Genome-Wide Identification and Expression Analysis of BAG Family in Sweet Potato and Its Two Diploid Relatives
Abstract
1. Introduction
2. Results
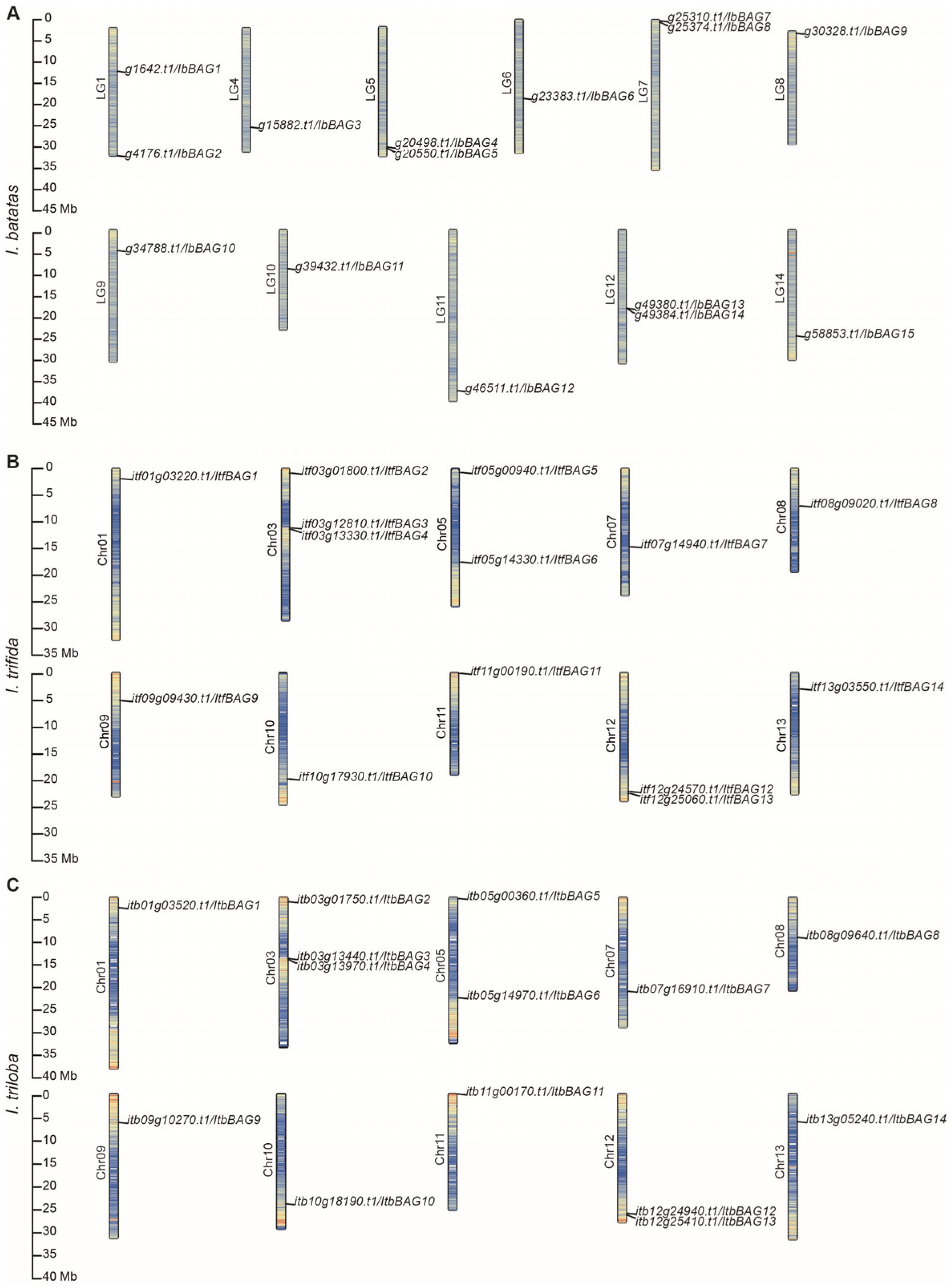
2.1. Identification and Characteristics of BAGs in Sweet Potato and Its Two Diploid Relatives
2.2. Collinearity and Ka/Ks Analysis of BAG Genes
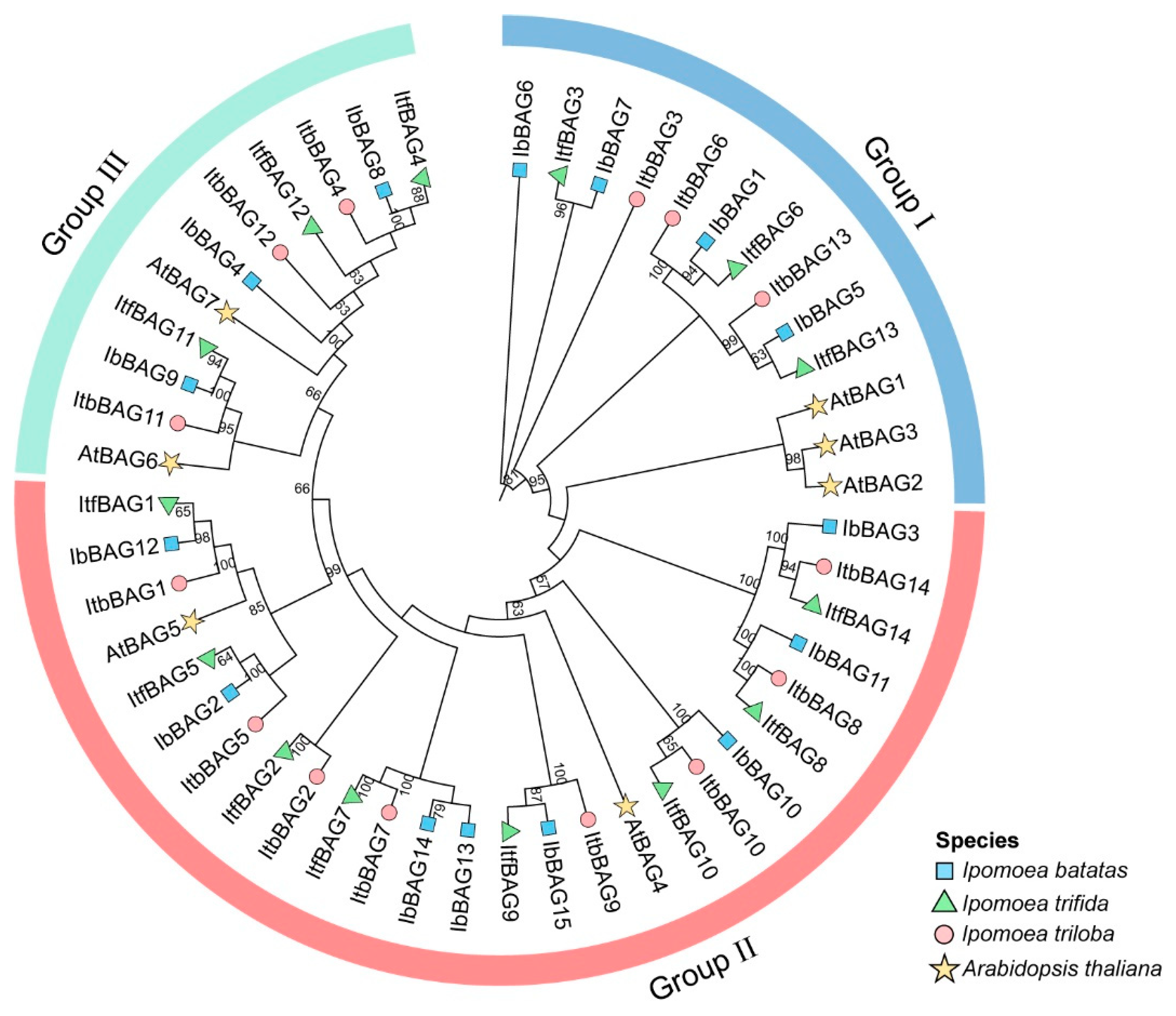
2.3. Phylogenetic Relationship of BAGs in Sweet Potato and Its Two Diploid Relatives
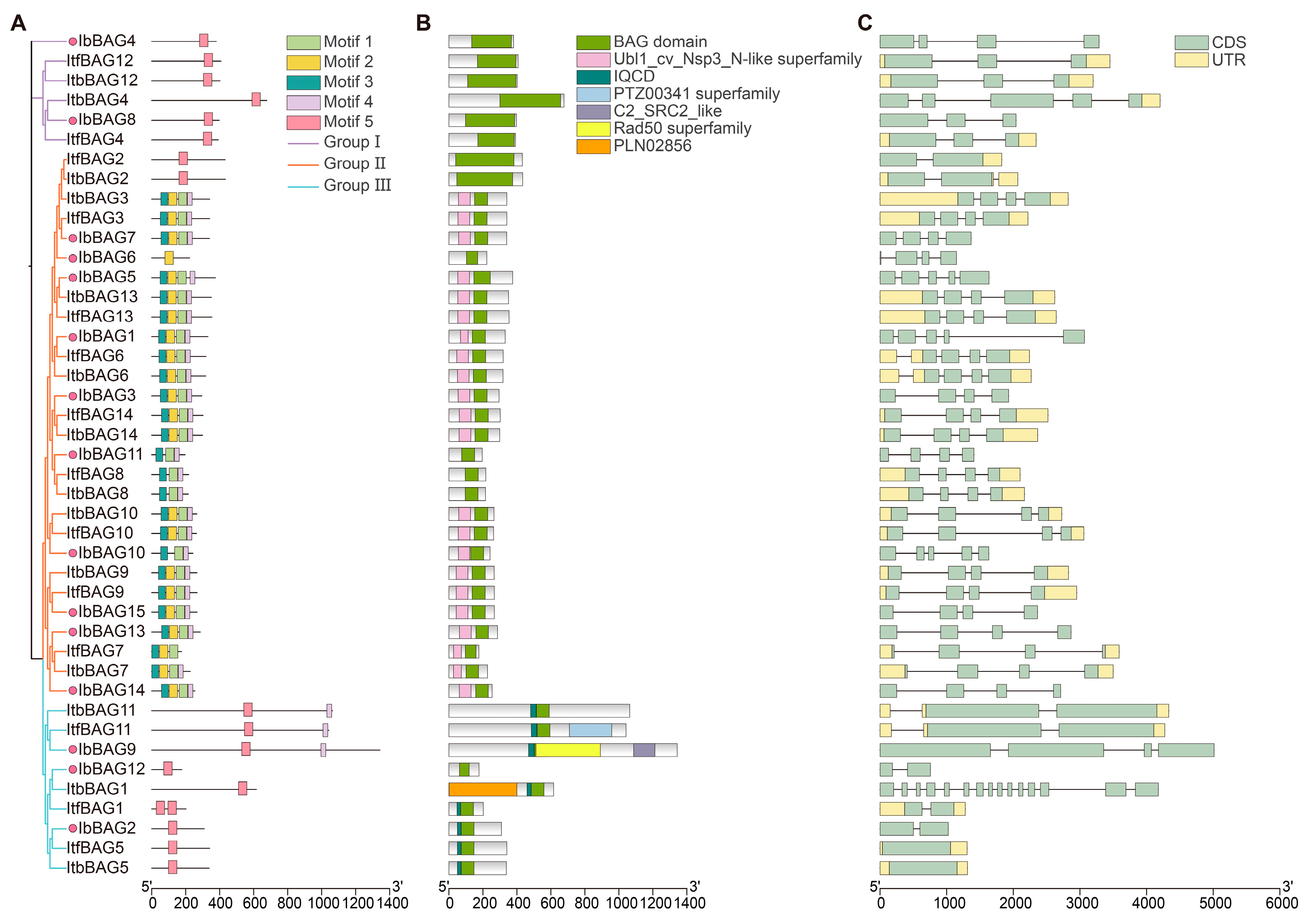
2.4. Conserved Motif and Exon–Intron Structure Analysis of BAGs in Sweet Potato and Its Two Diploid Relatives
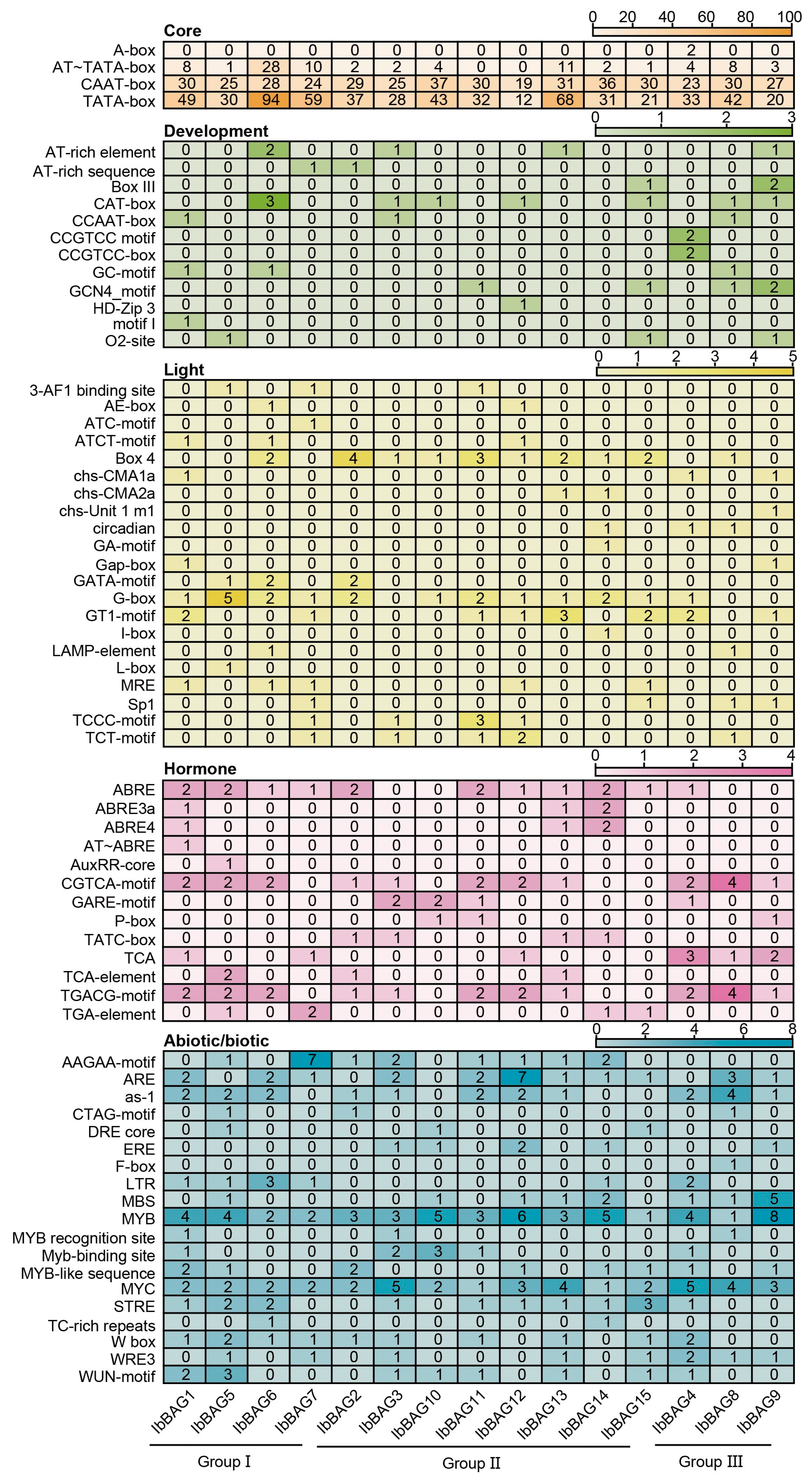
2.5. Cis-Element Analysis in the Promoter of IbBAGs in Sweet Potato
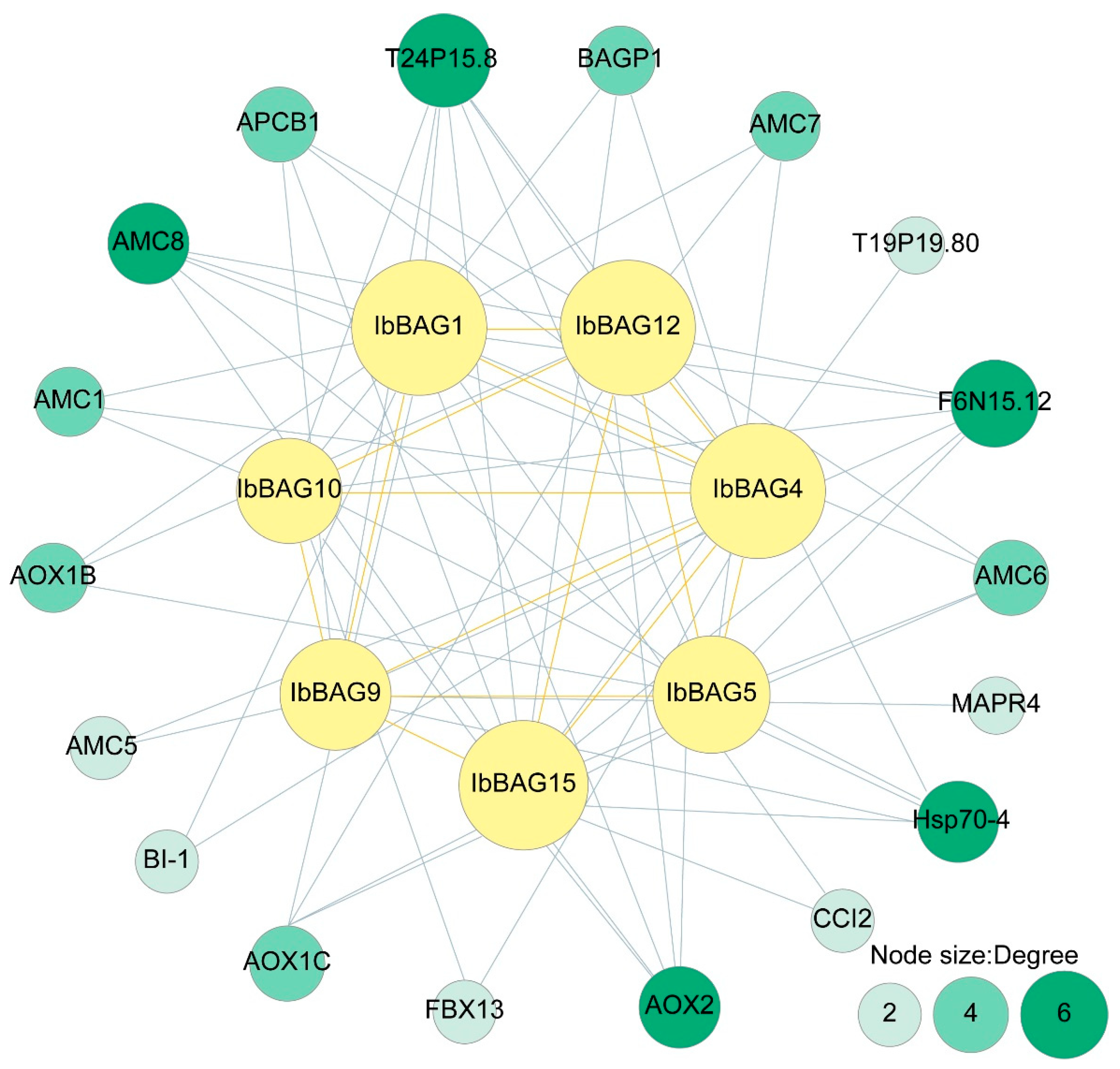
2.6. Protein Interaction Network of IbBAGs in Sweet Potato
2.7. Expression Analysis of BAGs in Sweet Potato and Its Two Diploid Relatives
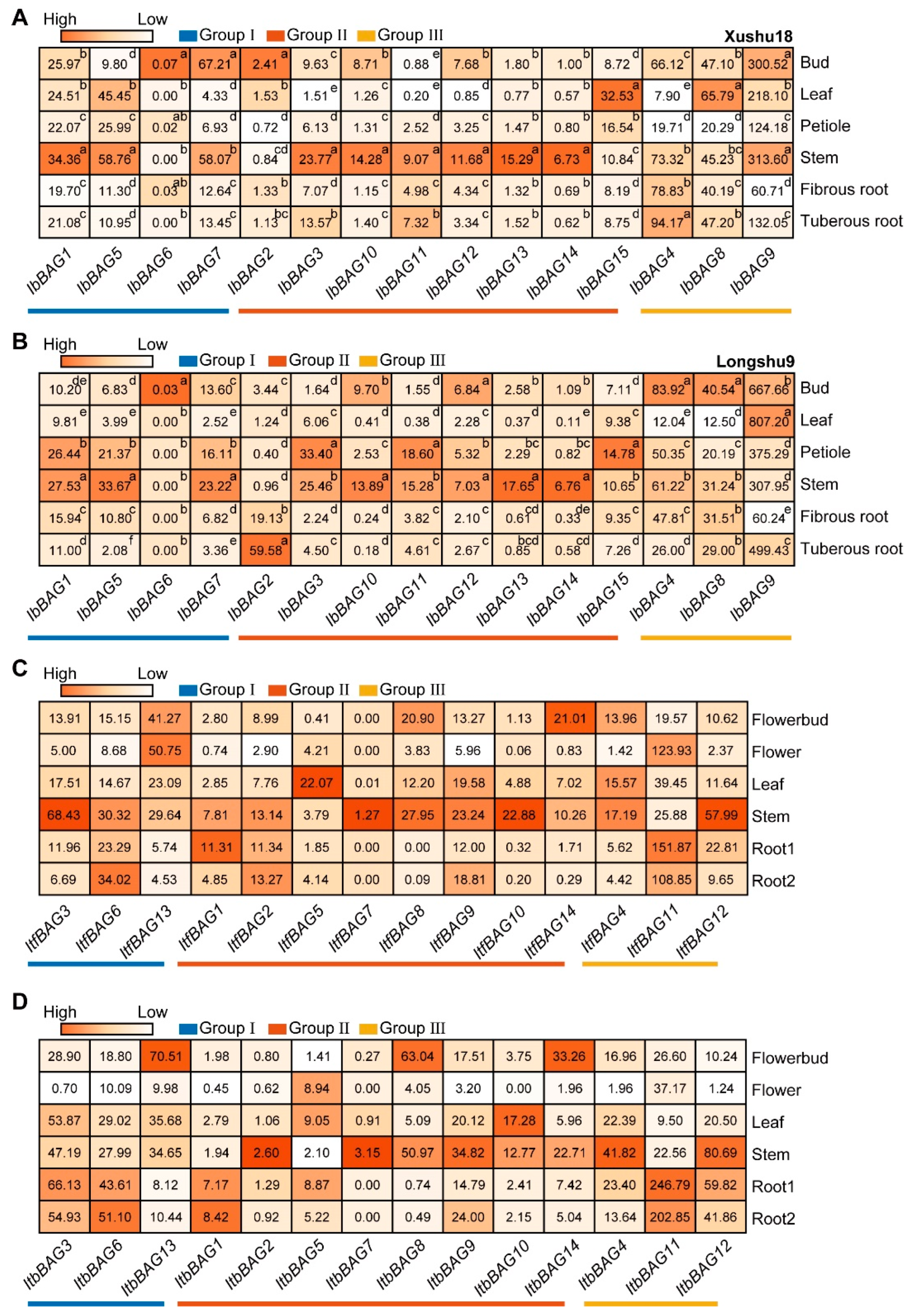
2.7.1. Expression Analysis in Various Tissues
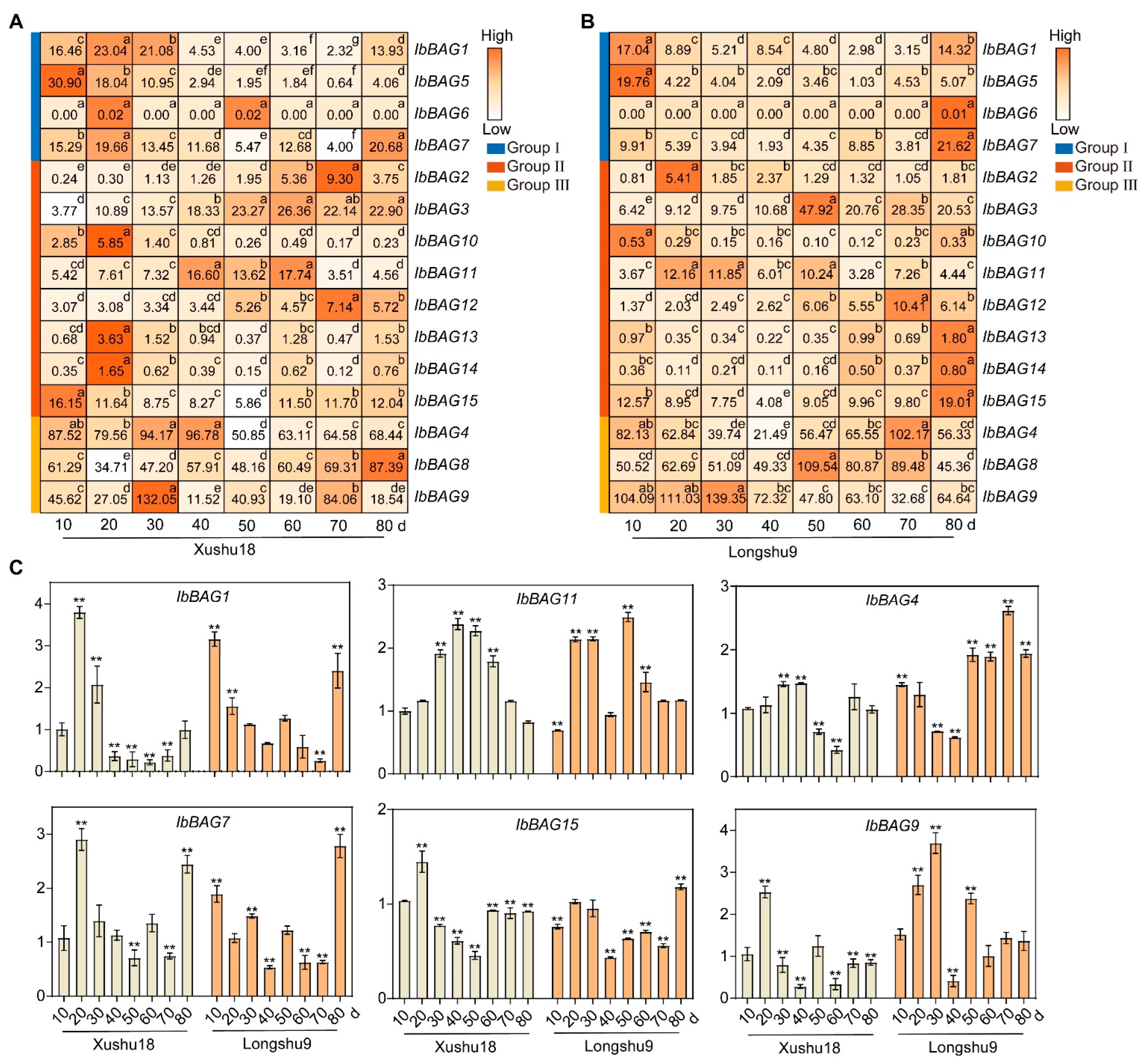
2.7.2. Expression Analysis in Different Development Stages
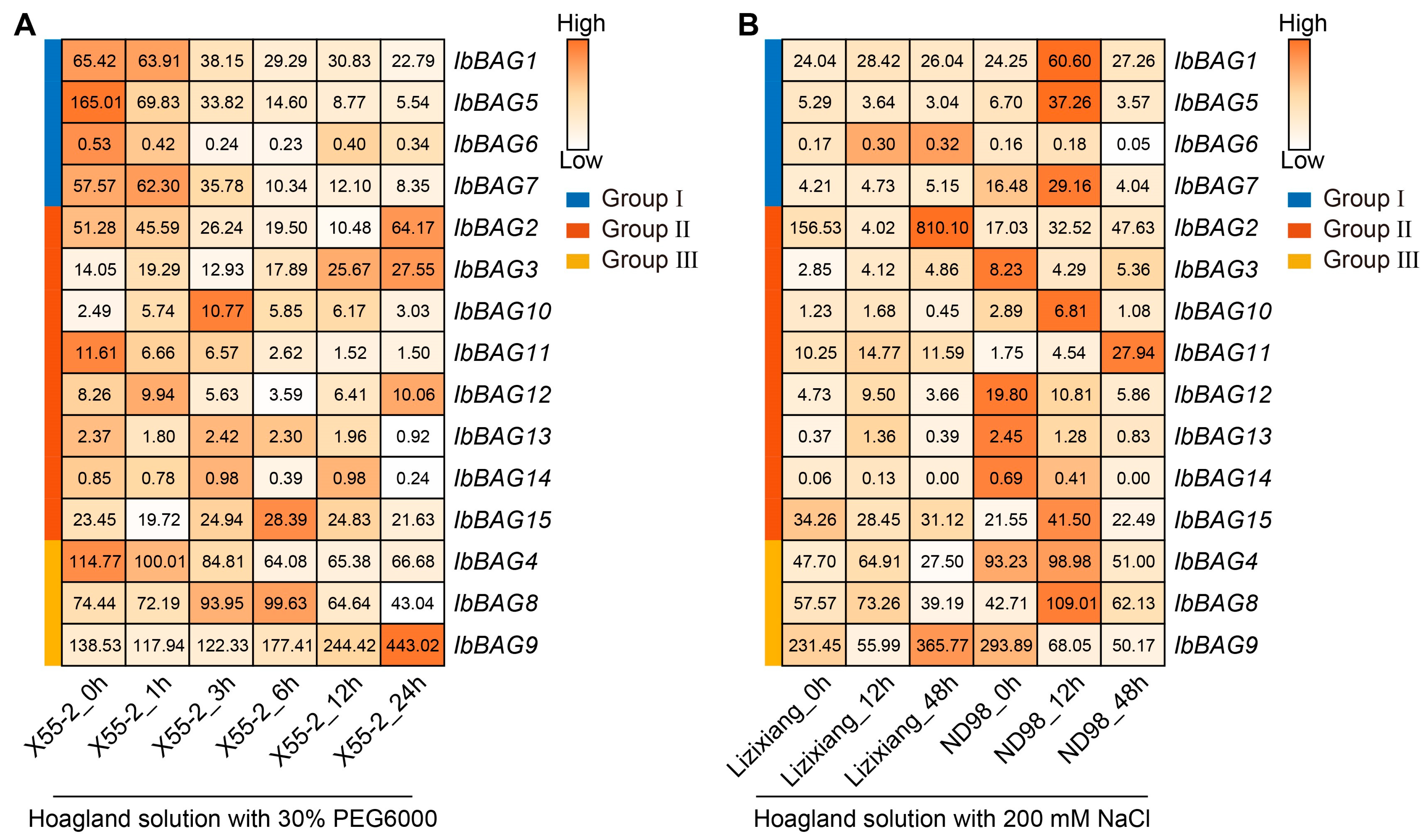
2.7.3. Expression Analysis Under Abiotic Stress
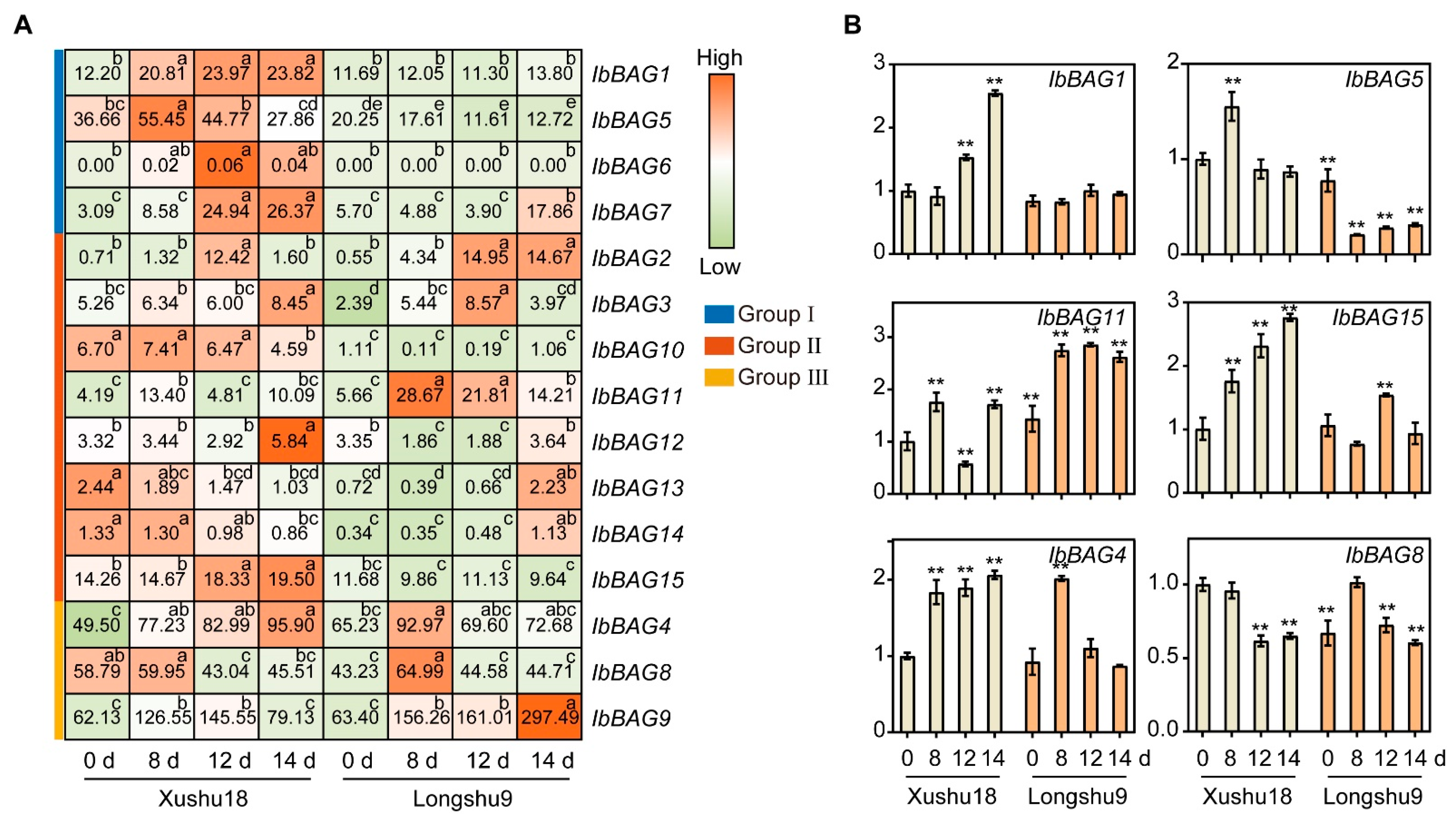
2.7.4. Expression Analysis in Different Varieties in a Root Rot Field
3. Discussion
3.1. Evolution of the BAGs Gene Family in Sweet Potato and Its Two Diploid Relatives
3.2. Different Expression Pattern of BAGs on Root Development Stage in Two Sweet Potato Varieties
3.3. Different Functions of BAGs on Abiotic Stress Response Between Two Sweet Potato Varieties and Its Two Diploid Relatives
3.4. Different Functions of BAGs on Biotic Stress Response Between Two Sweet Potato Varieties and Its Two Diploid Relatives
4. Materials and Methods
4.1. Identification of BAGs
4.2. Chromosomal Distribution of BAGs
4.3. Collinearity and Ka/Ks Ratios of BAG Genes
4.4. Phylogenetic Analysis of BAGs
4.5. Domain Identification and Conserved Motifs Analysis of BAGs
4.6. Exon–Intron Structures and Promoter Analysis of BAGs
4.7. Protein Interaction Network of BAGs
4.8. Transcriptome Analysis
4.9. qRT-PCR Analysis of BAGs
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Antoku, K.; Maser, R.S.; Scully, W.J.; Delach, S.M.; Johnson, D.E. Isolation of Bcl-2 Binding Proteins That Exhibit Homology with BAG-1 and Suppressor of Death Domains Protein. Biochem. Biophys. Res. Commun. 2001, 286, 1003–1010. [Google Scholar] [CrossRef]
- Takayama, S.; Reed, J.C. Molecular Chaperone Targeting and Regulation by BAG Family Proteins. Nat. Cell Biol. 2001, 3, E237–E241. [Google Scholar] [CrossRef] [PubMed]
- Doong, H.; Vrailas, A.; Kohn, E.C. What’s in the ‘BAG’?—A Functional Domain Analysis of the BAG-Family Proteins. Cancer Lett. 2002, 188, 25–32. [Google Scholar] [CrossRef]
- Li, Y.; Dickman, M. Processing of AtBAG6 Triggers Autophagy and Fungal Resistance. Plant Signal. Behav. 2016, 11, e1175699. [Google Scholar] [CrossRef] [PubMed]
- Doukhanina, E.V.; Chen, S.; van der Zalm, E.; Godzik, A.; Reed, J.; Dickman, M.B. Identification and Functional Characterization of the BAG Protein Family in Arabidopsis thaliana. J. Biol. Chem. 2006, 281, 18793–18801. [Google Scholar] [CrossRef]
- Kang, C.H.; Jung, W.Y.; Kang, Y.H.; Kim, J.Y.; Kim, D.G.; Jeong, J.C.; Baek, D.W.; Jin, J.B.; Lee, J.Y.; Kim, M.O.; et al. AtBAG6, a Novel Calmodulin-Binding Protein, Induces Programmed Cell Death in Yeast and Plants. Cell Death Differ. 2006, 13, 84–95. [Google Scholar] [CrossRef]
- Behl, C. Breaking BAG: The Co-Chaperone BAG3 in Health and Disease. Trends Pharmacol. Sci. 2016, 37, 672–688. [Google Scholar] [CrossRef]
- Froesch, B.A.; Takayama, S.; Reed, J.C. BAG-1L Protein Enhances Androgen Receptor Function. J. Biol. Chem. 1998, 273, 11660–11666. [Google Scholar] [CrossRef]
- Kabbage, M.; Dickman, M.B. The BAG Proteins: A Ubiquitous Family of Chaperone Regulators. Cell. Mol. Life Sci. 2008, 65, 1390–1402. [Google Scholar] [CrossRef]
- Nawkar, G.M.; Maibam, P.; Park, J.H.; Woo, S.G.; Kim, C.Y.; Lee, S.Y.; Kang, C.H. In Silico Study on Arabidopsis BAG Gene Expression in Response to Environmental Stresses. Protoplasma 2017, 254, 409–421. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Li, J.; Liu, X.; Wei, X.; He, Z.; Hu, L.; Wang, J.; Duan, M.; Xie, G.; Wang, J.; et al. The Divergent Roles of the Rice Bcl-2 Associated Athanogene (BAG) Genes in Plant Development and Environmental Responses. Plants 2021, 10, 2169. [Google Scholar] [CrossRef]
- Farid, B.; Saddique, M.A.B.; Tahir, M.H.N.; Ikram, R.M.; Ali, Z.; Akbar, W. Expression Divergence of BAG Gene Family in Maize under Heat Stress. BMC Plant Biol. 2025, 25, 16. [Google Scholar] [CrossRef]
- Fang, S.; Li, L.; Cui, B.; Men, S.; Shen, Y.; Yang, X. Structural Insight into Plant Programmed Cell Death Mediated by BAG Proteins in Arabidopsis thaliana. Acta Crystallogr. Sect. D Biol. Crystallogr. 2013, 69, 934–945. [Google Scholar] [CrossRef]
- Lee, D.W.; Kim, S.J.; Oh, Y.J.; Choi, B.; Lee, J.; Hwang, I. Arabidopsis BAG1 Functions as a Cofactor in Hsc70-Mediated Proteasomal Degradation of Unimported Plastid Proteins. Mol. Plant 2016, 9, 1428–1431. [Google Scholar] [CrossRef]
- Cui, B.; Fang, S.; Xing, Y.; Shen, Y.; Yang, X. Crystallographic analysis of the Arabidopsis thaliana BAG5-calmodulin protein complex. Acta Crystallogr. Sect. F Struct. Biol. Commun. 2015, 71, 870–875. [Google Scholar] [CrossRef]
- Arif, M.; Men, S.; Nawaz, A.F.; Li, X.; Xu, L.; Yang, X.; Fahad, S.; Ahmad, P.; Xu, R.; Li, L. Bcl-2-Associated Athanogene (BAG) Co-Chaperones: Key Players in Multiple Abiotic and Biotic Stress Tolerance in Plants. J. Plant Growth Regul. 2024. [Google Scholar] [CrossRef]
- Zhou, Y.; Yang, K.; Cheng, M.; Cheng, Y.; Li, Y.; Ai, G.; Bai, T.; Xu, R.; Duan, W.; Peng, H.; et al. Double-Faced Role of Bcl-2-Associated Athanogene 7 in Plant–Phytophthora Interaction. J. Exp. Bot. 2021, 72, 5751–5765. [Google Scholar] [CrossRef]
- You, Q.; Zhai, K.; Yang, D.; Yang, W.; Wu, J.; Liu, J.; Pan, W.; Wang, J.; Zhu, X.; Jian, Y.; et al. An E3 Ubiquitin Ligase-BAG Protein Module Controls Plant Innate Immunity and Broad-Spectrum Disease Resistance. Cell Host Microbe 2016, 20, 758–769. [Google Scholar] [CrossRef]
- Hoang, T.M.L.; Moghaddam, L.; Williams, B.; Khanna, H.; Dale, J.; Mundree, S.G. Development of Salinity Tolerance in Rice by Constitutive-Overexpression of Genes Involved in the Regulation of Programmed Cell Death. Front. Plant Sci. 2015, 6, 175. [Google Scholar] [CrossRef]
- Ge, S.; Kang, Z.; Li, Y.; Zhang, F.; Shen, Y.; Ge, R.; Huang, Z. Cloning and Function Analysis of BAG Family Genes in Wheat. Funct. Plant Biol. 2016, 43, 393. [Google Scholar] [CrossRef]
- Liu, Q.C. Improvement for Agronomically Important Traits by Gene Engineering in Sweetpotato. Breed. Sci. 2017, 67, 15–26. [Google Scholar] [CrossRef]
- El Sheikha, A.F.; Ray, R.C. Potential Impacts of Bioprocessing of Sweet Potato: Review. Crit. Rev. Food Sci. Nutr. 2017, 57, 455–471. [Google Scholar] [CrossRef] [PubMed]
- Bartels, D.; Sunkar, R. Drought and Salt Tolerance in Plants. Crit. Rev. Plant Sci. 2005, 24, 23–58. [Google Scholar] [CrossRef]
- Scruggs, A.C.; Quesada-Ocampo, L.M. Etiology and Epidemiological Conditions Promoting Fusarium Root Rot in Sweetpotato. Phytopathology 2016, 106, 909–919. [Google Scholar] [CrossRef]
- Kim, S.; Kim, T.H.; Chung, M.-N.; Lee, Y.; Lee, I.B.; Lee, H.; Park, W. Incidence Rates of Root Rot in Sweetpotato Caused by Cultivation Soil and Soil Microorganisms During Storage Periods. Front. Plant Sci. 2022, 13, 897590. [Google Scholar] [CrossRef]
- Wu, S.; Lau, K.H.; Cao, Q.H.; Hamilton, J.P.; Sun, H.H.; Zhou, C.X.; Eserman, L.; Gemenet, D.C.; Olukolu, B.A.; Wang, H.Y.; et al. Genome Sequences of Two Diploid Wild Relatives of Cultivated Sweetpotato Reveal Targets for Genetic Improvement. Nat. Commun. 2018, 9, 4580. [Google Scholar] [CrossRef]
- Yang, J.; Moeinzadeh, M.H.; Kuhl, H.; Helmuth, J.; Xiao, P.; Haas, S.; Liu, G.L.; Zheng, J.L.; Sun, Z.; Fan, W.J.; et al. Haplotype-resolved Sweet Potato Genome Traces Back Its Hexaploidization History. Nat. Plants 2017, 3, 696–703. [Google Scholar] [CrossRef]
- Wu, S.; Sun, H.; Zhao, X.; Hamilton, J.P.; Mollinari, M.; Gesteira, G.D.S.; Kitavi, M.; Yan, M.; Wang, H.; Yang, J.; et al. Phased Chromosome-Level Assembly Provides Insight into the Genome Architecture of Hexaploid Sweetpotato. Nat. Plants, 2025; epub ahead of print. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Zhang, Y.; Feng, S. Whole-Genome and Dispersed Duplication, Including Transposed Duplication, Jointly Advance the Evolution of TLP Genes in Seven Representative Poaceae Lineages. BMC Genom. 2023, 24, 290. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. TBtools-II: A “One for All, All for One” Bioinformatics Platform for Biological Big-Data Mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef]
- Brive, L.; Takayama, S.; Briknarová, K.; Homma, S.; Ishida, S.K.; Reed, J.C.; Ely, K.R. The Carboxyl-Terminal Lobe of Hsc70 ATPase Domain Is Sufficient for Binding to BAG1. Biochem. Biophys. Res. Commun. 2001, 289, 1099–1105. [Google Scholar] [CrossRef]
- Yan, J.; He, C.; Zhang, H. The BAG-Family Proteins in Arabidopsis thaliana. Plant Sci. 2003, 165, 1–7. [Google Scholar] [CrossRef]
- Seo, P.J.; Xiang, F.; Qiao, M.; Park, J.-Y.; Lee, Y.N.; Kim, S.-G.; Lee, Y.-H.; Park, W.J.; Park, C.-M. The MYB96 Transcription Factor Mediates Abscisic Acid Signaling during Drought Stress Response in Arabidopsis. Plant Physiol. 2009, 151, 275–289. [Google Scholar] [CrossRef] [PubMed]
- Yang, A.; Dai, X.; Zhang, W.-H. A R2R3-Type MYB Gene, OsMYB2, Is Involved in Salt, Cold, and Dehydration Tolerance in Rice. J. Exp. Bot. 2012, 63, 2541–2556. [Google Scholar] [CrossRef]
- Zhang, P.; Wang, R.; Yang, X.; Ju, Q.; Li, W.; Lü, S.; Tran, L.P.; Xu, J. The R2R3-MYB Transcription Factor AtMYB49 Modulates Salt Tolerance in Arabidopsis by Modulating the Cuticle Formation and Antioxidant Defence. Plant Cell Environ. 2020, 43, 1925–1943. [Google Scholar] [CrossRef] [PubMed]
- Abe, H.; Urao, T.; Ito, T.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) Function as Transcriptional Activators in Abscisic Acid Signaling. Plant Cell 2003, 15, 63–78. [Google Scholar] [CrossRef]
- Uji, Y.; Taniguchi, S.; Tamaoki, D.; Shishido, H.; Akimitsu, K.; Gomi, K. Overexpression of OsMYC2 Results in the Up-Regulation of Early JA-Rresponsive Genes and Bacterial Blight Resistance in Rice. Plant Cell Physiol. 2016, 57, 1814–1827. [Google Scholar] [CrossRef]
- Eulgem, T.; Somssich, I.E. Networks of WRKY Transcription Factors in Defense Signaling. Curr. Opin. Plant Biol. 2007, 10, 366–371. [Google Scholar] [CrossRef]
- Rushton, P.J.; Somssich, I.E.; Ringler, P.; Shen, Q.J. WRKY Transcription Factors. Trends Plant Sci. 2010, 15, 247–258. [Google Scholar] [CrossRef]
- Li, Y.; Kabbage, M.; Liu, W.; Dickman, M.B. Aspartyl Protease-Mediated Cleavage of BAG6 Is Necessary for Autophagy and Fungal Resistance in Plants. Plant Cell 2016, 28, 233–247. [Google Scholar] [CrossRef]
- Ishikawa, T.; Watanabe, N.; Nagano, M.; Kawai-Yamada, M.; Lam, E. Bax Inhibitor-1: A Highly Conserved Endoplasmic Reticulum-Resident Cell Death Suppressor. Cell Death Differ. 2011, 18, 1271–1278. [Google Scholar] [CrossRef] [PubMed]
- Albury, M.S.; Elliott, C.; Moore, A.L. Ubiquinol-Binding Site in the Alternative Oxidase: Mutagenesis Reveals Features Important for Substrate Binding and Inhibition. Biochim. Biophys. Acta (BBA)—Bioenerg. 2010, 1797, 1933–1939. [Google Scholar] [CrossRef]
- Zhang, H.; Dai, Z.; Zhang, X.; Shang, M.; Gao, X.; Ma, R.; Zhao, L.; Zhang, X.; Liu, Q.; Zhai, H.; et al. Natural Allelic Variations in IbCHYR1–IbZnFR Complex Regulate Fusarium Root Rot Resistance in Sweet Potato. Adv. Sci. 2025, 12, e15202. [Google Scholar] [CrossRef]
- Jiang, H.; Liu, X.; Xiao, P.; Wang, Y.; Xie, Q.; Wu, X.; Ding, H. Functional Insights of Plant Bcl-2-Associated Ahanogene (BAG) Proteins: Multi-Taskers in Diverse Cellular Signal Transduction Pathways. Front. Plant Sci. 2023, 14, 1136873. [Google Scholar] [CrossRef] [PubMed]
- Wan, R.; Liu, J.; Yang, Z.; Zhu, P.; Cao, Q.; Xu, T. Genome-Wide Identification, Characterisation and Expression Profile Analysis of DEAD-Box Family Genes in Sweet Potato Wild Ancestor Ipomoea trifida under Abiotic Stresses. Genes Genom. 2020, 42, 325–335. [Google Scholar] [CrossRef]
- Holland, S.; Blake, C. Proteins, Exons and Molecular Evolution. Biosystems 1987, 20, 181–206. [Google Scholar] [CrossRef]
- Morello, L.; Gianì, S.; Troina, F.; Breviario, D. Testing the IMEter on Rice Introns and Other Aspects of Intron-Mediated Enhancement of Gene Expression. J. Exp. Bot. 2011, 62, 533–544. [Google Scholar] [CrossRef]
- Mukherjee, D.; Saha, D.; Acharya, D.; Mukherjee, A.; Chakraborty, S.; Ghosh, T.C. The Role of Introns in the Conservation of the Metabolic Genes of Arabidopsis thaliana. Genomics 2018, 110, 310–317. [Google Scholar] [CrossRef]
- Cheniclet, C.; Rong, W.Y.; Causse, M.; Frangne, N.; Bolling, L.; Carde, J.-P.; Renaudin, J.-P. Cell Expansion and Endoreduplication Show a Large Genetic Variability in Pericarp and Contribute Strongly to Tomato Fruit Growth. Plant Physiol. 2005, 139, 1984–1994. [Google Scholar] [CrossRef]
- Butterfass, T. Patterns of chloroplast reproduction. A developmental approach to protoplasmic plant anatomy. In Cell Biology Monographs. Continuation of Protoplasmatologie; Heilbrunn, L.V., Beermann, W., Rudkin, G., Eds.; Springer: Wien, Austria; New York, NY, USA, 1979; Volume 6, pp. 1–205. [Google Scholar]
- Wang, J.; Nan, N.; Li, N.; Liu, Y.; Wang, T.-J.; Hwang, I.; Liu, B.; Xu, Z.-Y. A DNA Methylation Reader–Chaperone Regulator–Transcription Factor Complex Activates OsHKT1;5 Expression during Salinity Stress. Plant Cell 2020, 32, 3535–3558. [Google Scholar] [CrossRef] [PubMed]
- Kohl, M.; Wiese, S.; Warscheid, B. Cytoscape: Software for Visualization and Analysis of Biological Networks. Methods Mol. Biol. 2011, 696, 291–303. [Google Scholar] [CrossRef]
- Zhang, H.; Zhang, Q.; Zhai, H.; Li, Y.; Wang, X.; Liu, Q.; He, S. Transcript Profile Analysis Reveals Important Roles of Jasmonic Acid Signalling Pathway in the Response of Sweet Potato to Salt Stress. Sci. Rep. 2017, 7, 40819. [Google Scholar] [CrossRef]
- Zhu, H.; Zhou, Y.; Zhai, H.; He, S.; Zhao, N.; Liu, Q. Transcriptome Profiling Reveals Insights into the Molecular Mechanism of Drought Tolerance in Sweetpotato. J. Integr. Agric. 2019, 18, 9–23. [Google Scholar] [CrossRef]

| Gene ID | Gene Name | pI | Molecular Weight/kDa | Genomic Length/bp | CDS Length/bp | Phosphorylation Site | Protein Size/aa | Aliphatic Index | GRAVY | Subcellular Locations | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ser | Thr | Tyr | ||||||||||
| g1642 | IbBAG1 | 9.8 | 36.31 | 3757 | 993 | 13 | 8 | 7 | 330 | 85.33 | −0.353 | Plastid |
| g4176 | IbBAG2 | 9.3 | 35.47 | 1377 | 927 | 11 | 9 | 4 | 308 | 76.2 | −0.783 | Cytoplasmic |
| g15882 | IbBAG3 | 9.73 | 32.62 | 2263 | 885 | 9 | 7 | 7 | 294 | 83.88 | −0.438 | Cytoplasmic |
| g20498 | IbBAG4 | 9.03 | 43.21 | 3555 | 1140 | 8 | 3 | 6 | 379 | 85.99 | −0.539 | Plastid |
| g20550 | IbBAG5 | 9.42 | 41.26 | 2435 | 1125 | 16 | 9 | 3 | 374 | 77.38 | −0.651 | Nucleus |
| g23383 | IbBAG6 | 9.3 | 24.91 | 1395 | 669 | 4 | 3 | 2 | 222 | 88.6 | −0.314 | Plastid |
| g25310 | IbBAG7 | 9.43 | 37.42 | 2994 | 1020 | 10 | 10 | 6 | 339 | 73.3 | −0.634 | Nucleus |
| g25374 | IbBAG8 | 9.37 | 45.82 | 2401 | 1191 | 7 | 8 | 8 | 396 | 77.1 | −0.686 | Plastid |
| g30328 | IbBAG9 | 5.55 | 149.72 | 5977 | 4029 | 49 | 37 | 22 | 1342 | 63.92 | −0.814 | Nucleus |
| g34788 | IbBAG10 | 8.86 | 26.58 | 2143 | 726 | 6 | 5 | 1 | 241 | 78.13 | −0.502 | Plastid |
| g39432 | IbBAG11 | 9.49 | 21.96 | 1656 | 588 | 2 | 4 | 2 | 195 | 78 | −0.579 | Plastid |
| g46511 | IbBAG12 | 5.47 | 20.16 | 1182 | 531 | 9 | 3 | 1 | 176 | 75.28 | −0.517 | Plastid |
| g49380 | IbBAG13 | 8.55 | 31.61 | 3107 | 858 | 15 | 7 | 2 | 285 | 81.37 | −0.412 | Nucleus |
| g49384 | IbBAG14 | 9.01 | 28.01 | 3057 | 762 | 14 | 5 | 2 | 253 | 85.89 | −0.399 | Plastid |
| g58853 | IbBAG15 | 4.57 | 29.03 | 3191 | 801 | 9 | 7 | 3 | 266 | 81.95 | −0.489 | Nucleus |
| Seq_1 | Seq_2 | Ka | Ks | Ka/Ks | Effective Length | Average S-Sites | Average N-Sites | Type of Selection |
|---|---|---|---|---|---|---|---|---|
| IbBAG1 | IbBAG7 | 0.25 | 1.54 | 0.16 | 768 | 180.33 | 587.67 | Purify selection |
| IbBAG1 | IbBAG5 | 0.33 | 1.34 | 0.25 | 819 | 189 | 630 | Purify selection |
| IbBAG5 | IbBAG7 | 0.21 | 1.36 | 0.15 | 981 | 222.33 | 758.67 | Purify selection |
| IbBAG4 | IbBAG8 | 0.16 | 0.94 | 0.17 | 1080 | 247.42 | 832.58 | Purify selection |
| IbBAG7 | IbBAG11 | 0.41 | 2.67 | 0.15 | 555 | 120.08 | 434.92 | Purify selection |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, X.; Liu, Q.; Zhai, H.; Zhao, N.; Gao, S.; Zhang, H.; He, S. Genome-Wide Identification and Expression Analysis of BAG Family in Sweet Potato and Its Two Diploid Relatives. Int. J. Mol. Sci. 2025, 26, 9053. https://doi.org/10.3390/ijms26189053
Zhang X, Liu Q, Zhai H, Zhao N, Gao S, Zhang H, He S. Genome-Wide Identification and Expression Analysis of BAG Family in Sweet Potato and Its Two Diploid Relatives. International Journal of Molecular Sciences. 2025; 26(18):9053. https://doi.org/10.3390/ijms26189053
Chicago/Turabian StyleZhang, Xiaochen, Qingchang Liu, Hong Zhai, Ning Zhao, Shaopei Gao, Huan Zhang, and Shaozhen He. 2025. "Genome-Wide Identification and Expression Analysis of BAG Family in Sweet Potato and Its Two Diploid Relatives" International Journal of Molecular Sciences 26, no. 18: 9053. https://doi.org/10.3390/ijms26189053
APA StyleZhang, X., Liu, Q., Zhai, H., Zhao, N., Gao, S., Zhang, H., & He, S. (2025). Genome-Wide Identification and Expression Analysis of BAG Family in Sweet Potato and Its Two Diploid Relatives. International Journal of Molecular Sciences, 26(18), 9053. https://doi.org/10.3390/ijms26189053







