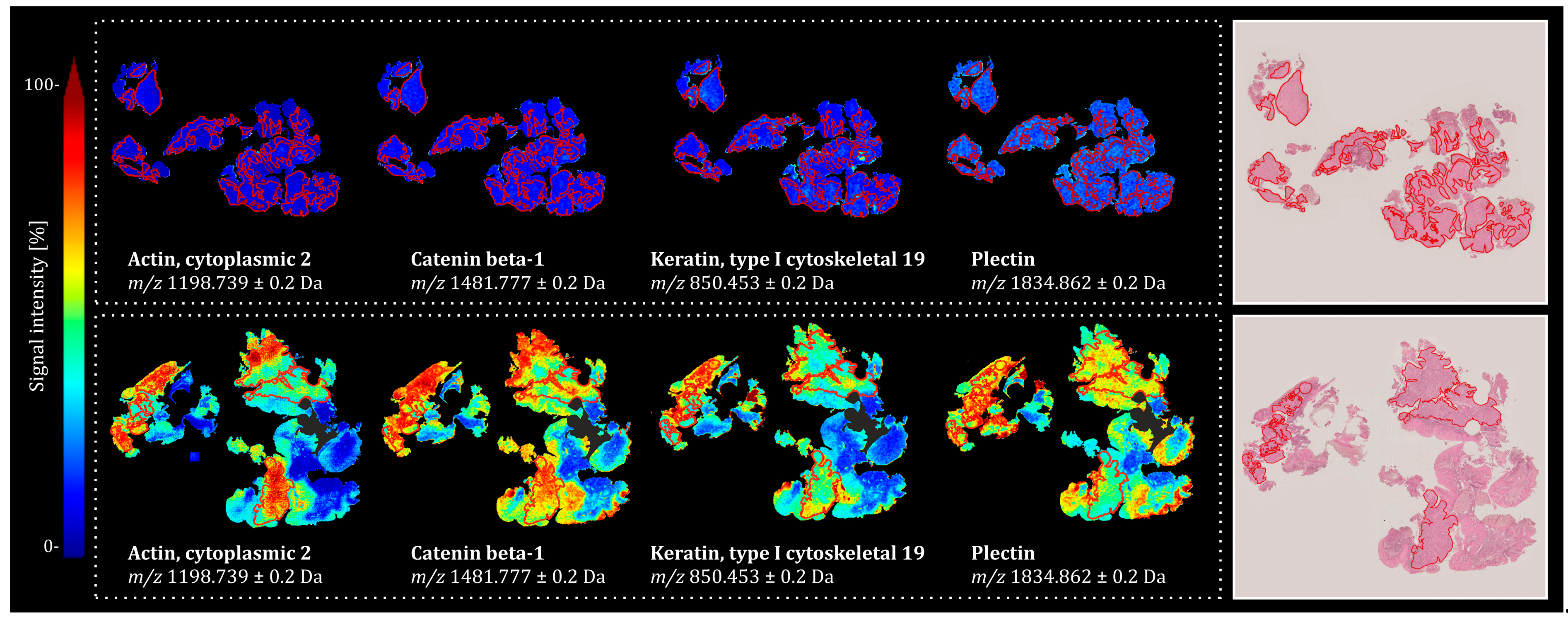
2.1. Univariate Statistics in Combination with Bottom-Up Proteomics Reveals Discriminative m/z Features Associated with Treatment Outcome in HNSCC
2.2. Predictive Value of Multivariate MALDI-MSI–Derived Proteomic Signatures for Two-Year Outcomes in CDDP-CRT-Treated HNSCC
2.2.1. Classification Performance Based on All MALDI-MSI m/z Features
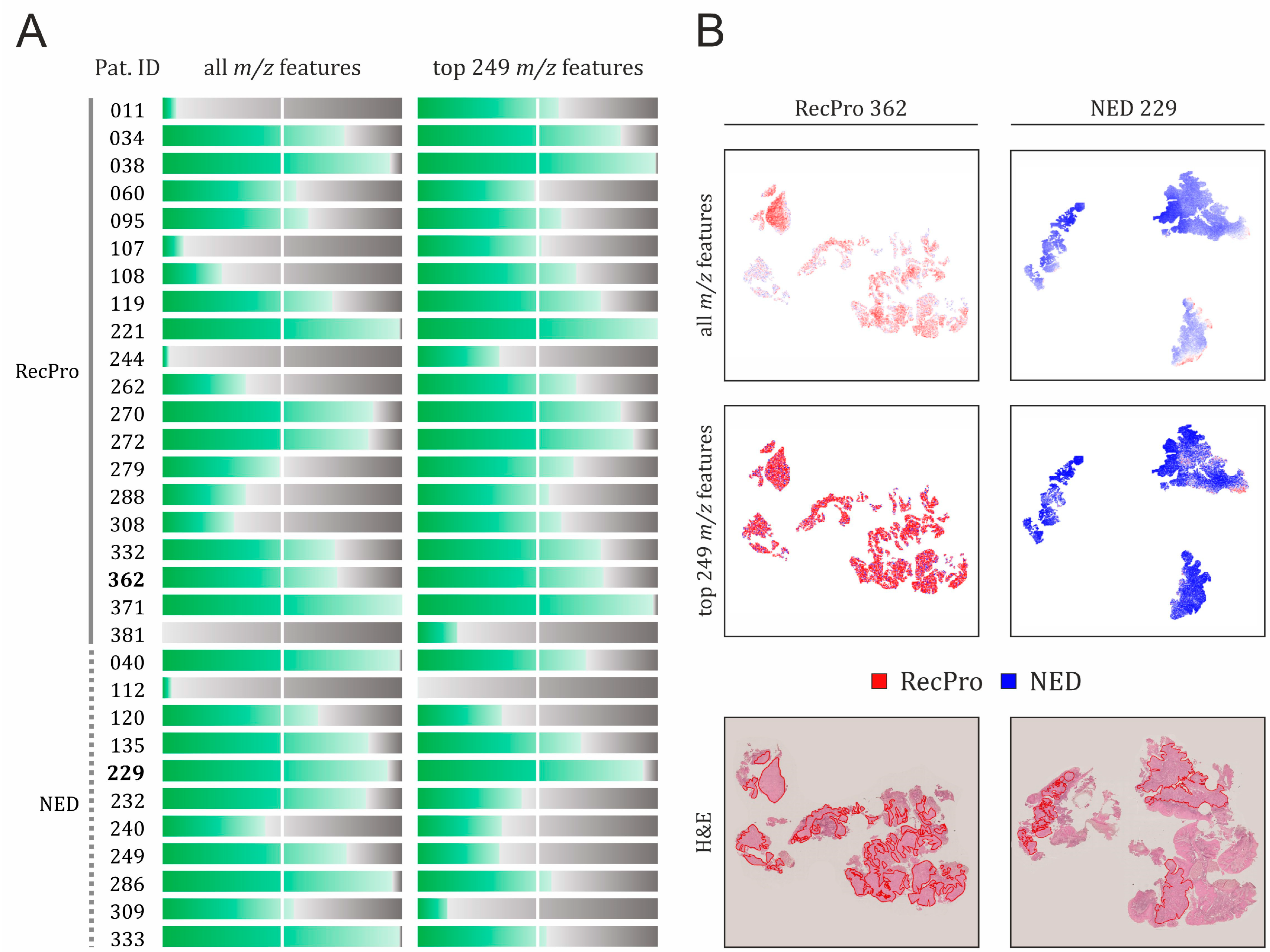
2.2.2. Feature Selection as a Means to Improve Specificity of the Classification Model
2.3. Performance of Classification Models in HNSCC Patient Cohort Subjected to Other Treatment Regimens
3. Discussion
4. Materials and Methods
4.1. Patient Material
4.2. Tissue Sample Preparation and MALDI-MSI Measurement
4.3. MALDI-MSI Data Processing
4.4. Univariate Statistical Testing
4.5. Machine Learning-Based Data Analysis
4.6. Protein Identification by Nano-LC-ESI-MS/MS
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| 5-FU | 5-fluorouracil |
| AUC | Area under the curve |
| CDDP | cis-Diammindichloridoplatin (cisplatin) |
| CHCA | α-Cyano-4-hydroxycinnamic acid |
| CRT | Chemoradiotherapy |
| FFPE | Formalin-fixed paraffin-embedded |
| H&E | Hematoxylin and eosin |
| HNSCC | Head and neck squamous cell carcinoma |
| HPV | Human papillomavirus |
| ITH | Intratumoral heterogeneity |
| ITO | Indium tin oxide |
| LC | Liquid chromatography |
| LCM | Laser capture microdissection |
| MALDI | Matrix-assisted laser desorption/ionization |
| ML | Machine learning |
| MMC | Mitomycin C |
| MSI | Mass spectrometry imaging |
| MS/MS | Tandem mass spectrometry |
| m/z | Mass-to-charge ratio |
| NED | No evidence of disease |
| RecPro | Recurrence/progression |
| ROC | Receiver operating characteristic |
| ROI | Region of interest |
| TNM | Tumor, node, metastasis |
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Ferlay, J.; Colombet, M.; Soerjomataram, I.; Mathers, C.; Parkin, D.M.; Pineros, M.; Znaor, A.; Bray, F. Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int. J. Cancer 2019, 144, 1941–1953. [Google Scholar] [CrossRef]
- Gormley, M.; Creaney, G.; Schache, A.; Ingarfield, K.; Conway, D.I. Reviewing the epidemiology of head and neck cancer: Definitions, trends and risk factors. Br. Dent. J. 2022, 233, 780–786. [Google Scholar] [CrossRef]
- Burri, R.J.; Lee, N.Y. Concurrent chemotherapy and radiotherapy for head and neck cancer. Expert. Rev. Anticancer Ther. 2009, 9, 293–302. [Google Scholar] [CrossRef]
- Pignon, J.P.; le Maitre, A.; Maillard, E.; Bourhis, J.; Group, M.-N.C. Meta-analysis of chemotherapy in head and neck cancer (MACH-NC): An update on 93 randomised trials and 17,346 patients. Radiother. Oncol. 2009, 92, 4–14. [Google Scholar] [CrossRef]
- Rettig, E.M.; D’Souza, G. Epidemiology of head and neck cancer. Surg. Oncol. Clin. N. Am. 2015, 24, 379–396. [Google Scholar] [CrossRef]
- Rivelli, T.G.; Mak, M.P.; Martins, R.E.; da Costa e Silva, V.T.; de Castro, G., Jr. Cisplatin based chemoradiation late toxicities in head and neck squamous cell carcinoma patients. Discov. Med. 2015, 20, 57–66. [Google Scholar] [PubMed]
- Budach, V.; Tinhofer, I. Novel prognostic clinical factors and biomarkers for outcome prediction in head and neck cancer: A systematic review. Lancet Oncol. 2019, 20, e313–e326. [Google Scholar] [CrossRef]
- Amin, M.B.; Greene, F.L.; Edge, S.B.; Compton, C.C.; Gershenwald, J.E.; Brookland, R.K.; Meyer, L.; Gress, D.M.; Byrd, D.R.; Winchester, D.P. The Eighth Edition AJCC Cancer Staging Manual: Continuing to build a bridge from a population-based to a more "personalized" approach to cancer staging. CA Cancer J. Clin. 2017, 67, 93–99. [Google Scholar] [CrossRef] [PubMed]
- Lydiatt, W.M.; Patel, S.G.; O’Sullivan, B.; Brandwein, M.S.; Ridge, J.A.; Migliacci, J.C.; Loomis, A.M.; Shah, J.P. Head and Neck cancers-major changes in the American Joint Committee on cancer eighth edition cancer staging manual. CA Cancer J. Clin. 2017, 67, 122–137. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.C.; Wu, C.T.; Ko, J.Y.; Yang, T.L.; Lou, P.J.; Wang, C.P.; Chang, Y.L. Clinical characteristics and treatment outcome of oropharyngeal squamous cell carcinoma in an endemic betel quid region. Sci. Rep. 2020, 10, 526. [Google Scholar] [CrossRef] [PubMed]
- Mroz, E.A.; Tward, A.D.; Pickering, C.R.; Myers, J.N.; Ferris, R.L.; Rocco, J.W. High intratumor genetic heterogeneity is related to worse outcome in patients with head and neck squamous cell carcinoma. Cancer 2013, 119, 3034–3042. [Google Scholar] [CrossRef]
- Mroz, E.A.; Tward, A.D.; Hammon, R.J.; Ren, Y.; Rocco, J.W. Intra-tumor genetic heterogeneity and mortality in head and neck cancer: Analysis of data from the Cancer Genome Atlas. PLoS Med. 2015, 12, e1001786, Erratum in PLoS Med. 2015, 12, e1001844. [Google Scholar] [CrossRef]
- Berghmans, E.; Boonen, K.; Maes, E.; Mertens, I.; Pauwels, P.; Baggerman, G. Implementation of MALDI Mass Spectrometry Imaging in Cancer Proteomics Research: Applications and Challenges. J. Pers. Med. 2020, 10, 54. [Google Scholar] [CrossRef] [PubMed]
- Schone, C.; Hofler, H.; Walch, A. MALDI imaging mass spectrometry in cancer research: Combining proteomic profiling and histological evaluation. Clin. Biochem. 2013, 46, 539–545. [Google Scholar] [CrossRef]
- Stillger, M.N.; Li, M.J.; Honscheid, P.; von Neubeck, C.; Foll, M.C. Advancing rare cancer research by MALDI mass spectrometry imaging: Applications, challenges, and future perspectives in sarcoma. Proteomics 2024, 24, e2300001. [Google Scholar] [CrossRef]
- Klein, O.; Kanter, F.; Kulbe, H.; Jank, P.; Denkert, C.; Nebrich, G.; Schmitt, W.D.; Wu, Z.; Kunze, C.A.; Sehouli, J.; et al. MALDI-Imaging for Classification of Epithelial Ovarian Cancer Histotypes from a Tissue Microarray Using Machine Learning Methods. Proteom. Clin. Appl. 2019, 13, e1700181. [Google Scholar] [CrossRef] [PubMed]
- Pertzborn, D.; Arolt, C.; Ernst, G.; Lechtenfeld, O.J.; Kaesler, J.; Pelzel, D.; Guntinas-Lichius, O.; von Eggeling, F.; Hoffmann, F. Multi-Class Cancer Subtyping in Salivary Gland Carcinomas with MALDI Imaging and Deep Learning. Cancers 2022, 14, 4342. [Google Scholar] [CrossRef]
- Patterson, N.H.; Alabdulkarim, B.; Lazaris, A.; Thomas, A.; Marcinkiewicz, M.M.; Gao, Z.H.; Vermeulen, P.B.; Chaurand, P.; Metrakos, P. Assessment of pathological response to therapy using lipid mass spectrometry imaging. Sci. Rep. 2016, 6, 36814. [Google Scholar] [CrossRef]
- Hardesty, W.M.; Kelley, M.C.; Mi, D.; Low, R.L.; Caprioli, R.M. Protein signatures for survival and recurrence in metastatic melanoma. J. Proteom. 2011, 74, 1002–1014. [Google Scholar] [CrossRef]
- Hoffmann, F.; Umbreit, C.; Kruger, T.; Pelzel, D.; Ernst, G.; Kniemeyer, O.; Guntinas-Lichius, O.; Berndt, A.; von Eggeling, F. Identification of Proteomic Markers in Head and Neck Cancer Using MALDI-MS Imaging, LC-MS/MS, and Immunohistochemistry. Proteom. Clin. Appl. 2019, 13, e1700173. [Google Scholar] [CrossRef]
- Kurczyk, A.; Gawin, M.; Paul, P.; Chmielik, E.; Rutkowski, T.; Pietrowska, M.; Widlak, P. Prognostic Value of Molecular Intratumor Heterogeneity in Primary Oral Cancer and Its Lymph Node Metastases Assessed by Mass Spectrometry Imaging. Molecules 2022, 27, 5458. [Google Scholar] [CrossRef]
- Boskamp, T.; Lachmund, D.; Oetjen, J.; Cordero Hernandez, Y.; Trede, D.; Maass, P.; Casadonte, R.; Kriegsmann, J.; Warth, A.; Dienemann, H.; et al. A new classification method for MALDI imaging mass spectrometry data acquired on formalin-fixed paraffin-embedded tissue samples. Biochim. Biophys. Acta Proteins Proteom. 2017, 1865, 916–926. [Google Scholar] [CrossRef]
- Galli, M.; Zoppis, I.; Smith, A.; Magni, F.; Mauri, G. Machine learning approaches in MALDI-MSI: Clinical applications. Expert Rev. Proteom. 2016, 13, 685–696. [Google Scholar] [CrossRef]
- Kanter, F.; Lellmann, J.; Thiele, H.; Kalloger, S.; Schaeffer, D.F.; Wellmann, A.; Klein, O. Classification of Pancreatic Ductal Adenocarcinoma Using MALDI Mass Spectrometry Imaging Combined with Neural Networks. Cancers 2023, 15, 686. [Google Scholar] [CrossRef]
- Ang, K.K.; Harris, J.; Wheeler, R.; Weber, R.; Rosenthal, D.I.; Nguyen-Tan, P.F.; Westra, W.H.; Chung, C.H.; Jordan, R.C.; Lu, C.; et al. Human papillomavirus and survival of patients with oropharyngeal cancer. N. Engl. J. Med. 2010, 363, 24–35. [Google Scholar] [CrossRef]
- Cillero-Pastor, B.; Heeren, R.M. Matrix-assisted laser desorption ionization mass spectrometry imaging for peptide and protein analyses: A critical review of on-tissue digestion. J. Proteome Res. 2014, 13, 325–335. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Hundsdoerfer, P.; Schulte, J.H.; Astrahantseff, K.; Boral, S.; Schmelz, K.; Eggert, A.; Klein, O. Discovery of Spatial Peptide Signatures for Neuroblastoma Risk Assessment by MALDI Mass Spectrometry Imaging. Cancers 2021, 13, 3184. [Google Scholar] [CrossRef] [PubMed]
- Kushner, I.K.; Clair, G.; Purvine, S.O.; Lee, J.Y.; Adkins, J.N.; Payne, S.H. Individual Variability of Protein Expression in Human Tissues. J. Proteome Res. 2018, 17, 3914–3922. [Google Scholar] [CrossRef] [PubMed]
- Rasmussen, J.H.; Olin, A.B.; Lelkaitis, G.; Hansen, A.E.; Andersen, F.L.; Johannesen, H.H.; Kjaer, A.; Fischer, B.M.; Specht, L.; Bentzen, S.M.; et al. Intratumor heterogeneity is biomarker specific and challenges the association with heterogeneity in multimodal functional imaging in head and neck squamous cell carcinoma. Eur. J. Radiol. 2021, 139, 109668. [Google Scholar] [CrossRef]
- McGranahan, N.; Swanton, C. Clonal Heterogeneity and Tumor Evolution: Past, Present, and the Future. Cell 2017, 168, 613–628. [Google Scholar] [CrossRef]
- Kundig, P.; Giesen, C.; Jackson, H.; Bodenmiller, B.; Papassotirolopus, B.; Freiberger, S.N.; Aquino, C.; Opitz, L.; Varga, Z. Limited utility of tissue micro-arrays in detecting intra-tumoral heterogeneity in stem cell characteristics and tumor progression markers in breast cancer. J. Transl. Med. 2018, 16, 118, Erratum in J. Transl. Med. 2018, 16, 180. [Google Scholar] [CrossRef]
- Balermpas, P.; Michel, Y.; Wagenblast, J.; Seitz, O.; Weiss, C.; Rodel, F.; Rodel, C.; Fokas, E. Tumour-infiltrating lymphocytes predict response to definitive chemoradiotherapy in head and neck cancer. Br. J. Cancer 2014, 110, 501–509, Erratum in Br. J. Cancer 2014, 110, 547. [Google Scholar] [CrossRef]
- Eckert, R.L. Sequence of the human 40-kDa keratin reveals an unusual structure with very high sequence identity to the corresponding bovine keratin. Proc. Natl. Acad. Sci. USA 1988, 85, 1114–1118. [Google Scholar] [CrossRef]
- Wiche, G. Role of plectin in cytoskeleton organization and dynamics. J. Cell Sci. 1998, 111 Pt 17, 2477–2486. [Google Scholar] [CrossRef]
- Calderwood, D.A.; Shattil, S.J.; Ginsberg, M.H. Integrins and actin filaments: Reciprocal regulation of cell adhesion and signaling. J. Biol. Chem. 2000, 275, 22607–22610. [Google Scholar] [CrossRef] [PubMed]
- Hailstones, D.L.; Gunning, P.W. Characterization of human myosin light chains 1sa and 3nm: Implications for isoform evolution and function. Mol. Cell Biol. 1990, 10, 1095–1104. [Google Scholar] [CrossRef]
- Montagna, G.N.; Matuschewski, K.; Buscaglia, C.A. Small heat shock proteins in cellular adhesion and migration: Evidence from Plasmodium genetics. Cell Adhes. Migr. 2012, 6, 78–84. [Google Scholar] [CrossRef] [PubMed]
- Gottardi, C.J.; Gumbiner, B.M. Adhesion signaling: How beta-catenin interacts with its partners. Curr. Biol. 2001, 11, R792–R794. [Google Scholar] [CrossRef]
- Dowling, J.; Yu, Q.C.; Fuchs, E. Beta4 integrin is required for hemidesmosome formation, cell adhesion and cell survival. J. Cell Biol. 1996, 134, 559–572. [Google Scholar] [CrossRef] [PubMed]
- Crowe, D.L.; Milo, G.E.; Shuler, C.F. Keratin 19 downregulation by oral squamous cell carcinoma lines increases invasive potential. J. Dent. Res. 1999, 78, 1256–1263. [Google Scholar] [CrossRef]
- Lu, X.; Mei, Y.; Fan, C.; Chen, P.; Li, X.; Zeng, Z.; Li, G.; Xiong, W.; Xiang, B.; Yi, M. Silencing AHNAK promotes nasopharyngeal carcinoma progression by upregulating the ANXA2 protein. Cell Oncol. 2024, 47, 833–850. [Google Scholar] [CrossRef] [PubMed]
- Katada, K.; Tomonaga, T.; Satoh, M.; Matsushita, K.; Tonoike, Y.; Kodera, Y.; Hanazawa, T.; Nomura, F.; Okamoto, Y. Plectin promotes migration and invasion of cancer cells and is a novel prognostic marker for head and neck squamous cell carcinoma. J. Proteom. 2012, 75, 1803–1815. [Google Scholar] [CrossRef] [PubMed]
- Rong, C.; Muller, M.F.; Xiang, F.; Jensen, A.; Weichert, W.; Major, G.; Plinkert, P.K.; Hess, J.; Affolter, A. Adaptive ERK signalling activation in response to therapy and in silico prognostic evaluation of EGFR-MAPK in HNSCC. Br. J. Cancer 2020, 123, 288–297. [Google Scholar] [CrossRef] [PubMed]
- Longuespee, R.; Alberts, D.; Baiwir, D.; Mazzucchelli, G.; Smargiasso, N.; De Pauw, E. MALDI Imaging Combined with Laser Microdissection-Based Microproteomics for Protein Identification: Application to Intratumor Heterogeneity Studies. Methods Mol. Biol. 2018, 1788, 297–312. [Google Scholar] [CrossRef]
- Hartl, J.; Kurth, F.; Kappert, K.; Horst, D.; Mulleder, M.; Hartmann, G.; Ralser, M. Quantitative protein biomarker panels: A path to improved clinical practice through proteomics. EMBO Mol. Med. 2023, 15, e16061. [Google Scholar] [CrossRef]
- Turck, N.; Vutskits, L.; Sanchez-Pena, P.; Robin, X.; Hainard, A.; Gex-Fabry, M.; Fouda, C.; Bassem, H.; Mueller, M.; Lisacek, F.; et al. A multiparameter panel method for outcome prediction following aneurysmal subarachnoid hemorrhage. Intensive Care Med. 2010, 36, 107–115. [Google Scholar] [CrossRef]
- Robin, X.; Turck, N.; Hainard, A.; Lisacek, F.; Sanchez, J.C.; Muller, M. Bioinformatics for protein biomarker panel classification: What is needed to bring biomarker panels into in vitro diagnostics? Expert Rev. Proteom. 2009, 6, 675–689. [Google Scholar] [CrossRef]
- Drucker, E.; Krapfenbauer, K. Pitfalls and limitations in translation from biomarker discovery to clinical utility in predictive and personalised medicine. EPMA J. 2013, 4, 7. [Google Scholar] [CrossRef]
- Hemphill, E.; Lindsay, J.; Lee, C.; Mandoiu, I.I.; Nelson, C.E. Feature selection and classifier performance on diverse bio- logical datasets. BMC Bioinform. 2014, 15 (Suppl. 13), S4. [Google Scholar] [CrossRef]
- Zhang, Y.; Bernau, C.; Parmigiani, G.; Waldron, L. The impact of different sources of heterogeneity on loss of accuracy from genomic prediction models. Biostatistics 2020, 21, 253–268. [Google Scholar] [CrossRef]
- Benkarim, O.; Paquola, C.; Park, B.Y.; Kebets, V.; Hong, S.J.; Vos de Wael, R.; Zhang, S.; Yeo, B.T.T.; Eickenberg, M.; Ge, T.; et al. Population heterogeneity in clinical cohorts affects the predictive accuracy of brain imaging. PLoS Biol. 2022, 20, e3001627. [Google Scholar] [CrossRef]
- Bednarczyk, K.; Gawin, M.; Chekan, M.; Kurczyk, A.; Mrukwa, G.; Pietrowska, M.; Polanska, J.; Widlak, P. Discrimination of normal oral mucosa from oral cancer by mass spectrometry imaging of proteins and lipids. J. Mol. Histol. 2019, 50, 1–10. [Google Scholar] [CrossRef]
- Wisniewski, J.R.; Dus, K.; Mann, M. Proteomic workflow for analysis of archival formalin-fixed and paraffin-embedded clinical samples to a depth of 10 000 proteins. Proteom. Clin. Appl. 2013, 7, 225–233. [Google Scholar] [CrossRef]
- Tinhofer, I.; Stenzinger, A.; Eder, T.; Konschak, R.; Niehr, F.; Endris, V.; Distel, L.; Hautmann, M.G.; Mandic, R.; Stromberger, C.; et al. Targeted next-generation sequencing identifies molecular subgroups in squamous cell carcinoma of the head and neck with distinct outcome after concurrent chemoradiation. Ann. Oncol. 2016, 27, 2262–2268. [Google Scholar] [CrossRef] [PubMed]
- Hess, A.K.; Muer, A.; Mairinger, F.D.; Weichert, W.; Stenzinger, A.; Hummel, M.; Budach, V.; Tinhofer, I. MiR-200b and miR-155 as predictive biomarkers for the efficacy of chemoradiation in locally advanced head and neck squamous cell carcinoma. Eur. J. Cancer 2017, 77, 3–12. [Google Scholar] [CrossRef] [PubMed]
- Bankhead, P.; Loughrey, M.B.; Fernandez, J.A.; Dombrowski, Y.; McArt, D.G.; Dunne, P.D.; McQuaid, S.; Gray, R.T.; Murray, L.J.; Coleman, H.G.; et al. QuPath: Open source software for digital pathology image analysis. Sci. Rep. 2017, 7, 16878. [Google Scholar] [CrossRef] [PubMed]
- Hosmer, D.; Lemeshow, S. Applied Logistic Regression, 2nd ed.; John Wiley and Sons: New York, NY, USA, 2000; Chapter 5. [Google Scholar]
- Sonego, P.; Kocsor, A.; Pongor, S. ROC analysis: Applications to the classification of biological sequences and 3D structures. Brief. Bioinform. 2008, 9, 198–209. [Google Scholar] [CrossRef]
- Zhang, J.; Xin, L.; Shan, B.; Chen, W.; Xie, M.; Yuen, D.; Zhang, W.; Zhang, Z.; Lajoie, G.A.; Ma, B. PEAKS DB: De novo sequencing assisted database search for sensitive and accurate peptide identification. Mol. Cell Proteom. 2012, 11, M111.010587. [Google Scholar] [CrossRef]

| CDDP-CRT | MMC-CRT | ||
|---|---|---|---|
| total number (n) | 31 | 29 | |
| Age at diagnosis mean (±SD) | 56.4 (±9.3) | 56.1 (±6.2) | |
| Sex | male | 24 (77%) | 23 (79%) |
| female | 7 (23%) | 6 (21%) | |
| HPV | negative | 33 (100%) | 29 (100%) |
| Stage of the disease | IVA | 27 (87%) | 26 (90%) |
| IVB | 4 (13%) | 3 (10%) | |
| Risk category 1 | high | 33 (100%) | 29 (100%) |
| Smoking | yes | 18 (58%) | 23 (79%) |
| no | 13 (42%) | 6 (21%) | |
| Localization | oropharynx | 15 (55%) | 16 (55%) |
| hypopharynx | 16 (45%) | 13 (45%) | |
| Outcome 2 | NED | 11 (35%) | 14 (48%) |
| RecPro | 20 (65%) | 15 (52%) |
| Model | Level | Metric | Split 1 | Split 2 | Split 3 | Split 4 | Split 5 | Mean |
|---|---|---|---|---|---|---|---|---|
| all m/z features | Spectra | b.acc. | 0.917 | 0.388 | 0.824 | 0.690 | 0.452 | 0.654 |
| AUC | 0.975 | 0.293 | 0.899 | 0.892 | 0.449 | 0.702 | ||
| sensitivity | 0.945 | 0.049 | 0.830 | 0.993 | 0.646 | 0.693 | ||
| specificity | 0.888 | 0.727 | 0.819 | 0.388 | 0.257 | 0.616 | ||
| Patient | b.acc. | 1.000 | 0.375 | 0.900 | 0.750 | 0.500 | 0.705 | |
| AUC | 1.000 | 0.500 | 1.000 | 0.833 | 0.438 | 0.754 | ||
| sensitivity | 1.000 | 0.000 | 1.000 | 1.000 | 0.750 | 0.750 | ||
| specificity | 1.000 | 0.750 | 0.800 | 0.500 | 0.250 | 0.660 | ||
| 249 discriminatory m/z features | Spectra | b.acc. | 0.903 | 0.414 | 0.679 | 0.544 | 0.471 | 0.602 |
| AUC | 0.967 | 0.470 | 0.791 | 0.555 | 0.433 | 0.643 | ||
| sensitivity | 0.947 | 0.033 | 0.489 | 0.554 | 0.345 | 0.474 | ||
| specificity | 0.859 | 0.794 | 0.869 | 0.535 | 0.597 | 0.731 | ||
| Patient | b.acc. | 1.000 | 0.500 | 0.667 | 0.917 | 0.500 | 0.717 | |
| AUC | 1.000 | 0.500 | 1.000 | 0.833 | 0.375 | 0.742 | ||
| sensitivity | 1.000 | 0.000 | 0.333 | 1.000 | 0.250 | 0.517 | ||
| specificity | 1.000 | 1.000 | 1.000 | 0.833 | 0.750 | 0.917 |
| Model | Level | Metric | Split 1 | Split 2 | Split 3 | Split 4 | Split 5 | Mean |
|---|---|---|---|---|---|---|---|---|
| all m/z features | Spectra | b.acc. | 0.509 | 0.568 | 0.564 | 0.538 | 0.534 | 0.543 |
| AUC | 0.524 | 0.586 | 0.585 | 0.575 | 0.546 | 0.563 | ||
| sensitivity | 0.399 | 0.469 | 0.454 | 0.573 | 0.480 | 0.475 | ||
| specificity | 0.618 | 0.667 | 0.674 | 0.504 | 0.589 | 0.610 | ||
| Patient | b.acc. | 0.440 | 0.540 | 0.474 | 0.445 | 0.443 | 0.469 | |
| AUC | 0.476 | 0.476 | 0.538 | 0.467 | 0.457 | 0.483 | ||
| sensitivity | 0.214 | 0.214 | 0.214 | 0.357 | 0.286 | 0.257 | ||
| specificity | 0.667 | 0.867 | 0.733 | 0.533 | 0.600 | 0.680 | ||
| 249 discriminatory m/z features | Spectra | b.acc. | 0.485 | 0.466 | 0.509 | 0.524 | 0.465 | 0.490 |
| AUC | 0.477 | 0.460 | 0.506 | 0.526 | 0.440 | 0.482 | ||
| sensitivity | 0.415 | 0.257 | 0.340 | 0.449 | 0.198 | 0.332 | ||
| specificity | 0.555 | 0.675 | 0.679 | 0.599 | 0.733 | 0.648 | ||
| Patient | b.acc. | 0.376 | 0.369 | 0.436 | 0.405 | 0.469 | 0.411 | |
| AUC | 0.438 | 0.390 | 0.424 | 0.419 | 0.448 | 0.424 | ||
| sensitivity | 0.286 | 0.071 | 0.071 | 0.143 | 0.071 | 0.129 | ||
| specificity | 0.467 | 0.667 | 0.800 | 0.667 | 0.867 | 0.693 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Grzeski, M.; Jensen, P.M.; Hempel, B.-F.; Thiele, H.; Lellmann, J.; Schallenberg, S.; Budach, V.; Keilholz, U.; Tinhofer, I.; Klein, O. Integrating MALDI-MSI-Based Spatial Proteomics and Machine Learning to Predict Chemoradiotherapy Outcomes in Head and Neck Cancer. Int. J. Mol. Sci. 2025, 26, 9084. https://doi.org/10.3390/ijms26189084
Grzeski M, Jensen PM, Hempel B-F, Thiele H, Lellmann J, Schallenberg S, Budach V, Keilholz U, Tinhofer I, Klein O. Integrating MALDI-MSI-Based Spatial Proteomics and Machine Learning to Predict Chemoradiotherapy Outcomes in Head and Neck Cancer. International Journal of Molecular Sciences. 2025; 26(18):9084. https://doi.org/10.3390/ijms26189084
Chicago/Turabian StyleGrzeski, Marta, Patrick Moeller Jensen, Benjamin-Florian Hempel, Herbert Thiele, Jan Lellmann, Simon Schallenberg, Volker Budach, Ulrich Keilholz, Ingeborg Tinhofer, and Oliver Klein. 2025. "Integrating MALDI-MSI-Based Spatial Proteomics and Machine Learning to Predict Chemoradiotherapy Outcomes in Head and Neck Cancer" International Journal of Molecular Sciences 26, no. 18: 9084. https://doi.org/10.3390/ijms26189084
APA StyleGrzeski, M., Jensen, P. M., Hempel, B.-F., Thiele, H., Lellmann, J., Schallenberg, S., Budach, V., Keilholz, U., Tinhofer, I., & Klein, O. (2025). Integrating MALDI-MSI-Based Spatial Proteomics and Machine Learning to Predict Chemoradiotherapy Outcomes in Head and Neck Cancer. International Journal of Molecular Sciences, 26(18), 9084. https://doi.org/10.3390/ijms26189084






