Simulation and Machine Learning Assessment of P-Glycoprotein Pharmacology in the Blood–Brain Barrier: Inhibition and Substrate Transport
Abstract
1. Introduction
2. Results
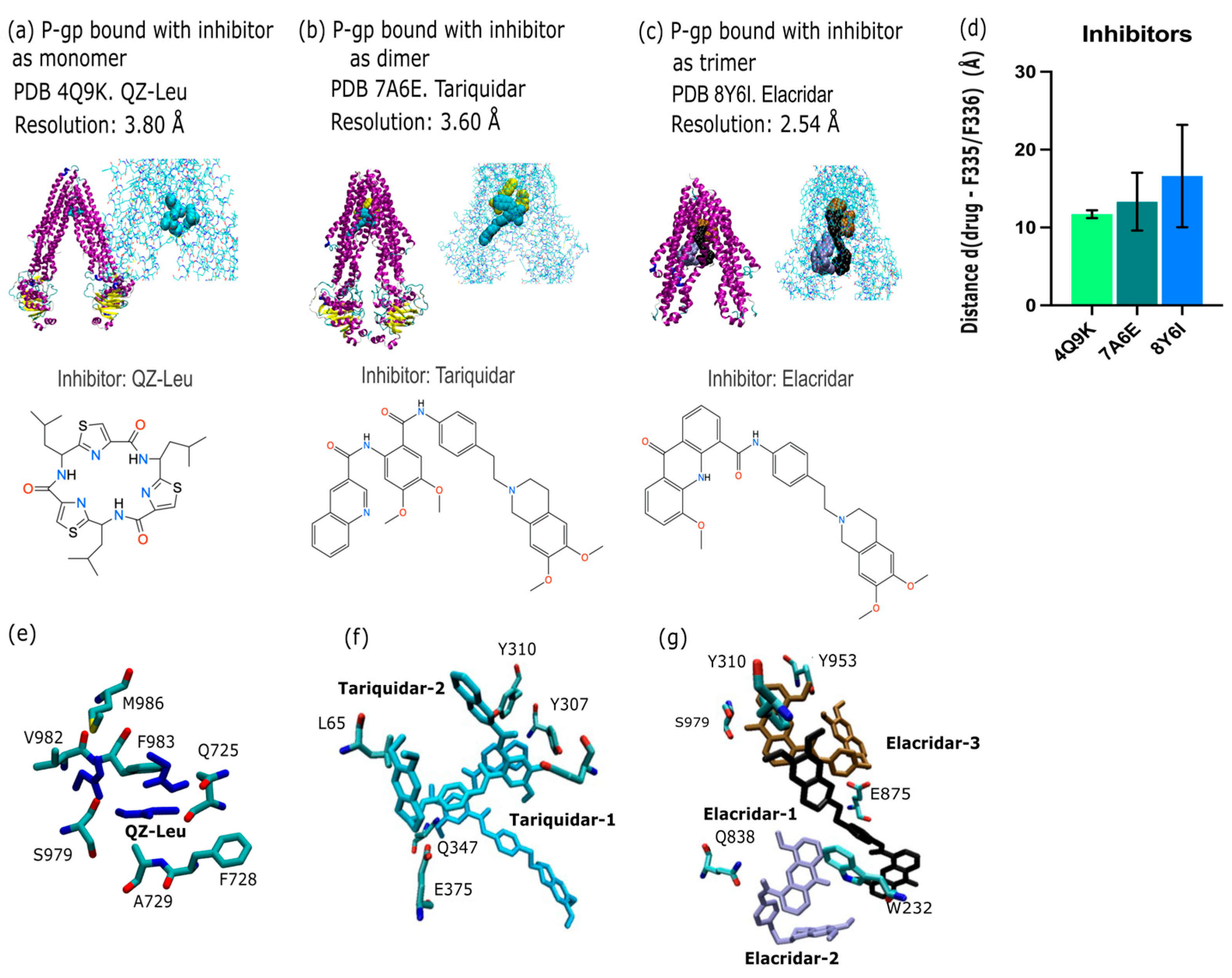
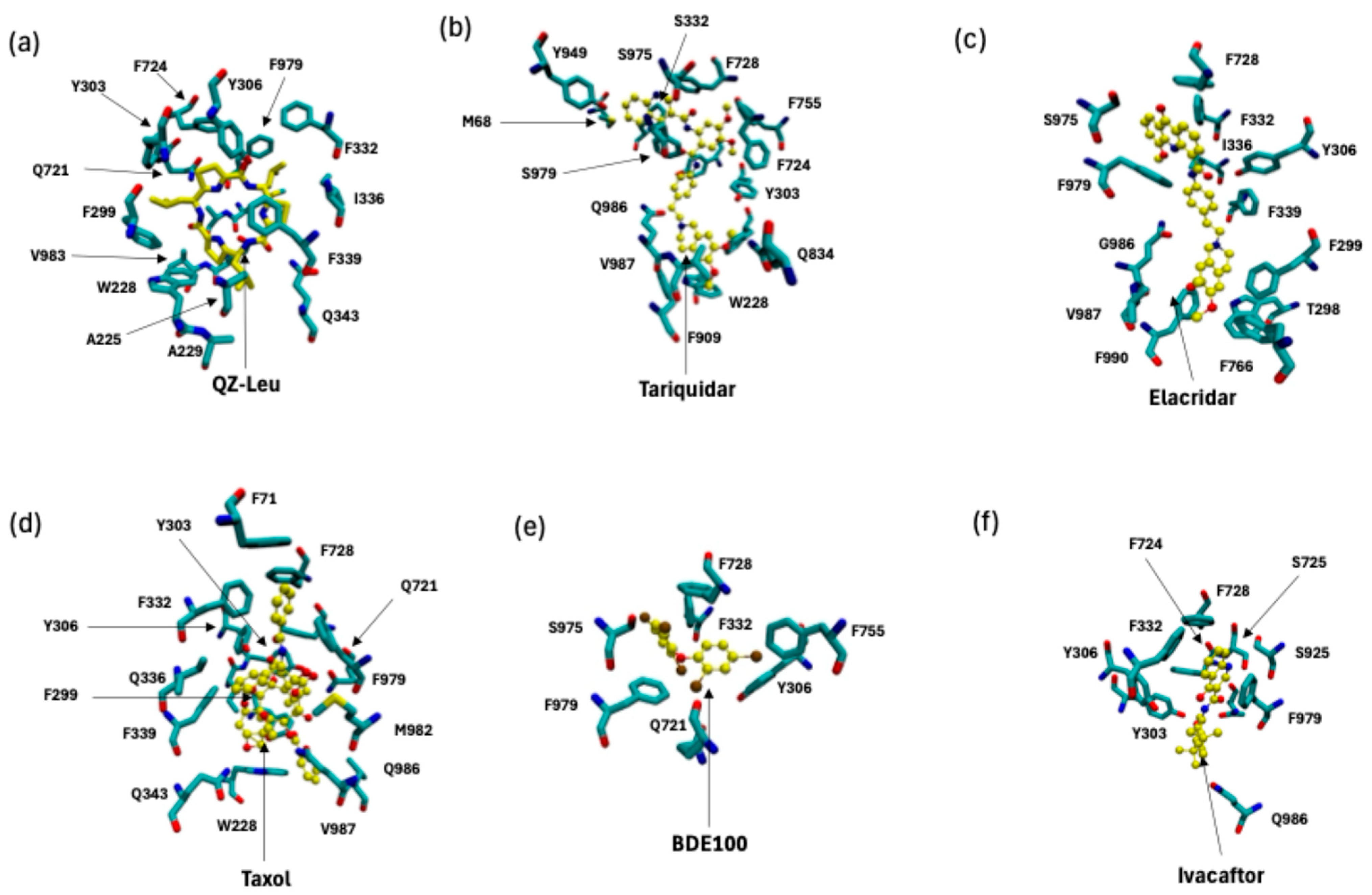
2.1. P-gp Inhibitor Pharmacology: Multimodality of Inhibition
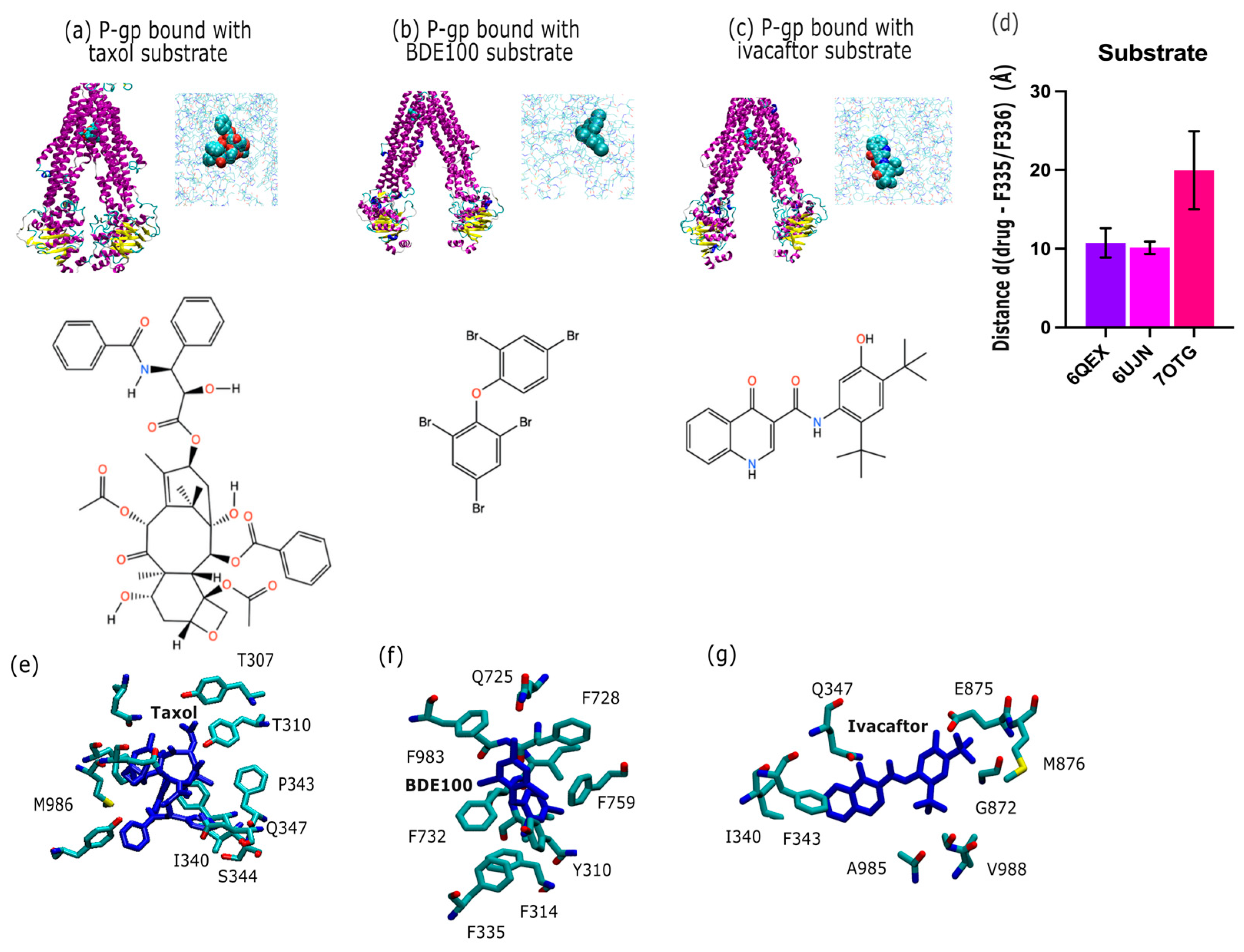
2.2. P-Glycoprotein Substrate Pharmacology: The Handling of Chemotherapeutics and Environmental Pollutants from PDB Database Analysis
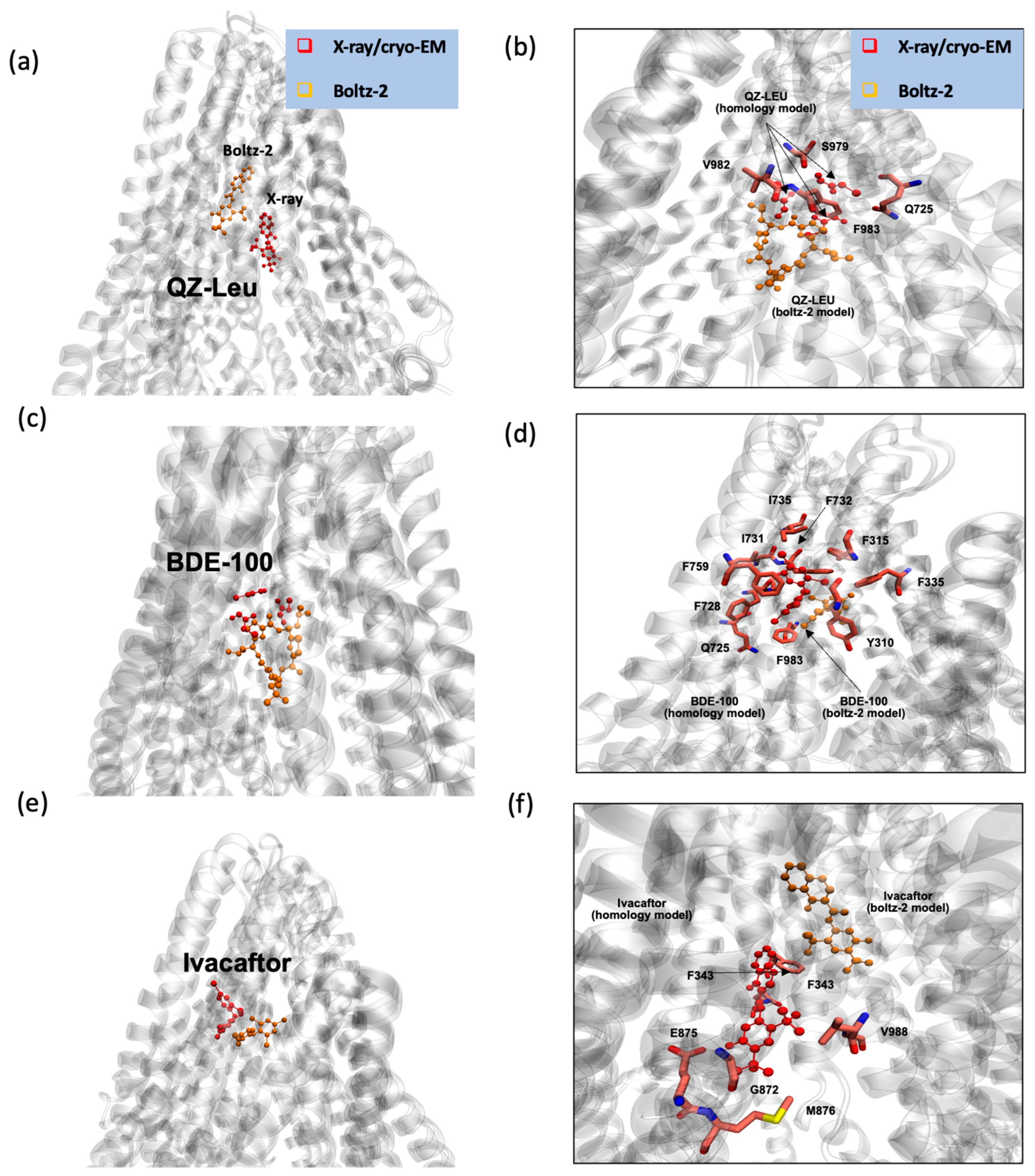
2.3. Assessing IC50 of P-gp Inhibitors and Substrates with AI
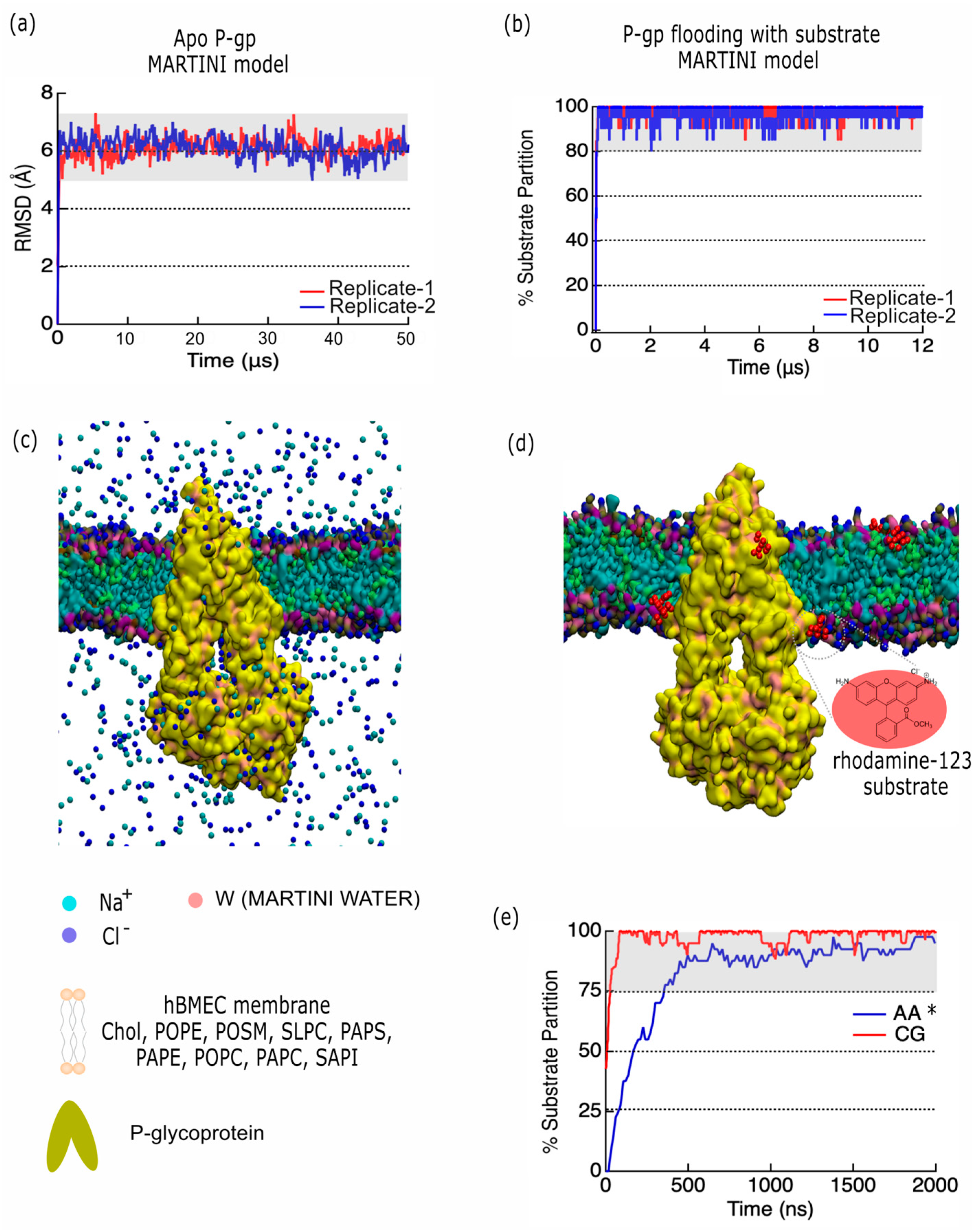
2.4. Developing a P-Glycoprotein Coarse-Grained Model to Explore Substrate Interactions
3. Discussion
3.1. P-gp Inhibition Exhibits Multimodal Character
3.2. P-gp Substrate Pharmacology: The Handling of Chemotherapeutics and Environmental Pollutants from PDB Database Analysis Shows Diversity in Character
3.3. Boltz-2 AI Suggests That Later Generation Inhibitors Bind More Efficiently
3.4. P-gp Coarse-Grained Model
3.5. Conclusions
4. Materials and Methods
4.1. P-gp Human Homology Models
4.2. Protein–Ligand Co-Folding Using Boltz-2
4.3. Coarse-Grained Protein–Membrane Model Setup
4.4. Coarse-Grained Molecular Dynamics
4.5. Software
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| P-gp | P-glycoprotein |
| ABC | ATP-binding cassette |
| BMEC | brain microvascular endothelial cell |
| BBB | blood–brain barrier |
| TMD | transmembrane domain |
| NBD | nucleotide binding domain |
| MD | molecular dynamics |
| CG | coarse-grain |
| POPC | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine |
| POPE | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine |
| POSM | palmitoyl sphingomyelin |
| SLPC | 1-stearoyl-2-linoleoyl-sn-glycero-3-phosphocholine |
| PAPS | 1-palmitoyl-2-arachidonoyl-sn-glycero-3-phosphoserine |
| PAPE | 1-palmitoyl-2-arachidonoyl-sn-glycero-3-phosphoethanolamine |
| PAPC | 1-palmitoyl-2-arachidonoyl-sn-glycero-3-phosphocholine |
| SAPI | stearoyl arachidonoyl phosphatidylinositol |
| SMILES | Simplified Molecular Input Line Entry System |
| PDB | Protein Data Bank |
| COM | center of mass |
| CFTR | cystic fibrosis transmembrane conductance regulator |
| pTM | predicted TM score |
| ipTM | interface predicted TM score |
| pLDDT | predicted local distance difference test |
| IC50 | half-maximal inhibitory concentration |
| pIC50 | negative log of IC50 |
| QMEAN | qualitative model energy analysis |
| MSA | multiple sequence alignment |
| PME | particle mesh Ewald |
| LINCS | linear constraint solver |
| GROMACS | Groningen Machine for Chemical Simulations |
References
- Wong, A.D.; Ye, M.; Levy, A.F.; Rothstein, J.D.; Bergles, D.E.; Searson, P.C. The Blood-Brain Barrier: An Engineering Perspective. Front. Neuroeng. 2013, 6, 7. [Google Scholar] [CrossRef]
- Gottesman, M.M.; Ling, V. The Molecular Basis of Multidrug Resistance in Cancer: The Early Years of P-Glycoprotein Research. FEBS Lett. 2006, 580, 998–1009. [Google Scholar] [CrossRef] [PubMed]
- van Veen, H.W.; Konings, W.N. Structure and Function of Multidrug Transporters. In Resolving the Antibiotic Paradox: Progress in Understanding Drug Resistance and Development of New Antibiotics; Kluwer Academic Publishers: Dordrecht, The Netherlands, 1998; pp. 145–158. [Google Scholar]
- Kim, Y.; Chen, J. Molecular Structure of Human P-Glycoprotein in the ATP-Bound, Outward-Facing Conformation. Science 2018, 359, 915–919. [Google Scholar] [CrossRef] [PubMed]
- Condic-Jurkic, K.; Subramanian, N.; Mark, A.E.; O’Mara, M.L. The Reliability of Molecular Dynamics Simulations of the Multidrug Transporter P-Glycoprotein in a Membrane Environment. PLoS ONE 2018, 13, e0191882. [Google Scholar] [CrossRef]
- Jorgensen, C.; Ulmschneider, M.B.; Searson, P.C. Modeling Substrate Entry into the P-Glycoprotein Efflux Pump at the Blood–Brain Barrier. J. Med. Chem. 2023, 66, 16615–16627. [Google Scholar] [CrossRef]
- Szewczyk, P.; Tao, H.; McGrath, A.P.; Villaluz, M.; Rees, S.D.; Lee, S.C.; Doshi, R.; Urbatsch, I.L.; Zhang, Q.; Chang, G. Snapshots of Ligand Entry, Malleable Binding and Induced Helical Movement in P-Glycoprotein. Biol. Crystallogr. 2015, 71, 732–741. [Google Scholar] [CrossRef]
- Jin, M.S.; Oldham, M.L.; Zhang, Q.; Chen, J. Crystal Structure of the Multidrug Transporter P-Glycoprotein from Caenorhabditis Elegans. Nature 2012, 490, 566–569. [Google Scholar] [CrossRef]
- Aller, S.G.; Yu, J.; Ward, A.; Weng, Y.; Chittaboina, S.; Zhuo, R.; Harrell, P.M.; Trinh, Y.T.; Zhang, Q.; Urbatsch, I.L. Structure of P-Glycoprotein Reveals a Molecular Basis for Poly-Specific Drug Binding. Science 2009, 323, 1718–1722. [Google Scholar] [CrossRef]
- Alam, A.; Kowal, J.; Broude, E.; Roninson, I.; Locher, K.P. Structural Insight into Substrate and Inhibitor Discrimination by Human P-Glycoprotein. Science 2019, 363, 753–756. [Google Scholar] [CrossRef]
- Xing, J.; Huang, S.; Heng, Y.; Mei, H.; Pan, X. Computational Insights into Allosteric Conformational Modulation of P-Glycoprotein by Substrate and Inhibitor Binding. Molecules 2020, 25, 6006. [Google Scholar] [CrossRef] [PubMed]
- Loo, T.W.; Bartlett, M.C.; Clarke, D.M. Drug Binding in Human P-Glycoprotein Causes Conformational Changes in Both Nucleotide-Binding Domains. J. Biol. Chem. 2003, 278, 1575–1578. [Google Scholar] [CrossRef]
- McCormick, J.W.; Vogel, P.D.; Wise, J.G. Multiple Drug Transport Pathways through Human P-Glycoprotein. Biochemistry 2015, 54, 4374–4390. [Google Scholar] [CrossRef]
- Ward, A.B.; Szewczyk, P.; Grimard, V.; Lee, C.-W.; Martinez, L.; Doshi, R.; Caya, A.; Villaluz, M.; Pardon, E.; Cregger, C. Structures of P-Glycoprotein Reveal Its Conformational Flexibility and an Epitope on the Nucleotide-Binding Domain. Proc. Natl. Acad. Sci. USA 2013, 110, 13386–13391. [Google Scholar] [CrossRef]
- Callaghan, R.; Ford, R.C.; Kerr, I.D. The Translocation Mechanism of P-Glycoprotein. FEBS Lett. 2006, 580, 1056–1063. [Google Scholar] [CrossRef] [PubMed]
- Prajapati, R.; Sangamwar, A.T. Translocation Mechanism of P-Glycoprotein and Conformational Changes Occurring at Drug-Binding Site: Insights from Multi-Targeted Molecular Dynamics. Biochim. Biophys. Acta (BBA)-Biomembr. 2014, 1838, 2882–2898. [Google Scholar] [CrossRef]
- Li, H.; Gong, W. Study of Allosteric Transitions of Human P-Glycoprotein by Using the Two-State Anisotropic Network Model. Front. Med. 2022, 9, 815355. [Google Scholar] [CrossRef] [PubMed]
- De Vecchis, D.; Schäfer, L.V. Coupling the Role of Lipids to the Conformational Dynamics of the ABC Transporter P-Glycoprotein. Biophys. J. 2024, 123, 2522–2536. [Google Scholar] [CrossRef] [PubMed]
- Venable, R.M.; Kramer, A.; Pastor, R.W. Molecular Dynamics Simulations of Membrane Permeability. Chem. Rev. 2019, 119, 5954–5997. [Google Scholar] [CrossRef] [PubMed]
- Van Breemen, R.B.; Li, Y. Caco-2 Cell Permeability Assays to Measure Drug Absorption. Expert. Opin. Drug Metab. Toxicol. 2005, 1, 175–185. [Google Scholar] [CrossRef]
- Bednarek, R. In Vitro Methods for Measuring the Permeability of Cell Monolayers. Methods Protoc. 2022, 5, 17. [Google Scholar] [CrossRef]
- Jorgensen, C.; Linville, R.M.; Galea, I.; Lambden, E.; Vögele, M.; Chen, C.; Troendle, E.P.; Ruggiu, F.; Ulmschneider, M.B.; Schiøtt, B. Permeability Benchmarking: Guidelines for Comparing in Silico, in Vitro, and in Vivo Measurements. J. Chem. Inf. Model. 2025, 65, 1067–1084. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, K.B.; Borges-Araújo, L.; Stange, A.D.; Souza, P.C.T.; Marrink, S.J.; Schiøtt, B. OLIVES: A Go-like Model for Stabilizing Protein Structure via Hydrogen Bonding Native Contacts in the Martini 3 Coarse-Grained Force Field. J. Chem. Theory. Comput. 2024, 20, 8049–8070. [Google Scholar] [CrossRef]
- Pedersen, K.B.; Ingólfsson, H.I.; Ramirez-Echemendia, D.P.; Borges-Araújo, L.; Andreasen, M.D.; Empereur-Mot, C.; Melcr, J.; Ozturk, T.N.; Bennett, W.F.D.; Kjølbye, L.R. The Martini 3 Lipidome: Expanded and Refined Parameters Improve Lipid Phase Behavior. ACS Cent. Sci. 2025. [Google Scholar] [CrossRef]
- Ozturk, T.N.; König, M.; Carpenter, T.S.; Pedersen, K.B.; Wassenaar, T.A.; Ingólfsson, H.I.; Marrink, S.J. Building Complex Membranes with Martini 3. Methods Enzym. 2024, 701, 237–285. [Google Scholar]
- Alessandri, R.; Barnoud, J.; Gertsen, A.S.; Patmanidis, I.; de Vries, A.H.; Souza, P.C.T.; Marrink, S.J. Martini 3 Coarse-grained Force Field: Small Molecules. Adv. Theory. Simul. 2022, 5, 2100391. [Google Scholar] [CrossRef]
- Rehman, M.U.; to Chong, K.; Tayara, H. Drugs Inhibition Prediction in P-Gp Enzyme: A Comparative Study of Machine Learning and Graph Neural Network. Comput. Toxicol. 2025, 34, 100344. [Google Scholar] [CrossRef]
- Doniger, S.; Hofmann, T.; Yeh, J. Predicting CNS Permeability of Drug Molecules: Comparison of Neural Network and Support Vector Machine Algorithms. J. Comput. Biol. 2002, 9, 849–864. [Google Scholar] [CrossRef] [PubMed]
- Xue, Y.; Yap, C.W.; Sun, L.Z.; Cao, Z.W.; Wang, J.F.; Chen, Y.Z. Prediction of P-Glycoprotein Substrates by a Support Vector Machine Approach. J. Chem. Inf. Comput. Sci. 2004, 44, 1497–1505. [Google Scholar] [CrossRef]
- Neela, M.M.V.; Peramss, S.R. A Novel Ligand-Based Convolutional Neural Network for Identification of P-Glycoprotein Ligands in Drug Discovery. Mol. Divers. 2025, 1–18. [Google Scholar] [CrossRef]
- Hsu, K.-C.; Wang, P.-H.; Su, B.-H.; Tseng, Y.J. A Robust and Interpretable Graph Neural Network-Based Protocol for Predicting p-Glycoprotein Substrates. Brief. Bioinform. 2025, 26, bbaf392. [Google Scholar] [CrossRef]
- Nosol, K.; Romane, K.; Irobalieva, R.N.; Alam, A.; Kowal, J.; Fujita, N.; Locher, K.P. Cryo-EM Structures Reveal Distinct Mechanisms of Inhibition of the Human Multidrug Transporter ABCB1. Proc. Natl. Acad. Sci. USA 2020, 117, 26245–26253. [Google Scholar] [CrossRef] [PubMed]
- Hamaguchi-Suzuki, N.; Adachi, N.; Moriya, T.; Yasuda, S.; Kawasaki, M.; Suzuki, K.; Ogasawara, S.; Anzai, N.; Senda, T.; Murata, T. Cryo-EM Structure of P-Glycoprotein Bound to Triple Elacridar Inhibitor Molecules. Biochem. Biophys. Res. Commun. 2024, 709, 149855. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual Molecular Dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef] [PubMed]
- Le, C.A.; Harvey, D.S.; Aller, S.G. Structural Definition of Polyspecific Compensatory Ligand Recognition by P-Glycoprotein. IUCrJ 2020, 7, 663–672. [Google Scholar] [CrossRef] [PubMed]
- Barbieri, A.; Thonghin, N.; Shafi, T.; Prince, S.M.; Collins, R.F.; Ford, R.C. Structure of ABCB1/P-Glycoprotein in the Presence of the CFTR Potentiator Ivacaftor. Membranes 2021, 11, 923. [Google Scholar] [CrossRef]
- Passaro, S.; Corso, G.; Wohlwend, J.; Reveiz, M.; Thaler, S.; Somnath, V.R.; Getz, N.; Portnoi, T.; Roy, J.; Stark, H. Boltz-2: Towards Accurate and Efficient Binding Affinity Prediction. bioRxiv 2025. [Google Scholar] [CrossRef]
- Jorgensen, C.; Ulmschneider, M.B.; Searson, P.C. Atomistic Model of Solute Transport across the Blood–Brain Barrier. ACS Omega 2021, 7, 1100–1112. [Google Scholar] [CrossRef]
- Jorgensen, C.; Troendle, E.P.; Ulmschneider, J.P.; Searson, P.C.; Ulmschneider, M.B. A Least-Squares-Fitting Procedure for an Efficient Preclinical Ranking of Passive Transport across the Blood–Brain Barrier Endothelium. J. Comput. Aided. Mol. Des. 2023, 37, 537–549. [Google Scholar] [CrossRef]
- Wassenaar, T.A.; Ingólfsson, H.I.; Bockmann, R.A.; Tieleman, D.P.; Marrink, S.J. Computational Lipidomics with Insane: A Versatile Tool for Generating Custom Membranes for Molecular Simulations. J. Chem. Theory. Comput. 2015, 11, 2144–2155. [Google Scholar] [CrossRef]
- Souza, P.C.T.; Alessandri, R.; Barnoud, J.; Thallmair, S.; Faustino, I.; Grünewald, F.; Patmanidis, I.; Abdizadeh, H.; Bruininks, B.M.H.; Wassenaar, T.A. Martini 3: A General Purpose Force Field for Coarse-Grained Molecular Dynamics. Nat. Methods 2021, 18, 382–388. [Google Scholar] [CrossRef]
- Periole, X.; Cavalli, M.; Marrink, S.-J.; Ceruso, M.A. Combining an Elastic Network with a Coarse-Grained Molecular Force Field: Structure, Dynamics, and Intermolecular Recognition. J. Chem. Theory. Comput. 2009, 5, 2531–2543. [Google Scholar] [CrossRef]
- Monticelli, L.; Kandasamy, S.K.; Periole, X.; Larson, R.G.; Tieleman, D.P.; Marrink, S.-J. The MARTINI Coarse-Grained Force Field: Extension to Proteins. J. Chem. Theory. Comput. 2008, 4, 819–834. [Google Scholar] [CrossRef]
- Vanommeslaeghe, K.; Hatcher, E.; Acharya, C.; Kundu, S.; Zhong, S.; Shim, J.; Darian, E.; Guvench, O.; Lopes, P.; Vorobyov, I. CHARMM General Force Field: A Force Field for Drug-like Molecules Compatible with the CHARMM All-atom Additive Biological Force Fields. J. Comput. Chem. 2010, 31, 671–690. [Google Scholar] [CrossRef]
- Nørholm, M.H.H.; von Heijne, G.; Draheim, R.R. Forcing the Issue: Aromatic Tuning Facilitates Stimulus-Independent Modulation of a Two-Component Signaling Circuit. ACS Synth. Biol. 2015, 4, 474–481. [Google Scholar] [CrossRef] [PubMed]
- Yusuf, R.; Yau, J.J.Y.; Baker-Valla, G.W.; Lawrence, R.J.; Draheim, R.R. EnvZ Signal Output Correlates with Capacity for Transmembrane Water Encapsulation. bioRxiv 2025. [Google Scholar] [CrossRef]
- Weidner, L.D.; Fung, K.L.; Kannan, P.; Moen, J.K.; Kumar, J.S.; Mulder, J.; Innis, R.B.; Gottesman, M.M.; Hall, M.D. Tariquidar Is an Inhibitor and Not a Substrate of Human and Mouse P-Glycoprotein. Drug Metab. Dispos. 2016, 44, 275–282. [Google Scholar] [CrossRef]
- Ma, J.; Biggin, P.C. Substrate versus Inhibitor Dynamics of P-glycoprotein. Proteins Struct. Funct. Bioinform. 2013, 81, 1653–1668. [Google Scholar] [CrossRef] [PubMed]
- Klepsch, F.; Vasanthanathan, P.; Ecker, G.F. Ligand and Structure-Based Classification Models for Prediction of P-Glycoprotein Inhibitors. J. Chem. Inf. Model. 2014, 54, 218–229. [Google Scholar] [CrossRef] [PubMed]
- Martin, C.; Berridge, G.; Mistry, P.; Higgins, C.; Charlton, P.; Callaghan, R. The Molecular Interaction of the High Affinity Reversal Agent XR9576 with P-glycoprotein. Br. J. Pharmacol. 1999, 128, 403–411. [Google Scholar] [CrossRef]
- Waghray, D.; Zhang, Q. Inhibit or Evade Multidrug Resistance P-Glycoprotein in Cancer Treatment: Miniperspective. J. Med. Chem. 2017, 61, 5108–5121. [Google Scholar] [CrossRef]
- Domicevica, L.; Biggin, P.C. Homology Modelling of Human P-Glycoprotein. Biochem. Soc. Trans. 2015, 43, 952–958. [Google Scholar] [CrossRef]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L. SWISS-MODEL: Homology Modelling of Protein Structures and Complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef]
- Kiefer, F.; Arnold, K.; Künzli, M.; Bordoli, L.; Schwede, T. The SWISS-MODEL Repository and Associated Resources. Nucleic Acids Res. 2009, 37, D387–D392. [Google Scholar] [CrossRef]
- Eargle, J.; Wright, D.; Luthey-Schulten, Z. Multiple Alignment of Protein Structures and Sequences for VMD. Bioinformatics 2006, 22, 504–506. [Google Scholar] [CrossRef]
- UniProt Consortium. UniProt: The Universal Protein Knowledgebase in 2021. Nucleic Acids Res. 2021, 49, D480–D489. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Thiessen, P.A.; Bolton, E.E.; Chen, J.; Fu, G.; Gindulyte, A.; Han, L.; He, J.; He, S.; Shoemaker, B.A. PubChem Substance and Compound Databases. Nucleic Acids Res. 2016, 44, D1202–D1213. [Google Scholar] [CrossRef] [PubMed]
- Van Der Spoel, D.; Lindahl, E.; Hess, B.; Groenhof, G.; Mark, A.E.; Berendsen, H.J.C. GROMACS: Fast, Flexible, and Free. J. Comput. Chem. 2005, 26, 1701–1718. [Google Scholar] [CrossRef]
- Berendsen, H.J.C.; Postma, J.P.M.; Van Gunsteren, W.F.; DiNola, A.; Haak, J.R. Molecular Dynamics with Coupling to an External Bath. J. Chem. Phys. 1984, 81, 3684–3690. [Google Scholar] [CrossRef]
- Bussi, G.; Donadio, D.; Parrinello, M. Canonical Sampling through Velocity Rescaling. J. Chem. Phys. 2007, 126, 014101. [Google Scholar] [CrossRef]
- Hess, B.; Bekker, H.; Berendsen, H.J.C.; Fraaije, J.G.E.M. LINCS: A Linear Constraint Solver for Molecular Simulations. J. Comput. Chem. 1997, 18, 1463–1472. [Google Scholar] [CrossRef]

| Inhibitors | Substrates | |||||
|---|---|---|---|---|---|---|
| QZ-Leu | Tariquidar | Elacridar | Taxol | BDE100 | Ivacaftor | |
| Predicted TM score (pTM) | 0.763 | 0.772 | 0.781 | 0.780 | 0.792 | 0.774 |
| Interface predicted TM score (ipTM) | 0.890 | 0.911 | 0.933 | 0.892 | 0.779 | 0.915 |
| Confidence score | 0.743 | 0.785 | 0.784 | 0.755 | 0.781 | 0.777 |
| Average predicted local distance difference test (pLDDT) | 0.706 | 0.753 | 0.746 | 0.721 | 0.782 | 0.743 |
| Affinity probability | 0.402 | 0.667 | 0.672 | 0.545 | 0.503 | 0.663 |
| Predicted pIC50 | 6.565 | 7.243 | 6.447 | 6.928 | 5.589 | 6.237 |
| Predicted IC50 (nM) | 272.3 | 57.1 | 357.3 | 118.0 | 2580 | 579.4 |
| Lipid | Total Bilayer (%) |
|---|---|
| CHOL | 30 (96) |
| POPE | 6 (20) |
| POSM | 19 (61) |
| SLPC | 8 (27) |
| PAPS | 8 (25) |
| PAPE | 15 (47) |
| POPC | 4 (13) |
| PAPC | 8 (27) |
| SAPI | 2 (6) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jorgensen, C.; Oliphant, E.; Barker, M.; López Martínez, E.; Thulasi, S.; Prior, H.; Franey, B.W.; Gregory, C.; Oluwasegun, J.; Ajay, A.; et al. Simulation and Machine Learning Assessment of P-Glycoprotein Pharmacology in the Blood–Brain Barrier: Inhibition and Substrate Transport. Int. J. Mol. Sci. 2025, 26, 9050. https://doi.org/10.3390/ijms26189050
Jorgensen C, Oliphant E, Barker M, López Martínez E, Thulasi S, Prior H, Franey BW, Gregory C, Oluwasegun J, Ajay A, et al. Simulation and Machine Learning Assessment of P-Glycoprotein Pharmacology in the Blood–Brain Barrier: Inhibition and Substrate Transport. International Journal of Molecular Sciences. 2025; 26(18):9050. https://doi.org/10.3390/ijms26189050
Chicago/Turabian StyleJorgensen, Christian, Elizabeth Oliphant, Milly Barker, Eduardo López Martínez, Saaihasamreen Thulasi, Holly Prior, Ben William Franey, Charley Gregory, Jerry Oluwasegun, Anjalee Ajay, and et al. 2025. "Simulation and Machine Learning Assessment of P-Glycoprotein Pharmacology in the Blood–Brain Barrier: Inhibition and Substrate Transport" International Journal of Molecular Sciences 26, no. 18: 9050. https://doi.org/10.3390/ijms26189050
APA StyleJorgensen, C., Oliphant, E., Barker, M., López Martínez, E., Thulasi, S., Prior, H., Franey, B. W., Gregory, C., Oluwasegun, J., Ajay, A., & Draheim, R. R. (2025). Simulation and Machine Learning Assessment of P-Glycoprotein Pharmacology in the Blood–Brain Barrier: Inhibition and Substrate Transport. International Journal of Molecular Sciences, 26(18), 9050. https://doi.org/10.3390/ijms26189050








