Structural and Evolutionary Analysis of Saci2-Like LTR Retrotransposons in Diphyllobothriidean Tapeworms
Abstract
1. Introduction
2. Results
2.1. Retrievals of Eg_lennie Pol Homologs in S. erinaceieuropaei
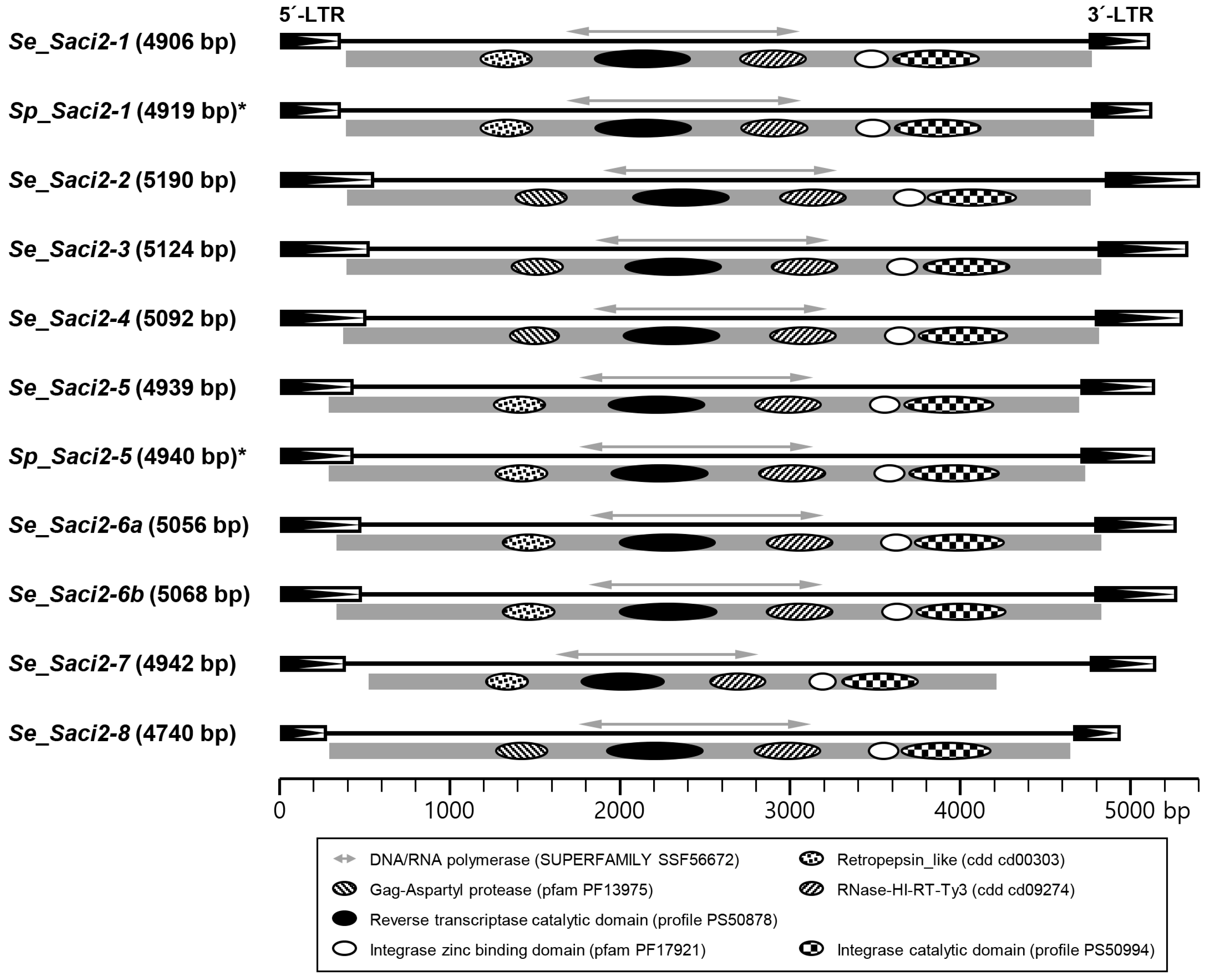
2.2. Se_Saci2 Retrotransposons in S. erinaceieuropaei
2.3. Se_Saci2 Orthologs in Other Parasitic Cestodes
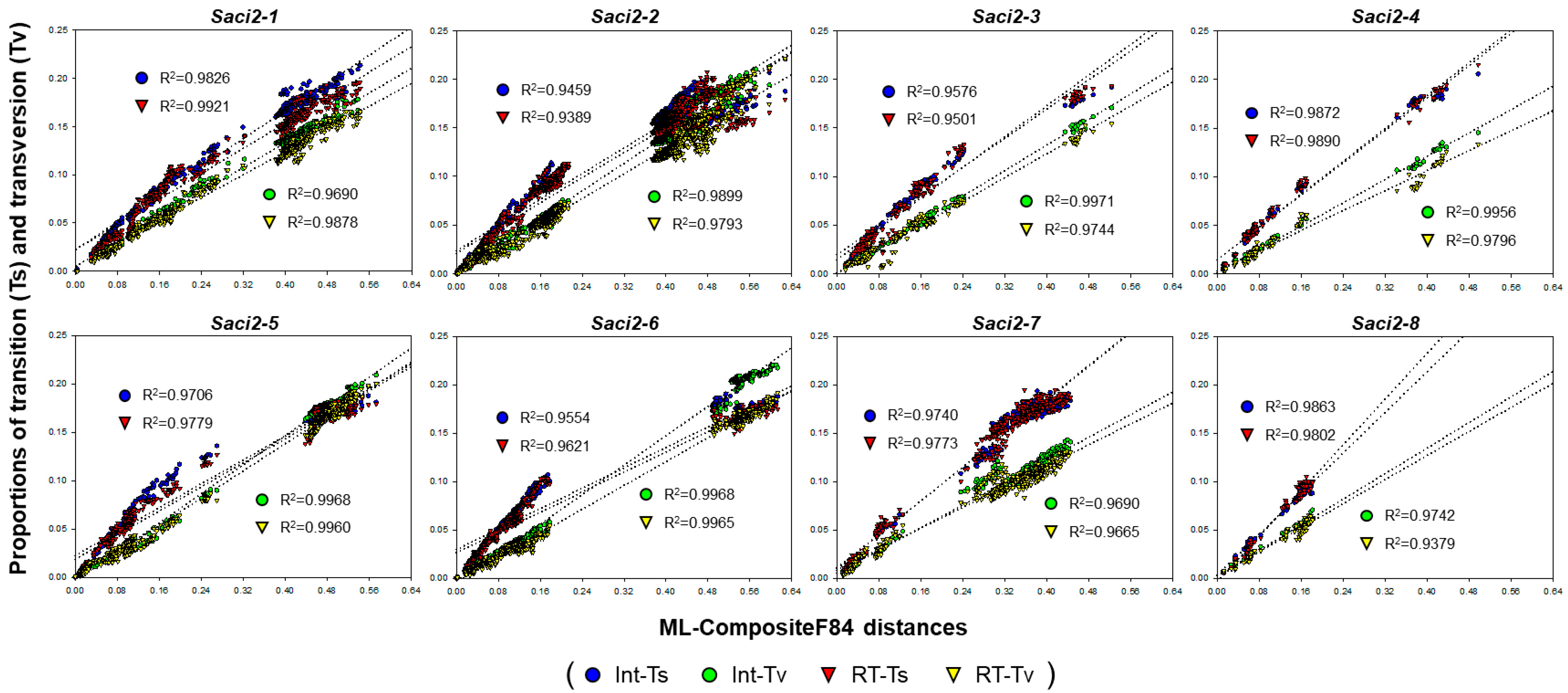
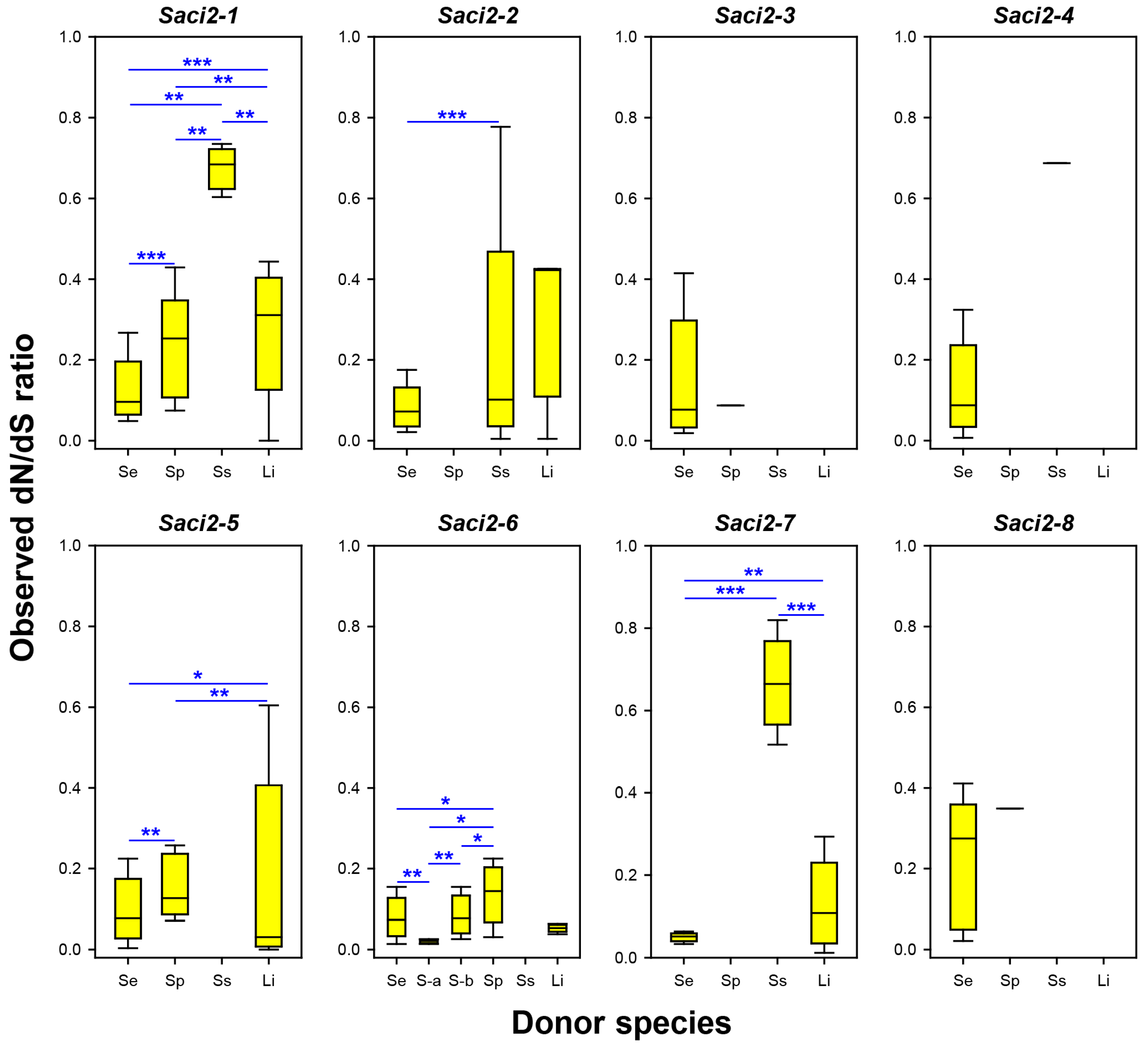
2.4. Evolutionary Statistics Among Cestode Saci2 Element Copies
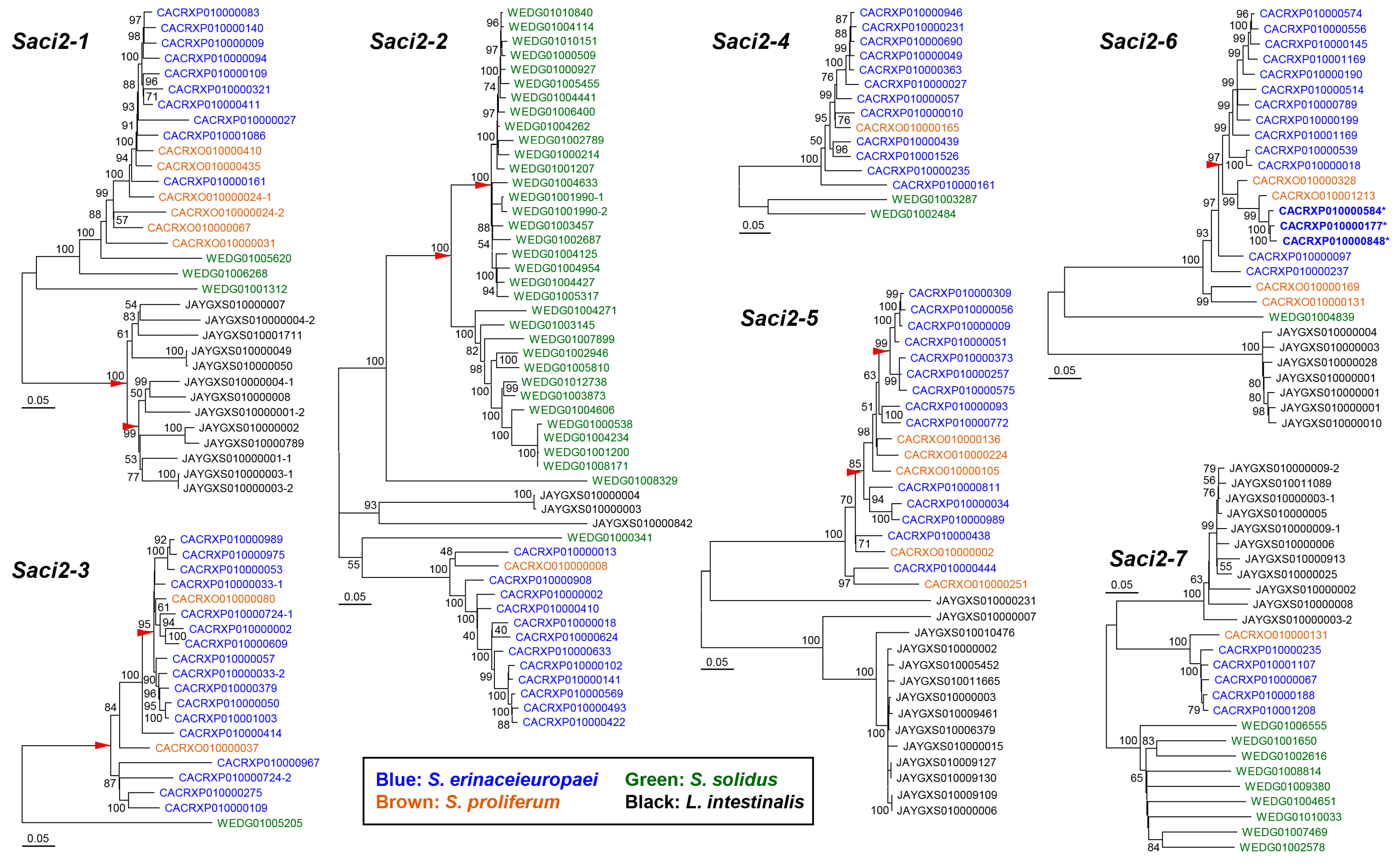
2.5. Phylogenies of Cestode Saci2 Elements
3. Discussion
4. Materials and Methods
4.1. Retrieval of Amino Acid Sequences Homologous to the Pol Protein of E. granulosus Lennie
4.2. Identification of Full-Unit LTR Retrotransposons in S. erinaceieuropaei Genome
4.3. Sequence Analyses
4.4. Phylogenetic Analyses
5. Conclusions
Supplementary Materials
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| LTR | Long terminal repeat |
| TE | Transposable element |
| CDS | Coding DNA sequence |
| PBS | Primer binding site |
| TSD | Target site duplication |
| ORF | Open reading frame |
| Pol | Polyprotein |
| RT | Reverse transcriptase |
| Ts | Transition |
| Tv | Transversion |
References
- Hoberg, E.P.; Gardner, S.L.; Campbell, R.A. Systematics of the Eucestoda: Advances toward a new phylogenetic paradigm, and observations on the early diversification of tapeworms and vertebrates. Syst. Parasitol. 1999, 42, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Goater, T.M.; Goater, C.P.; Esch, G.W. Platyhelminthes: The flatworms. In Parasitism: The Diversity and Ecology of Animal Parasites, 2nd ed.; Goater, T.M., Goater, C.P., Esch, G.W., Eds.; Cambridge University Press: Cambridge, UK, 2014; pp. 113–169. [Google Scholar]
- Liu, Q.; Li, M.W.; Wang, Z.D.; Zhao, H.; Zhu, X.Q. Human sparganosis, a neglected food borne zoonosis. Lancet Infect. Dis. 2015, 15, 1226–1235. [Google Scholar] [CrossRef] [PubMed]
- Moore, J.; Brooks, D.R. Asexual reproduction in cestodes (Cyclophyllidea: Taeniidae): Ecological and phylogenetic influences. Evolution 1987, 41, 882–891. [Google Scholar] [CrossRef] [PubMed]
- de Noya, B.A.; Torres, J.R.; Noya, O. Maintenance of Sparganum proliferum in vitro and in experimental animals. Int. J. Parasitol. 1992, 22, 835–838. [Google Scholar] [CrossRef]
- Kikuchi, T.; Dayi, M.; Hunt, V.L.; Ishiwata, K.; Toyoda, A.; Kounosu, A.; Sun, S.; Maeda, Y.; Kondo, Y.; de Noya, B.A.; et al. Genome of the fatal tapeworm Sparganum proliferum uncovers mechanisms for cryptic life cycle and aberrant larval proliferation. Commun. Biol. 2021, 4, 649. [Google Scholar] [CrossRef]
- Tsai, I.J.; Zarowiecki, M.; Holroyd, N.; Garciarrubio, A.; Sanchez-Flores, A.; Brooks, K.L.; Tracey, A.; Bobes, R.J.; Fragoso, G.; Sciutto, E.; et al. The genomes of four tapeworm species reveal adaptations to parasitism. Nature 2013, 496, 57–63. [Google Scholar] [CrossRef]
- Boeke, J.D.; Stoye, J.P. Retrotransposons, endogenous retroviruses, and the evolution of retroelements. In Retroviruses; Coffin, J.M., Hughes, S.H., Varmus, H.E., Eds.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 1997; pp. 343–435. [Google Scholar]
- Eickbush, T.H.; Jamburuthugoda, V.K. The diversity of retrotransposons and the properties of their reverse transcriptases. Virus Res. 2008, 134, 221–234. [Google Scholar] [CrossRef]
- Chuong, E.B.; Elde, N.C.; Feschotte, C. Regulatory activities of transposable elements: From conflicts to benefits. Nat. Rev. Genet. 2017, 18, 71–86. [Google Scholar] [CrossRef]
- Chen, M.; Huang, X.; Wang, C.; Wang, S.; Jia, L.; Li, L. Endogenous retroviral solo-LTRs in human genome. Front. Genet. 2024, 15, 1358078. [Google Scholar] [CrossRef]
- Vitte, C.; Panaud, O. Formation of solo-LTRs through unequal homologous recombination counterbalances amplifications of LTR retrotransposons in rice Oryza sativa L. Mol. Biol. Evol. 2003, 20, 528–540. [Google Scholar] [CrossRef]
- Belshaw, R.; Watson, J.; Katzourakis, A.; Howe, A.; Woolven-Allen, J.; Burt, A.; Tristem, M. Rate of recombinational deletion among human endogenous retroviruses. J. Virol. 2007, 81, 9437–9442. [Google Scholar] [CrossRef] [PubMed]
- Devos, K.M.; Brown, J.K.; Bennetzen, J.L. Genome size reduction through illegitimate recombination counteracts genome expansion in Arabidopsis. Genome Res. 2002, 12, 1075–1079. [Google Scholar] [CrossRef] [PubMed]
- Li, S.F.; She, H.B.; Yang, L.L.; Lan, L.N.; Zhang, X.Y.; Wang, L.Y.; Zhang, Y.L.; Li, N.; Deng, C.L.; Qian, W.; et al. Impact of LTR-Retrotransposons on genome structure, evolution, and function in Cucurbitaceae species. Int. J. Mol. Sci. 2022, 23, 10158. [Google Scholar] [CrossRef] [PubMed]
- Bae, Y.A. Evolutionary characterization of Ty3/gypsy-like LTR retrotransposons in the parasitic cestode Echinococcus granulosus. Parasitology 2016, 143, 1691–1702. [Google Scholar] [CrossRef]
- Koziol, U.; Radio, S.; Smircich, P.; Zarowiecki, M.; Fernández, C.; Brehm, K. A novel terminal-repeat retrotransposon in miniature (TRIM) is massively expressed in Echinococcus multilocularis stem cells. Genome Biol. Evol. 2015, 7, 2136–2153. [Google Scholar] [CrossRef][Green Version]
- Brookfield, J.F. The ecology of the Genome—Mobile DNA elements and their hosts. Nat. Rev. Genet. 2005, 6, 128–136. [Google Scholar] [CrossRef]
- Petrov, D.A.; Fiston-Lavier, A.S.; Lipatov, M.; Lenkov, K.; González, J. Population genomics of transposable elements in Drosophila melanogaster. Mol. Biol. Evol. 2011, 28, 1633–1644. [Google Scholar] [CrossRef]
- Preston, B.D. Error-prone retrotransposition: Rime of the ancient mutators. Proc. Natl. Acad. Sci. USA 1996, 93, 7427–7431. [Google Scholar] [CrossRef]
- Le Rouzic, A.; Capy, P. The first steps of transposable elements invasion: Parasitic strategy vs. genetic drift. Genetica 2005, 123, 115–131. [Google Scholar]
- Doolittle, W.F.; Sapienza, C. Selfish genes, the phenotype paradigm and genome evolution. Nature 1980, 284, 601–603. [Google Scholar] [CrossRef]
- Charlesworth, B.; Langley, C.H. The population genetics of Drosophila transposable elements. Annu. Rev. Genet. 1989, 23, 251–287. [Google Scholar] [CrossRef] [PubMed]
- Sassaman, D.M.; Dombroski, B.A.; Moran, J.V.; Kimberland, M.L.; Naas, T.P.; DeBerardinis, R.J.; Gabriel, A.; Swergold, G.D.; Kazazian, H.H., Jr. Many human L1 elements are capable of retrotransposition. Nat. Genet. 1997, 16, 37–43. [Google Scholar] [CrossRef] [PubMed]
- Biémont, C.; Vieira, C. Genetics: Junk DNA as an evolutionary force. Nature 2006, 443, 521–524. [Google Scholar] [CrossRef] [PubMed]
- Feschotte, C.; Pritham, E.J. DNA transposons and the evolution of eukaryotic genomes. Annu. Rev. Genet. 2007, 41, 331–368. [Google Scholar] [CrossRef]
- Warren, I.A.; Naville, M.; Chalopin, D.; Levin, P.; Berger, C.S.; Galiana, D.; Volff, J.N. Evolutionary impact of transposable elements on genomic diversity and lineage-specific innovation in vertebrates. Chromosom. Res. 2015, 23, 505–531. [Google Scholar] [CrossRef]
- Thompson, P.J.; Macfarlan, T.S.; Lorincz, M.C. Long terminal repeats: From parasitic elements to building blocks of the transcriptional regulatory repertoire. Mol. Cell 2016, 62, 766–776. [Google Scholar] [CrossRef]
- Fueyo, R.; Judd, J.; Feschotte, C.; Wysocka, J. Roles of transposable elements in the regulation of mammalian transcription. Nat. Rev. Mol. Cell Biol. 2022, 23, 481–497. [Google Scholar] [CrossRef]
- Gerdes, P.; Richardson, S.R.; Mager, D.L.; Faulkner, G.J. Transposable elements in the mammalian embryo: Pioneers surviving through stealth and service. Genome Biol. 2016, 17, 100. [Google Scholar] [CrossRef]
- Zhang, L.; Yan, L.; Jiang, J.; Wang, Y.; Jiang, Y.; Yan, T.; Cao, Y. The structure and retrotransposition mechanism of LTR retrotransposons in the asexual yeast Candida albicans. Virulence 2014, 5, 655–664. [Google Scholar] [CrossRef][Green Version]
- Drost, H.-G.; Sanchez, D.H. Becoming a selfish clan: Recombination associated to reverse-transcription in LTR Retrotransposons. Genome Biol. Evol. 2019, 11, 3382–3392. [Google Scholar] [CrossRef]
- Borredá, C.; Pérez-Román, E.; Ibanez, V.; Terol, J.; Talon, M. Reprogramming of retrotransposon activity during speciation of the genus Citrus. Genome Biol. Evol. 2019, 11, 3478–3495. [Google Scholar] [CrossRef]
- Carr, M.; Soloway, J.R.; Robinson, T.E.; Brookfield, J.F.Y. Mechanisms regulating the copy numbers of six LTR retrotransposons in the genome of Drosophila melanogaster. Chromosoma 2002, 110, 511–518. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.N.; Pei, M.S.; Ampomah-Dwamena, C.; He, G.Q.; Wei, T.L.; Shi, Q.F.; Yu, Y.H.; Guo, D.L. Genome-wide characterization of long terminal repeat retrotransposons provides insights into trait evolution of four cucurbit species. Funct. Integr. Genom. 2023, 23, 218, Erratum in Funct. Integr. Genom. 2023, 23, 229. [Google Scholar] [CrossRef] [PubMed]
- Marsano, R.M.; Dimitri, P. Constitutive heterochromatin in eukaryotic genomes: A mine of transposable elements. Cells 2022, 11, 761. [Google Scholar] [CrossRef] [PubMed]
- Hoskins, R.A.; Smith, C.D.; Carlson, J.W.; Carvalho, A.B.; Halpern, A.; Kaminker, J.S.; Kennedy, C.; Mungall, C.J.; Sullivan, B.A.; Sutton, G.G.; et al. Heterochromatic sequences in a Drosophila whole-genome shotgun assembly. Genome Biol. 2002, 3, research0085. [Google Scholar] [CrossRef]
- Allshire, R.C.; Madhani, H.D. Ten principles of heterochromatin formation and function. Nat. Rev. Mol. Cell Biol. 2017, 19, 229–244. [Google Scholar] [CrossRef]
- Pereira, V. Insertion bias and purifying selection of retrotransposons in the Arabidopsis thaliana genome. Genome Biol. 2004, 5, R79. [Google Scholar] [CrossRef]
- Cordaux, R.; Batzer, M.A. The impact of retrotransposons on human genome evolution. Nat. Rev. Genet. 2009, 10, 691–703. [Google Scholar] [CrossRef]
- An, W.; Dai, L.; Niewiadomska, A.M.; Yetil, A.; O’Donnell, K.A.; Han, J.S.; Boeke, J.D. Characterization of a synthetic human LINE-1 retrotransposon ORFeus-Hs. Mob. DNA 2011, 2, 2. [Google Scholar] [CrossRef]
- Schorn, A.J.; Martienssen, R. Tie-Break: Host and retrotransposons play tRNA. Trends Cell Biol. 2018, 28, 793–806. [Google Scholar] [CrossRef]
- Iwasaki, Y.W.; Shoji, K.; Nakagwa, S.; Miyoshi, T.; Tomari, Y. Transposon-host arms race: A saga of genome evolution. Trends Genet. 2025, 41, 369–389. [Google Scholar] [CrossRef]
- Chan, P.P.; Lowe, T.M. GtRNAdb 2.0: An expanded database of transfer RNA genes identified in complete and draft genomes. Nucleic Acids Res. 2016, 44, D184–D189. [Google Scholar] [CrossRef] [PubMed]
- Wakefield, J.K.; Morrow, C.D. Mutations within the primer binding site of the human immunodeficiency virus type 1 define sequence requirements essential for reverse transcription. Virology 1996, 220, 290–298. [Google Scholar] [CrossRef] [PubMed]
- Ni, N.; Morrow, C.D. Impact of forced selection of tRNAs on HIV-1 replication and genome stability highlight preferences for selection of certain tRNAs. Virus Res. 2007, 124, 29–37. [Google Scholar] [CrossRef]
- Yang, Z. Maximum likelihood phylogenetic estimation from DNA sequences with variable rates over sites: Approximate methods. J. Mol. Evol. 1994, 39, 306–314. [Google Scholar] [CrossRef] [PubMed]
- Tajima, F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 1989, 123, 585–595. [Google Scholar] [CrossRef]
- Fu, Y.-X.; Li, W.-H. Statistical tests of neutrality of mutations. Genetics 1993, 133, 693–709. [Google Scholar] [CrossRef]
- Arkhipova, I.R. Neutral theory, transposable elements, and eukaryotic genome evolution. Mol. Biol. Evol. 2018, 35, 1332–1337. [Google Scholar] [CrossRef]
- Boissinot, S.; Furano, A.V. Adaptive evolution in LINE-1 retrotransposons. Mol. Biol. Evol. 2001, 18, 2186–2194. [Google Scholar] [CrossRef][Green Version]
- Xiang, H.; Pan, G.; Zhang, R.; Xu, J.; Li, T.; Li, W.; Zhou, Z.; Xiang, Z. Natural selection maintains the transcribed LTR retrotransposons in Nosema bombycis. J. Genet. Genom. 2010, 37, 305–314. [Google Scholar] [CrossRef]
- Gershan, J.A.; Karrer, K.M. A family of developmentally excised DNA elements in Tetrahymena is under selective pressure to maintain an open reading frame encoding an integrase-like protein. Nucleic Acids Res. 2000, 28, 4105–4112. [Google Scholar] [CrossRef][Green Version]
- Belshaw, R.; Pereira, V.; Katzourakis, A.; Talbot, G.; Paces, J.; Burt, A.; Tristem, M. Long-term reinfection of the human genome by endogenous retroviruses. Proc. Natl. Acad. Sci. USA 2004, 101, 4894–4899. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Li, G.M. DNA mismatch repair preferentially safeguards actively transcribed genes. DNA Repair 2018, 71, 82–86. [Google Scholar] [CrossRef] [PubMed]
- Strick, T.R.; Portman, J.R. Transcription-coupled repair: From cells to single molecules and back Again. J. Mol. Biol. 2019, 431, 4093–4102. [Google Scholar] [CrossRef]
- Aska, E.M.; Dermadi, D.; Kauppi, L. Single-cell sequencing of mouse thymocytes reveals mutational landscape shaped by replication errors, mismatch repair, and H3K36me3. iScience 2020, 23, 101452. [Google Scholar] [CrossRef] [PubMed]
- Putnam, C.D. Strand discrimination in DNA mismatch repair. DNA Repair 2021, 105, 103161. [Google Scholar] [CrossRef]
- Kim, S.H.; Cho, H.J.; Sohn, W.M.; Ahn, C.S.; Kong, Y.; Yang, H.J.; Bae, Y.A. Egg-specific expression of protein with DNA methyltransferase activity in the biocarcinogenic liver fluke Clonorchis sinensis. Parasitology 2015, 142, 1228–1238. [Google Scholar] [CrossRef]
- Fontenla, S.; Rinaldi, G.; Tort, J.F. Lost and found: Piwi and Argonaute pathways in flatworms. Front. Cell. Infect. Microbiol. 2021, 11, 653695. [Google Scholar] [CrossRef]
- Malik, H.S.; Eickbush, T.H. Modular evolution of the integrase domain in the Ty3/Gypsy class of LTR retrotransposons. J. Virol. 1999, 73, 5186–5190. [Google Scholar] [CrossRef]
- Guindon, S.; Dufayard, J.F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef]
- Xia, X.; Lemey, P. Assessing substitution saturation with DAMBE. In The Phylogenetic Handbook: A Practical Approach to DNA and Protein Phylogeny, 2nd ed.; Salemi, M., Vandamme, A.-M., Lemey, P., Eds.; Cambridge University Press: Cambridge, UK, 2009; pp. 615–630. [Google Scholar]
- Rozas, J.; Ferrer-Mata, A.; Sánchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sánchez-Gracia, A. DnaSP 6: DNA sequence polymorphism analysis of large datasets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef]
- Nei, M.; Gojobori, T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 1986, 3, 418–426. [Google Scholar] [CrossRef]

| Alignment | Donor Organism (Number of Sequences) a | Iss | 95% Confidence Interval | Iss.cSym | P1 b | Iss.cAsym | P2 b |
|---|---|---|---|---|---|---|---|
| Saci2-1 | All cesdtodes examined (32) | 0.4122 | 0.3962–0.4282 | 0.8091 | 0.0000 | 0.5562 | 0.0000 |
| Spirometra erinaceieuropaei (10) | 0.1102 | 0.1010–0.1194 | 0.8306 | 0.0000 | 0.7236 | 0.0000 | |
| Sparganum proliferum (6) | 0.1566 | 0.1448–0.1684 | 0.8377 | 0.0000 | 0.7877 | 0.0000 | |
| Ligula intestinalis (13) | 0.3531 | 0.3359–0.3702 | 0.8250 | 0.0000 | 0.6895 | 0.0000 | |
| Saci2-2 | All cesdtodes examined (51) | 0.3602 | 0.3415–0.3790 | 0.8044 | 0.0000 | 0.5426 | 0.0000 |
| S. erinaceieuropaei (12) | 0.1700 | 0.1579–0.1822 | 0.8192 | 0.0000 | 0.6855 | 0.0000 | |
| Schistocephalus solidus (35) | 0.2354 | 0.2207–0.2500 | 0.8077 | 0.0000 | 0.6181 | 0.0000 | |
| Saci2-3 | All cesdtodes examined (20) | 0.2729 | 0.2594–0.2863 | 0.8086 | 0.0000 | 0.6235 | 0.0000 |
| S. erinaceieuropaei (17) | 0.2517 | 0.2383–0.2562 | 0.8118 | 0.0000 | 0.6419 | 0.0000 | |
| Saci2-4 | All cesdtodes examined (15) | 0.3016 | 0.2874–0.3158 | 0.8183 | 0.0000 | 0.6643 | 0.0000 |
| S. erinaceieuropaei (12) | 0.1880 | 0.1753–0.2006 | 0.8233 | 0.0000 | 0.6927 | 0.0000 | |
| Saci2-5 | All cesdtodes examined (33) | 0.3673 | 0.3477–0.3869 | 0.8040 | 0.0000 | 0.5419 | 0.0000 |
| S. erinaceieuropaei (14) | 0.2858 | 0.2691–0.3025 | 0.8154 | 0.0000 | 0.1596 | 0.0000 | |
| S. proliferum (5) | 0.1392 | 0.1263–0.1522 | 0.8355 | 0.0000 | 0.7982 | 0.0000 | |
| L. intestinalis (14) | 0.1514 | 0.1481–0.1711 | 0.8170 | 0.0000 | 0.6742 | 0.0000 | |
| Saci2-6 | All cesdtodes examined (28) | 0.3332 | 0.3157–0.3507 | 0.8028 | 0.0000 | 0.5879 | 0.0000 |
| S. erinaceieuropaei (16) c | 0.1550 | 0.1435-0.1666 | 0.8274 | 0.0000 | 0.6696 | 0.0000 | |
| S. erinaceieuropaei_b (13) | 0.1543 | 0.1414–0.1673 | 0.8246 | 0.0000 | 0.6886 | 0.0000 | |
| L. intestinalis (7) | 0.1146 | 0.1039–0.1253 | 0.8356 | 0.0000 | 0.7686 | 0.0000 | |
| Saci2-7 | All cesdtodes examined (26) | 0.4435 | 0.4248–0.4622 | 0.8034 | 0.0000 | 0.5908 | 0.0000 |
| S. erinaceieuropaei (5) | 0.2558 | 0.2352–0.2763 | 0.8362 | 0.0000 | 0.7998 | 0.0000 | |
| S. solidus (9) | 0.4214 | 0.4026–0.4402 | 0.8260 | 0.0000 | 0.7260 | 0.0000 | |
| L. intestinalis (11) | 0.1701 | 0.1568–0.1835 | 0.8219 | 0.0000 | 0.6987 | 0.0000 | |
| Saci2-8 | All cesdtodes examined (12) | 0.2314 | 0.2172–0.2455 | 0.8205 | 0.0000 | 0.6877 | 0.0000 |
| S. erinaceieuropaei (10) | 0.2278 | 0.2135–0.2422 | 0.8243 | 0.0000 | 0.7123 | 0.0000 |
| Alignment | Donor Organism (Number of Squences) a | Int Region | RT Region | ||||
|---|---|---|---|---|---|---|---|
| Tajima’s D | Fu and Li’s D | Fu and Li’s F | Tajima’s D | Fu and Li’s D | Fu and Li’s F | ||
| Saci2-1 | All cesdtodes examined (32) | −1.31144 * | −1.73527 *,b | −1.46136 * | −1.40987 * | −1.85638 * | −2.02127 ** |
| Spirometra erinaceieuropaei (10) | −1.92425 *** | −2.17508 *** | −2.38869 *** | −1.94333 *** | −2.23464 **** | −2.44426 **** | |
| Sparganum proliferum (6) | −1.09044 * | −1.02791 * | −1.14655 * | −1.22400 * | −1.17210 * | −1.30227 * | |
| Ligula intestinalis (13) | −1.29726 * | −0.93173 * | −1.18111 * | −1.29678* | −0.90270 * | −1.15661 * | |
| Saci2-2 | All cesdtodes examined (51) | −1.20565 * | −1.68992 * | −1.80102 * | −1.27304 * | −1.88385 ** | −1.97508 ** |
| S. erinaceieuropaei (12) | −1.37539 * | −1.64454 * | −1.79592 * | −1.34268 * | −1.71113 * | −1.84185 * | |
| Schistocephalus solidus (35) | −1.84749 *** | −2.86640 *** | −2.98393 *** | −1.88691 *** | −3.03760 *** | −3.13260 *** | |
| Saci2-3 | All cesdtodes examined (20) | −1.87764 *** | −2.43667 *** | −2.64791 *** | −1.84227 *** | −2.34903 ** | −2.56387 ** |
| S. erinaceieuropaei (17) | −1.61478 ** | −2.02830 ** | −2.21366 ** | −1.65100 ** | −2.07445 ** | −2.26358 ** | |
| Saci2-4 | All cesdtodes examined (15) | −1.79439 *** | −1.84293* | −2.11068 ** | −1.78919 ** | −1.83971 * | −2.10621 ** |
| S. erinaceieuropaei (12) | −1.78627 *** | −2.17931 *** | −2.36896 *** | −1.78175 *** | −2.17335 *** | −2.36249 *** | |
| Saci2-5 | All cesdtodes examined (33) | −0.68779 * | −1.80233 * | −1.68241 * | −0.80231 * | −1.99026 ** | −1.87602 * |
| S. erinaceieuropaei (14) | −1.29237 * | −1.60660 * | −1.74782 * | −1.24387 * | −1.64771 * | −1.76641 * | |
| S. proliferum (5) | −0.87003 * | −0.80076 * | −0.88873 * | −0.93071 * | −0.88925 * | −0.97847 * | |
| L. intestinalis (14) | −2.16686 **** | −2.49157 *** | −2.76247 **** | −2.15851 **** | −2.48925 *** | −2.75779 **** | |
| Saci2-6 | All cesdtodes examined (28) | −0.66645 * | −1.24721 * | −1.24641 * | −0.73956 * | −1.46510 * | −1.44722 * |
| S. erinaceieuropaei (16) c | −1.40545 * | −1.61597 * | −1.80025 * | −1.52392 * | −1.79126 * | −1.98406 * | |
| S. erinaceieuropaei_b (13) | −1.60743 ** | −1,80309 * | −2.00568 * | −1.69063 ** | −1.93048 ** | −2.13776 ** | |
| L. intestinalis (7) | −1.27810 * | −1.34293 * | −1.47382 * | −1.40954 * | −1.51122 * | −1.65026 * | |
| Saci2-7 | All cesdtodes examined (26) | −1.11942 * | −1.69876 * | −1.78098 * | −1.15422 * | −1.73427 * | −1.82259 * |
| S. erinaceieuropaei (5) | −0.93593 * | −0.93593 * | −1.01835 * | −1.00076 * | −1.00076 * | −1.08369 * | |
| S. solidus (9) | −1.42583 * | −1.23910 * | −1.44474 * | −1.41018 * | −1.23146 * | −1.43390 * | |
| L. intestinalis (11) | −1.92872 *** | −2.21988 *** | −2.43739 *** | −1.95195 *** | −2.23629 *** | −2.45789 *** | |
| Saci2-8 | All cesdtodes examined (12) | −1.41048 * | −1.45125 * | −1.64427 * | −1.39480 * | −1.35879 * | −1.56165 * |
| S. erinaceieuropaei (10) | −1.21924 * | −1.14093 * | −1.31248 * | −1.23207 * | −1.12958 * | −1.30622 * | |
| Element | Paralogous Group a | Int Region | RT Region | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Se_Saci2 b | Sp_Saci2 | Ss_Saci2 | Li_Saci2 | Se_Saci2 | Sp_Saci2 | Ss_Saci2 | Li_Saci2 | ||
| Saci2-1 0.396 ± 0.032 c.1 0.348 ± 0.016 c.2 | Se_Saci2 | 0.073 ± 0.003 | 0.121 ± 0.006 | 0.390 ± 0.032 | 0.572 ± 0.059 | 0.069 ± 0.004 | 0.111 ± 0.005 | 0.357 ± 0.018 | 0.489 ± 0.030 |
| Sp_Saci2 | 0.140 ± 0.006 | 0.390 ± 0.032 | 0.593 ± 0.062 | 0.130 ± 0.008 | 0.363 ± 0.018 | 0.509 ± 0.029 | |||
| Ss_Saci2 | 0.585 ± 0.045 | 0.728 ± 0.081 | 0.547 ± 0.033 | 0.643 ± 0.033 | |||||
| Li_Saci2 | 0.180 ± 0.008 | 0.174 ± 0.007 | |||||||
| Saci2-2 0.379 ± 0.020 0.338 ± 0.017 | Se_Saci2 | 0.090 ± 0.004 | 0.182 ± 0.009 | 0.565 ± 0.030 | 0.688 ± 0.039 | 0.084 ± 0.005 | 0.182 ± 0.013 | 0.492 ± 0.030 | 0.629 ± 0.042 |
| Sp_Saci2 | NC d | 0.597 ± 0.033 | 0.729 ± 0.044 | NC | 0.543 ± 0.037 | 0.700 ± 0.049 | |||
| Ss_Saci2 | 0.186 ± 0.007 | 0.741 ± 0.043 | 0.172 ± 0.008 | 0.653 ± 0.041 | |||||
| Li_Saci2 | 0.544 ± 0.034 | 0.476 ± 0.039 | |||||||
| Saci2-3 0.173 ± 0.006 0.164 ± 0.008 | Se_Saci2 | 0.122 ± 0.004 | 0.113 ± 0.004 | 0.651 ± 0.029 | 0.116 ± 0.006 | 0.108 ± 0.006 | 0.618 ± 0.043 | ||
| Sp_Saci2 | 0.114 ± 0.008 | 0.633 ± 0.030 | 0.109 ± 0.011 | 0.603 ± 0.044 | |||||
| Ss_Saci2 | NC | NC | |||||||
| Saci2-4 0.202 ± 0.006 0.192 ± 0.010 | Se_Saci2 | 0.085 ± 0.003 | 0.076 ± 0.004 | 0.552 ± 0.022 | 0.086 ± 0.005 | 0.070 ± 0.006 | 0.510 ± 0.031 | ||
| Sp_Saci2 | NC | 0.523 ± 0.022 | NC | 0.457 ± 0.030 | |||||
| Ss_Saci2 | 0.446 ± 0.025 | 0.429 ± 0.038 | |||||||
| Saci2-5 0.415 ± 0.017 0.383 ± 0.024 | Se_Saci2 | 0.093 ± 0.004 | 0.117 ± 0.004 | 0.689 ± 0.030 | 0.087 ± 0.006 | 0.108 ± 0.006 | 0.631 ± 0.046 | ||
| Sp_Saci2 | 0.129 ± 0.005 | 0.722 ± 0.031 | 0.118 ± 0.007 | 0.677 ± 0.049 | |||||
| Li_Saci2 | 0.172 ± 0.007 | 0.161 ± 0.010 | |||||||
| Saci2-6 0.425 ± 0.015 0.337 ± 0.019 | Se_Saci2 | 0.090 ± 0.003 | 0.133 ± 0.004 | 0.721 ± 0.031 | 0.840 ± 0.037 | 0.080 ± 0.005 | 0.120 ± 0.007 | 0.621 ± 0.048 | 0.643 ± 0.046 |
| Sp_Saci2 | 0.148 ± 0.006 | 0.735 ± 0.031 | 0.879 ± 0.038 | 0.136 ± 0.010 | 0.635 ± 0.048 | 0.674 ± 0.047 | |||
| Ss_Saci2 | NC | 0.932 ± 0.044 | NC | 0.635 ± 0.048 | |||||
| Li_Saci2 | 0.031 ± 0.002 | 0.033 ± 0.003 | |||||||
| Saci2-7 0.382 ± 0.011 0.370 ± 0.016 | Se_Saci2 | 0.026 ± 0.002 | 0.083 ± 0.006 | 0.474 ± 0.017 | 0.412 ± 0.019 | 0.027 ± 0.003 | 0.101 ± 0.010 | 0.479 ± 0.026 | 0.402 ± 0.028 |
| Sp_Saci2 | NC | 0.507 ± 0.019 | 0.443 ± 0.021 | NC | 0.489 ± 0.027 | 0.418 ± 0.029 | |||
| Ss_Saci2 | 0.361 ± 0.010 | 0.542 ± 0.019 | 0.359± 0.015 | 0.508 ± 0.027 | |||||
| Li_Saci2 | 0.065 ± 0.003 | 0.068 ± 0.004 | |||||||
| Saci2-8 0.137 ± 0.005 0.136 ± 0.007 | Se_Saci2 | 0.123 ± 0.004 | 0.167 ± 0.006 | 0.120 ± 0.007 | 0.170 ± 0.010 | ||||
| Sp_Saci2 | 0.170 ± 0.010 | 0.180 ± 0.015 | |||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bae, Y.-A. Structural and Evolutionary Analysis of Saci2-Like LTR Retrotransposons in Diphyllobothriidean Tapeworms. Int. J. Mol. Sci. 2025, 26, 9061. https://doi.org/10.3390/ijms26189061
Bae Y-A. Structural and Evolutionary Analysis of Saci2-Like LTR Retrotransposons in Diphyllobothriidean Tapeworms. International Journal of Molecular Sciences. 2025; 26(18):9061. https://doi.org/10.3390/ijms26189061
Chicago/Turabian StyleBae, Young-An. 2025. "Structural and Evolutionary Analysis of Saci2-Like LTR Retrotransposons in Diphyllobothriidean Tapeworms" International Journal of Molecular Sciences 26, no. 18: 9061. https://doi.org/10.3390/ijms26189061
APA StyleBae, Y.-A. (2025). Structural and Evolutionary Analysis of Saci2-Like LTR Retrotransposons in Diphyllobothriidean Tapeworms. International Journal of Molecular Sciences, 26(18), 9061. https://doi.org/10.3390/ijms26189061






