BARD1: A Friend or Foe in Pancreatic Ductal Adenocarcinoma?
Abstract
1. Introduction
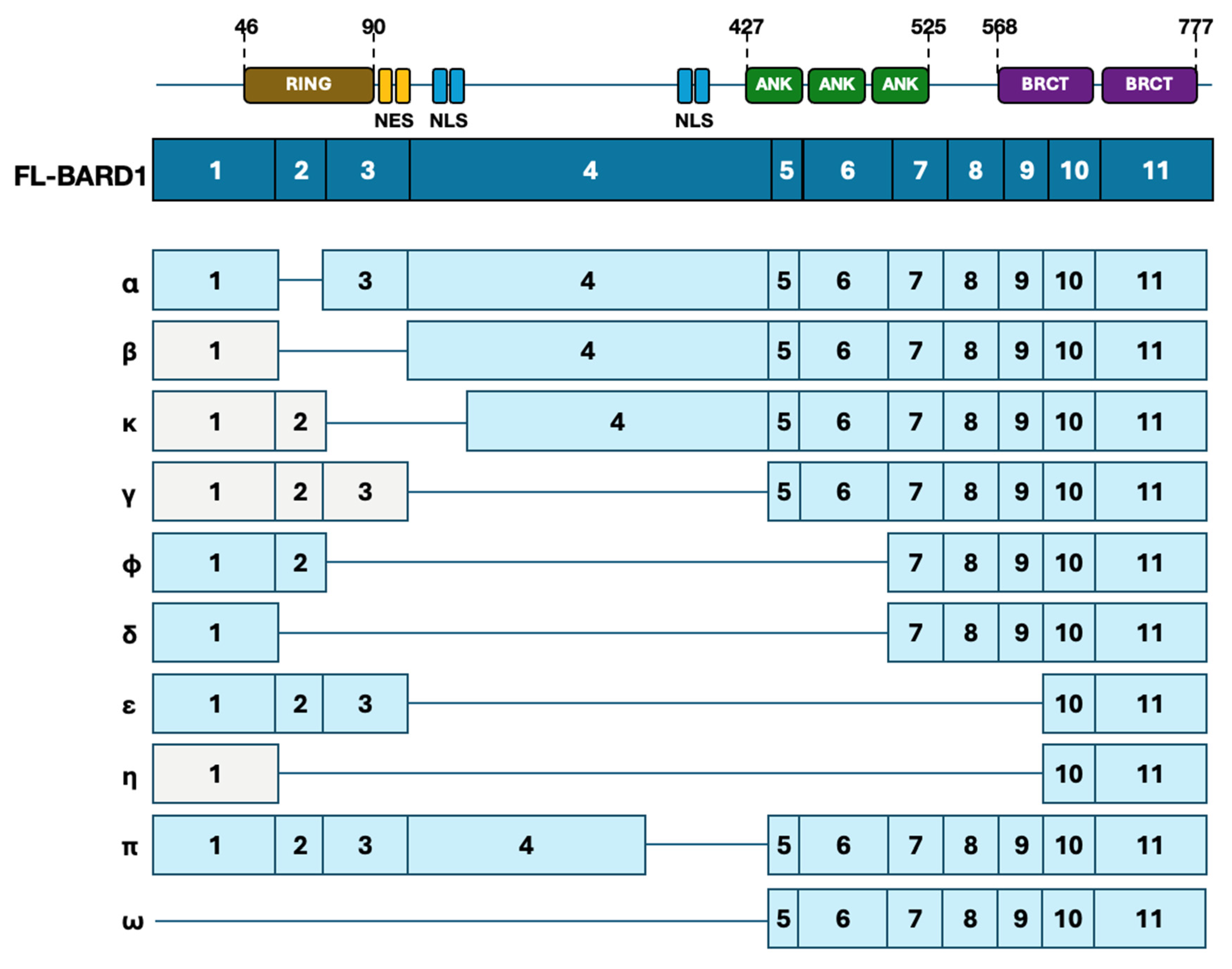
2. Overview of BARD1
3. Tumor Suppressor Effects of BARD1
4. Mechanisms Affecting Tumor-Suppressive Activity of BARD1
4.1. Genomic Alterations
4.2. Post-Translational Modulation of BARD1
4.3. Promoter Methylation
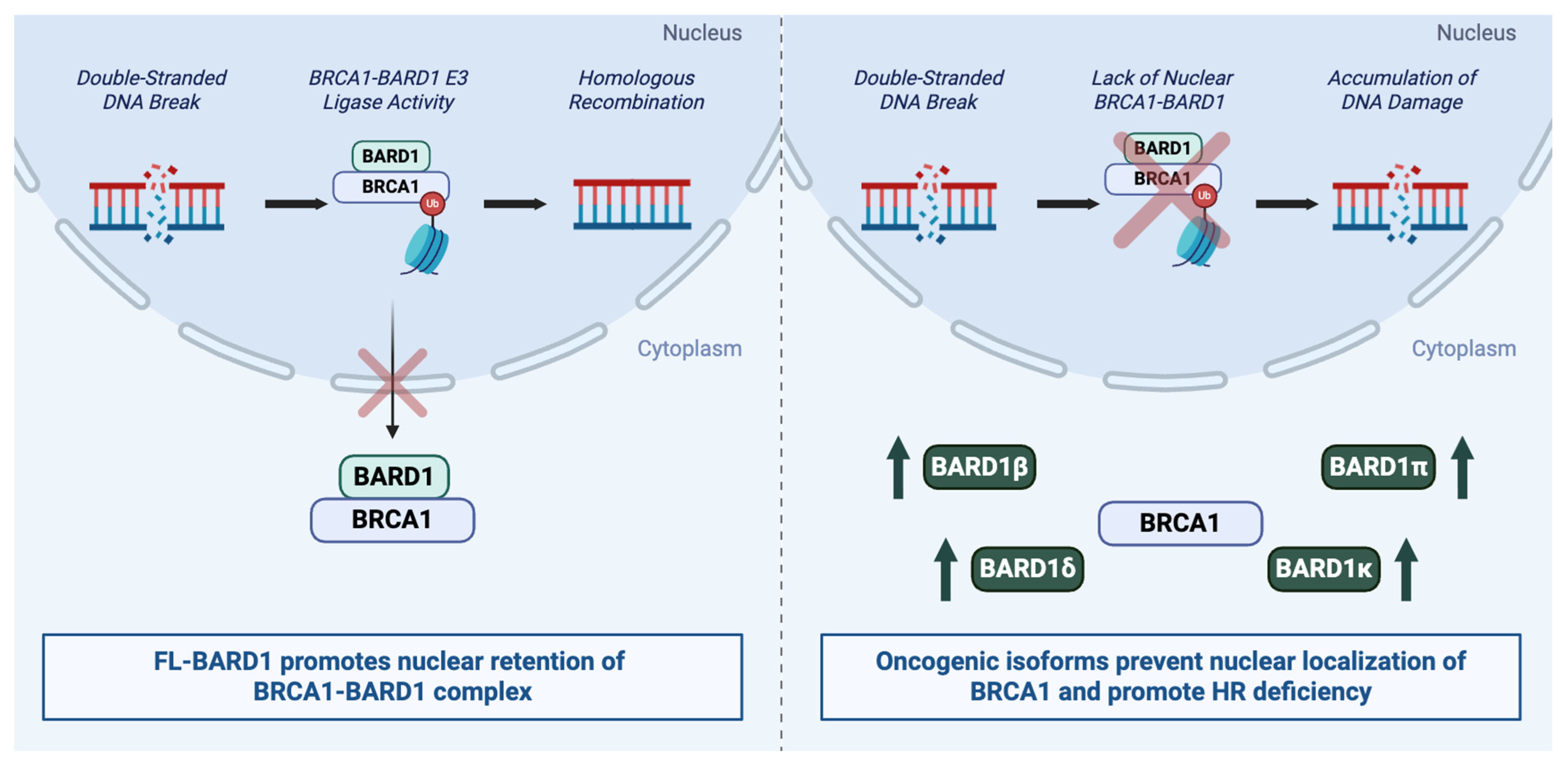
5. Oncogenic Effects of BARD1
5.1. Isoform-Specific Oncogenic Effects
5.1.1. BARD1β and BARD1δ Isoforms
5.1.2. Additional Isoforms and Paradoxical Roles
6. Mechanisms Affecting Oncogenic Activity of BARD1
6.1. MicroRNA-Mediated Post-Transcriptional Upregulation of BARD1 Isoforms
6.2. RNA-Binding Protein (RBP) Mediated Stabilization of BARD1 Isoforms
6.3. BARD1-Mediated Oncogenic Transcriptional Networks
7. Role of BARD1 in Therapeutic Resistance
8. Clinical Trials with DNA Damage Response (DDR) Agents
9. Conclusions and Perspectives on Research Strategies
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Kleeff, J.; Korc, M.; Apte, M.; La Vecchia, C.; Johnson, C.D.; Biankin, A.V.; Neale, R.E.; Tempero, M.; Tuveson, D.A.; Hruban, R.H.; et al. Pancreatic cancer. Nat. Rev. Dis. Primers 2016, 2, 16022. [Google Scholar] [CrossRef]
- Park, W.; Chawla, A.; O’Reilly, E.M. Pancreatic Cancer: A Review. JAMA 2021, 326, 851–862. [Google Scholar] [CrossRef] [PubMed]
- Stoof, J.; Harrold, E.; Mariottino, S.; Lowery, M.A.; Walsh, N. DNA Damage Repair Deficiency in Pancreatic Ductal Adenocarcinoma: Preclinical Models and Clinical Perspectives. Front. Cell Dev. Biol. 2021, 9, 749490. [Google Scholar] [CrossRef]
- Varghese, A.M.; Perry, M.A.; Chou, J.F.; Nandakumar, S.; Muldoon, D.; Erakky, A.; Zucker, A.; Fong, C.; Mehine, M.; Nguyen, B.; et al. Clinicogenomic landscape of pancreatic adenocarcinoma identifies KRAS mutant dosage as prognostic of overall survival. Nat. Med. 2025, 31, 466–477. [Google Scholar] [CrossRef] [PubMed]
- Orth, M.; Metzger, P.; Gerum, S.; Mayerle, J.; Schneider, G.; Belka, C.; Schnurr, M.; Lauber, K. Pancreatic ductal adenocarcinoma: Biological hallmarks, current status, and future perspectives of combined modality treatment approaches. Radiat. Oncol. 2019, 14, 141. [Google Scholar] [CrossRef]
- Barati Bagherabad, M.; Afzaljavan, F.; ShahidSales, S.; Hassanian, S.M.; Avan, A. Targeted therapies in pancreatic cancer: Promises and failures. J. Cell. Biochem. 2019, 120, 2726–2741. [Google Scholar] [CrossRef]
- Siegel, R.L.; Giaquinto, A.N.; Jemal, A. Cancer statistics, 2024. CA Cancer J. Clin. 2024, 74, 12–49. [Google Scholar] [CrossRef]
- Perkhofer, L.; Golan, T.; Cuyle, P.J.; Matysiak-Budnik, T.; Van Laethem, J.L.; Macarulla, T.; Cauchin, E.; Kleger, A.; Beutel, A.K.; Gout, J.; et al. Targeting DNA Damage Repair Mechanisms in Pancreas Cancer. Cancers 2021, 13, 4259. [Google Scholar] [CrossRef]
- Pishvaian, M.J.; Blais, E.M.; Brody, J.R.; Rahib, L.; Lyons, E.; De Arbeloa, P.; Hendifar, A.; Mikhail, S.; Chung, V.; Sohal, D.P.S.; et al. Outcomes in Patients With Pancreatic Adenocarcinoma With Genetic Mutations in DNA Damage Response Pathways: Results From the Know Your Tumor Program. JCO Precis. Oncol. 2019, 3, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Maginn, E.N.; de Sousa, C.H.; Wasan, H.S.; Stronach, E.A. Opportunities for translation: Targeting DNA repair pathways in pancreatic cancer. Biochim. Biophys. Acta 2014, 1846, 45–54. [Google Scholar] [CrossRef]
- Singhi, A.D.; George, B.; Greenbowe, J.R.; Chung, J.; Suh, J.; Maitra, A.; Klempner, S.J.; Hendifar, A.; Milind, J.M.; Golan, T.; et al. Real-Time Targeted Genome Profile Analysis of Pancreatic Ductal Adenocarcinomas Identifies Genetic Alterations That Might Be Targeted With Existing Drugs or Used as Biomarkers. Gastroenterology 2019, 156, 2242–2253.e4. [Google Scholar]
- Tarsounas, M.; Sung, P. The antitumorigenic roles of BRCA1-BARD1 in DNA repair and replication. Nat. Rev. Mol. Cell Biol. 2020, 21, 284–299. [Google Scholar] [CrossRef]
- Prakash, R.; Zhang, Y.; Feng, W.; Jasin, M. Homologous recombination and human health: The roles of BRCA1, BRCA2, and associated proteins. Cold Spring Harb. Perspect. Biol. 2015, 7, a016600. [Google Scholar] [CrossRef]
- Salunkhe, S.; Daley, J.M.; Kaur, H.; Tomimatsu, N.; Xue, C.; Raina, V.B.; Jasper, A.M.; Rogers, C.M.; Li, W.; Zhou, S.; et al. Promotion of DNA end resection by BRCA1-BARD1 in homologous recombination. Nature 2024, 634, 482–491. [Google Scholar] [CrossRef]
- Ceppi, I.; Dello Stritto, M.R.; Mutze, M.; Braunshier, S.; Mengoli, V.; Reginato, G.; Vo, H.M.P.; Jimeno, S.; Acharya, A.; Roy, M.; et al. Mechanism of BRCA1-BARD1 function in DNA end resection and DNA protection. Nature 2024, 634, 492–500. [Google Scholar] [PubMed]
- Dai, L.; Dai, Y.; Han, J.; Huang, Y.; Wang, L.; Huang, J.; Zhou, Z. Structural insight into BRCA1-BARD1 complex recruitment to damaged chromatin. Mol. Cell 2021, 81, 2765–2777.e2766. [Google Scholar] [CrossRef]
- Zhao, W.; Steinfeld, J.B.; Liang, F.; Chen, X.; Maranon, D.G.; Jian Ma, C.; Kwon, Y.; Rao, T.; Wang, W.; Sheng, C.; et al. BRCA1-BARD1 promotes RAD51-mediated homologous DNA pairing. Nature 2017, 550, 360–365. [Google Scholar] [CrossRef] [PubMed]
- Krais, J.J.; Wang, Y.; Patel, P.; Basu, J.; Bernhardy, A.J.; Johnson, N. RNF168-mediated localization of BARD1 recruits the BRCA1-PALB2 complex to DNA damage. Nat. Commun. 2021, 12, 5016. [Google Scholar] [PubMed]
- Foo, T.K.; Xia, B. BRCA1-Dependent and Independent Recruitment of PALB2-BRCA2-RAD51 in the DNA Damage Response and Cancer. Cancer Res. 2022, 82, 3191–3197. [Google Scholar] [CrossRef]
- Guo, M.; Wang, S.M. The BRCAness Landscape of Cancer. Cells 2022, 11, 3877. [Google Scholar] [CrossRef]
- Bryant, H.E.; Schultz, N.; Thomas, H.D.; Parker, K.M.; Flower, D.; Lopez, E.; Kyle, S.; Meuth, M.; Curtin, N.J.; Helleday, T. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature 2005, 434, 913–917. [Google Scholar] [PubMed]
- Farmer, H.; McCabe, N.; Lord, C.J.; Tutt, A.N.; Johnson, D.A.; Richardson, T.B.; Santarosa, M.; Dillon, K.J.; Hickson, I.; Knights, C.; et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 2005, 434, 917–921. [Google Scholar] [CrossRef] [PubMed]
- Golan, T.; Hammel, P.; Reni, M.; Van Cutsem, E.; Macarulla, T.; Hall, M.J.; Park, J.O.; Hochhauser, D.; Arnold, D.; Oh, D.Y.; et al. Maintenance Olaparib for Germline BRCA-Mutated Metastatic Pancreatic Cancer. N. Engl. J. Med. 2019, 381, 317–327. [Google Scholar] [CrossRef]
- Dias, M.P.; Moser, S.C.; Ganesan, S.; Jonkers, J. Understanding and overcoming resistance to PARP inhibitors in cancer therapy. Nat. Rev. Clin. Oncol. 2021, 18, 773–791. [Google Scholar] [CrossRef]
- Wu, L.C.; Wang, Z.W.; Tsan, J.T.; Spillman, M.A.; Phung, A.; Xu, X.L.; Yang, M.C.; Hwang, L.Y.; Bowcock, A.M.; Baer, R. Identification of a RING protein that can interact in vivo with the BRCA1 gene product. Nat. Genet. 1996, 14, 430–440. [Google Scholar] [CrossRef]
- Birrane, G.; Varma, A.K.; Soni, A.; Ladias, J.A.A. Crystal Structure of the BARD1 BRCT Domains. Biochemistry 2007, 46, 7706–7712. [Google Scholar] [CrossRef]
- Brzovic, P.S.; Rajagopal, P.; Hoyt, D.W.; King, M.C.; Klevit, R.E. Structure of a BRCA1-BARD1 heterodimeric RING-RING complex. Nat. Struct. Biol. 2001, 8, 833–837. [Google Scholar]
- Edwards, R.A.; Lee, M.S.; Tsutakawa, S.E.; Williams, R.S.; Nazeer, I.; Kleiman, F.E.; Tainer, J.A.; Glover, J.N. The BARD1 C-terminal domain structure and interactions with polyadenylation factor CstF-50. Biochemistry 2008, 47, 11446–11456. [Google Scholar] [CrossRef]
- Schuchner, S.; Tembe, V.; Rodriguez, J.A.; Henderson, B.R. Nuclear targeting and cell cycle regulatory function of human BARD1. J. Biol. Chem. 2005, 280, 8855–8861. [Google Scholar] [CrossRef]
- Rodriguez, J.A.; Schuchner, S.; Au, W.W.; Fabbro, M.; Henderson, B.R. Nuclear-cytoplasmic shuttling of BARD1 contributes to its proapoptotic activity and is regulated by dimerization with BRCA1. Oncogene 2004, 23, 1809–1820. [Google Scholar] [CrossRef]
- Brzovic, P.S.; Keeffe, J.R.; Nishikawa, H.; Miyamoto, K.; Fox, D., 3rd; Fukuda, M.; Ohta, T.; Klevit, R. Binding and recognition in the assembly of an active BRCA1/BARD1 ubiquitin-ligase complex. Proc. Natl. Acad. Sci. USA 2003, 100, 5646–5651. [Google Scholar] [CrossRef]
- Wang, M.; Li, W.; Tomimatsu, N.; Yu, C.H.; Ji, J.H.; Alejo, S.; Witus, S.R.; Alimbetov, D.; Fitzgerald, O.; Wu, B.; et al. Crucial roles of the BRCA1-BARD1 E3 ubiquitin ligase activity in homology-directed DNA repair. Mol. Cell 2023, 83, 3679–3691.e3678. [Google Scholar] [CrossRef] [PubMed]
- Fabbro, M.; Rodriguez, J.A.; Baer, R.; Henderson, B.R. BARD1 induces BRCA1 intranuclear foci formation by increasing RING-dependent BRCA1 nuclear import and inhibiting BRCA1 nuclear export. J. Biol. Chem. 2002, 277, 21315–21324. [Google Scholar] [CrossRef]
- Witus, S.R.; Stewart, M.D.; Klevit, R.E. The BRCA1/BARD1 ubiquitin ligase and its substrates. Biochem. J. 2021, 478, 3467–3483. [Google Scholar] [CrossRef]
- Eakin, C.M.; Maccoss, M.J.; Finney, G.L.; Klevit, R.E. Estrogen receptor alpha is a putative substrate for the BRCA1 ubiquitin ligase. Proc. Natl. Acad. Sci. USA 2007, 104, 5794–5799. [Google Scholar] [CrossRef] [PubMed]
- Yalcin, Z.; Koot, D.; Bezstarosti, K.; Salas-Lloret, D.; Bleijerveld, O.B.; Boersma, V.; Falcone, M.; Gonzalez-Prieto, R.; Altelaar, M.; Demmers, J.A.A.; et al. Ubiquitinome Profiling Reveals in Vivo UBE2D3 Targets and Implicates UBE2D3 in Protein Quality Control. Mol. Cell. Proteom. 2023, 22, 100548. [Google Scholar] [CrossRef] [PubMed]
- Ruffner, H.; Joazeiro, C.A.; Hemmati, D.; Hunter, T.; Verma, I.M. Cancer-predisposing mutations within the RING domain of BRCA1: Loss of ubiquitin protein ligase activity and protection from radiation hypersensitivity. Proc. Natl. Acad. Sci. USA 2001, 98, 5134–5139. [Google Scholar] [CrossRef]
- Mallery, D.L.; Vandenberg, C.J.; Hiom, K. Activation of the E3 ligase function of the BRCA1/BARD1 complex by polyubiquitin chains. EMBO J. 2002, 21, 6755–6762. [Google Scholar] [CrossRef]
- Hashizume, R.; Fukuda, M.; Maeda, I.; Nishikawa, H.; Oyake, D.; Yabuki, Y.; Ogata, H.; Ohta, T. The RING heterodimer BRCA1-BARD1 is a ubiquitin ligase inactivated by a breast cancer-derived mutation. J. Biol. Chem. 2001, 276, 14537–14540. [Google Scholar] [CrossRef]
- McCarthy, E.E.; Celebi, J.T.; Baer, R.; Ludwig, T. Loss of Bard1, the heterodimeric partner of the Brca1 tumor suppressor, results in early embryonic lethality and chromosomal instability. Mol. Cell. Biol. 2003, 23, 5056–5063. [Google Scholar] [CrossRef]
- Brady, C.A.; Jiang, D.; Mello, S.S.; Johnson, T.M.; Jarvis, L.A.; Kozak, M.M.; Kenzelmann Broz, D.; Basak, S.; Park, E.J.; McLaughlin, M.E.; et al. Distinct p53 transcriptional programs dictate acute DNA-damage responses and tumor suppression. Cell 2011, 145, 571–583. [Google Scholar] [CrossRef]
- Irminger-Finger, I.; Leung, W.C.; Li, J.; Dubois-Dauphin, M.; Harb, J.; Feki, A.; Jefford, C.E.; Soriano, J.V.; Jaconi, M.; Montesano, R.; et al. Identification of BARD1 as mediator between proapoptotic stress and p53-dependent apoptosis. Mol. Cell 2001, 8, 1255–1266. [Google Scholar] [CrossRef]
- Feki, A.; Jefford, C.E.; Berardi, P.; Wu, J.Y.; Cartier, L.; Krause, K.H.; Irminger-Finger, I. BARD1 induces apoptosis by catalysing phosphorylation of p53 by DNA-damage response kinase. Oncogene 2005, 24, 3726–3736. [Google Scholar] [CrossRef]
- Fabbro, M.; Schuechner, S.; Au, W.W.; Henderson, B.R. BARD1 regulates BRCA1 apoptotic function by a mechanism involving nuclear retention. Exp. Cell Res. 2004, 298, 661–673. [Google Scholar] [CrossRef]
- Joukov, V.; Groen, A.C.; Prokhorova, T.; Gerson, R.; White, E.; Rodriguez, A.; Walter, J.C.; Livingston, D.M. The BRCA1/BARD1 heterodimer modulates ran-dependent mitotic spindle assembly. Cell 2006, 127, 539–552. [Google Scholar] [CrossRef]
- Hu, C.; Hart, S.N.; Bamlet, W.R.; Moore, R.M.; Nandakumar, K.; Eckloff, B.W.; Lee, Y.K.; Petersen, G.M.; McWilliams, R.R.; Couch, F.J. Prevalence of Pathogenic Mutations in Cancer Predisposition Genes among Pancreatic Cancer Patients. Cancer Epidemiol. Biomark. Prev. 2016, 25, 207–211. [Google Scholar] [CrossRef] [PubMed]
- Chaffee, K.G.; Oberg, A.L.; McWilliams, R.R.; Majithia, N.; Allen, B.A.; Kidd, J.; Singh, N.; Hartman, A.R.; Wenstrup, R.J.; Petersen, G.M. Prevalence of germ-line mutations in cancer genes among pancreatic cancer patients with a positive family history. Genet. Med. 2018, 20, 119–127. [Google Scholar] [CrossRef]
- Thai, T.H.; Du, F.; Tsan, J.T.; Jin, Y.; Phung, A.; Spillman, M.A.; Massa, H.F.; Muller, C.Y.; Ashfaq, R.; Mathis, J.M.; et al. Mutations in the BRCA1-associated RING domain (BARD1) gene in primary breast, ovarian and uterine cancers. Hum. Mol. Genet. 1998, 7, 195–202. [Google Scholar] [CrossRef] [PubMed]
- Ghimenti, C.; Sensi, E.; Presciuttini, S.; Brunetti, I.M.; Conte, P.; Bevilacqua, G.; Caligo, M.A. Germline mutations of the BRCA1-associated ring domain (BARD1) gene in breast and breast/ovarian families negative for BRCA1 and BRCA2 alterations. Genes Chromosomes Cancer 2002, 33, 235–242. [Google Scholar] [CrossRef]
- Ishitobi, M.; Miyoshi, Y.; Hasegawa, S.; Egawa, C.; Tamaki, Y.; Monden, M.; Noguchi, S. Mutational analysis of BARD1 in familial breast cancer patients in Japan. Cancer Lett. 2003, 200, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Jefford, C.E.; Feki, A.; Harb, J.; Krause, K.H.; Irminger-Finger, I. Nuclear-cytoplasmic translocation of BARD1 is linked to its apoptotic activity. Oncogene 2004, 23, 3509–3520. [Google Scholar] [CrossRef]
- Tsuzuki, M.; Wu, W.; Nishikawa, H.; Hayami, R.; Oyake, D.; Yabuki, Y.; Fukuda, M.; Ohta, T. A truncated splice variant of human BARD1 that lacks the RING finger and ankyrin repeats. Cancer Lett. 2006, 233, 108–116. [Google Scholar] [CrossRef]
- Choudhary, R.K.; Siddiqui, M.Q.; Gadewal, N.; Kumar, N.S.; Kuligina, E.S.; Varma, A.K. Biophysical evaluation to categorize pathogenicity of cancer-predisposing mutations identified in the BARD1 BRCT domain. RSC Adv. 2018, 8, 34056–34068. [Google Scholar] [CrossRef]
- Adamovich, A.I.; Banerjee, T.; Wingo, M.; Duncan, K.; Ning, J.; Martins Rodrigues, F.; Huang, K.L.; Lee, C.; Chen, F.; Ding, L.; et al. Functional analysis of BARD1 missense variants in homology-directed repair and damage sensitivity. PLoS Genet. 2019, 15, e1008049. [Google Scholar] [CrossRef]
- Densham, R.M.; Garvin, A.J.; Stone, H.R.; Strachan, J.; Baldock, R.A.; Daza-Martin, M.; Fletcher, A.; Blair-Reid, S.; Beesley, J.; Johal, B.; et al. Human BRCA1-BARD1 ubiquitin ligase activity counteracts chromatin barriers to DNA resection. Nat. Struct. Mol. Biol. 2016, 23, 647–655. [Google Scholar] [CrossRef]
- Nakamura, K.; Saredi, G.; Becker, J.R.; Foster, B.M.; Nguyen, N.V.; Beyer, T.E.; Cesa, L.C.; Faull, P.A.; Lukauskas, S.; Frimurer, T.; et al. H4K20me0 recognition by BRCA1-BARD1 directs homologous recombination to sister chromatids. Nat. Cell Biol. 2019, 21, 311–318. [Google Scholar] [CrossRef] [PubMed]
- Kleiman, F.E.; Manley, J.L. The BARD1-CstF-50 interaction links mRNA 3′ end formation to DNA damage and tumor suppression. Cell 2001, 104, 743–753. [Google Scholar] [CrossRef] [PubMed]
- Fu, W.; Zhu, J.; Xiong, S.W.; Jia, W.; Zhao, Z.; Zhu, S.B.; Hu, J.H.; Wang, F.H.; Xia, H.; He, J.; et al. BARD1 Gene Polymorphisms Confer Nephroblastoma Susceptibility. EBioMedicine 2017, 16, 101–105. [Google Scholar] [CrossRef] [PubMed]
- Alshatwi, A.A.; Hasan, T.N.; Syed, N.A.; Shafi, G.; Grace, B.L. Identification of functional SNPs in BARD1 gene and in silico analysis of damaging SNPs: Based on data procured from dbSNP database. PLoS ONE 2012, 7, e43939. [Google Scholar] [CrossRef]
- Cimmino, F.; Avitabile, M.; Lasorsa, V.A.; Pezone, L.; Cardinale, A.; Montella, A.; Cantalupo, S.; Iolascon, A.; Capasso, M. Functional characterization of full-length BARD1 strengthens its role as a tumor suppressor in neuroblastoma. J. Cancer 2020, 11, 1495–1504. [Google Scholar] [CrossRef]
- Huo, X.; Hu, Z.; Zhai, X.; Wang, Y.; Wang, S.; Wang, X.; Qin, J.; Chen, W.; Jin, G.; Liu, J.; et al. Common non-synonymous polymorphisms in the BRCA1 Associated RING Domain (BARD1) gene are associated with breast cancer susceptibility: A case-control analysis. Breast Cancer Res. Treat. 2007, 102, 329–337. [Google Scholar] [CrossRef] [PubMed]
- Weber-Lassalle, N.; Borde, J.; Weber-Lassalle, K.; Horvath, J.; Niederacher, D.; Arnold, N.; Kaulfuss, S.; Ernst, C.; Paul, V.G.; Honisch, E.; et al. Germline loss-of-function variants in the BARD1 gene are associated with early-onset familial breast cancer but not ovarian cancer. Breast Cancer Res. 2019, 21, 55. [Google Scholar] [PubMed]
- Randall, M.P.; Egolf, L.E.; Vaksman, Z.; Samanta, M.; Tsang, M.; Groff, D.; Evans, J.P.; Rokita, J.L.; Layeghifard, M.; Shlien, A.; et al. BARD1 germline variants induce haploinsufficiency and DNA repair defects in neuroblastoma. bioRxiv 2023. [Google Scholar] [CrossRef]
- Takaiso, N.; Imoto, I.; Yoshimura, A.; Ouchi, A.; Komori, K.; Iwata, H.; Shimizu, Y. BARD1 deletion in a patient with suspected hereditary colorectal cancer. Hum. Genome Var. 2024, 11, 11. [Google Scholar] [CrossRef]
- Amemiya, Y.; Bacopulos, S.; Al-Shawarby, M.; Al-Tamimi, D.; Naser, W.; Ahmed, A.; Khalifa, M.; Slodkowska, E.; Seth, A. A Comparative Analysis of Breast and Ovarian Cancer-related Gene Mutations in Canadian and Saudi Arabian Patients with Breast Cancer. Anticancer Res. 2015, 35, 2601–2610. [Google Scholar]
- Zhou, X.; Han, S.; Wang, S.; Chen, X.; Dong, J.; Shi, X.; Xia, Y.; Wang, X.; Hu, Z.; Shen, H. Polymorphisms in HPV E6/E7 protein interacted genes and risk of cervical cancer in Chinese women: A case-control analysis. Gynecol. Oncol. 2009, 114, 327–331. [Google Scholar] [CrossRef]
- Sauer, M.K.; Andrulis, I.L. Identification and characterization of missense alterations in the BRCA1 associated RING domain (BARD1) gene in breast and ovarian cancer. J. Med. Genet. 2005, 42, 633–638. [Google Scholar]
- Stacey, S.N.; Sulem, P.; Johannsson, O.T.; Helgason, A.; Gudmundsson, J.; Kostic, J.P.; Kristjansson, K.; Jonsdottir, T.; Sigurdsson, H.; Hrafnkelsson, J.; et al. The BARD1 Cys557Ser variant and breast cancer risk in Iceland. PLoS Med. 2006, 3, e217. [Google Scholar] [CrossRef]
- Ramus, S.J.; Song, H.; Dicks, E.; Tyrer, J.P.; Rosenthal, A.N.; Intermaggio, M.P.; Fraser, L.; Gentry-Maharaj, A.; Hayward, J.; Philpott, S.; et al. Germline Mutations in the BRIP1, BARD1, PALB2, and NBN Genes in Women With Ovarian Cancer. J. Natl. Cancer Inst. 2015, 107, djv214. [Google Scholar] [PubMed]
- De Brakeleer, S.; de Grève, J.; Desmedt, C.; Joris, S.; Sotiriou, C.; Piccart, M.; Pauwels, I.; Teugels, E. Frequent incidence of BARD1-truncating mutations in germline DNA from triple-negative breast cancer patients. Clin. Genet. 2016, 89, 336–340. [Google Scholar] [CrossRef]
- De Brakeleer, S.; de Grève, J.; Loris, R.; Janin, N.; Lissens, W.; Sermijn, E.; Teugels, E. Cancer predisposing missense and protein truncating BARD1 mutations in non-BRCA1 or BRCA2 breast cancer families. Hum. Mutat. 2010, 31, E1175–E1185. [Google Scholar] [CrossRef]
- Klonowska, K.; Ratajska, M.; Czubak, K.; Kuzniacka, A.; Brozek, I.; Koczkowska, M.; Sniadecki, M.; Debniak, J.; Wydra, D.; Balut, M.; et al. Analysis of large mutations in BARD1 in patients with breast and/or ovarian cancer: The Polish population as an example. Sci. Rep. 2015, 5, 10424. [Google Scholar] [CrossRef]
- Gorringe, K.L.; Choong, D.Y.H.; Visvader, J.E.; Lindeman, G.J.; Campbell, I.G. BARD1 variants are not associated with breast cancer risk in Australian familial breast cancer. Breast Cancer Res. Treat. 2008, 111, 505–509. [Google Scholar] [CrossRef]
- Andre, P.A.; Prele, C.M.; Vierkotten, S.; Carnesecchi, S.; Donati, Y.; Chambers, R.C.; Pache, J.C.; Crestani, B.; Barazzone-Argiroffo, C.; Konigshoff, M.; et al. BARD1 mediates TGF-beta signaling in pulmonary fibrosis. Respir. Res. 2015, 16, 118. [Google Scholar] [CrossRef]
- Choudhury, A.D.; Xu, H.; Modi, A.P.; Zhang, W.; Ludwig, T.; Baer, R. Hyperphosphorylation of the BARD1 tumor suppressor in mitotic cells. J. Biol. Chem. 2005, 280, 24669–24679. [Google Scholar] [CrossRef]
- Han, Z.J.; Feng, Y.H.; Gu, B.H.; Li, Y.M.; Chen, H. The post-translational modification, SUMOylation, and cancer (Review). Int. J. Oncol. 2018, 52, 1081–1094. [Google Scholar] [CrossRef]
- Xiao, Z.; Chang, J.G.; Hendriks, I.A.; Sigurethsson, J.O.; Olsen, J.V.; Vertegaal, A.C. System-wide Analysis of SUMOylation Dynamics in Response to Replication Stress Reveals Novel Small Ubiquitin-like Modified Target Proteins and Acceptor Lysines Relevant for Genome Stability. Mol. Cell. Proteom. 2015, 14, 1419–1434. [Google Scholar] [CrossRef]
- Hendriks, I.A.; Lyon, D.; Young, C.; Jensen, L.J.; Vertegaal, A.C.; Nielsen, M.L. Site-specific mapping of the human SUMO proteome reveals co-modification with phosphorylation. Nat. Struct. Mol. Biol. 2017, 24, 325–336. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Sato, K.; Koike, A.; Nishikawa, H.; Koizumi, H.; Venkitaraman, A.R.; Ohta, T. HERC2 is an E3 ligase that targets BRCA1 for degradation. Cancer Res. 2010, 70, 6384–6392. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Lu, G.; Li, L.; Yi, J.; Yan, K.; Wang, Y.; Zhu, B.; Kuang, J.; Lin, M.; Zhang, S.; et al. HUWE1 interacts with BRCA1 and promotes its degradation in the ubiquitin-proteasome pathway. Biochem. Biophys. Res. Commun. 2014, 444, 549–554. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Li, J.; Cheng, D.; Parameswaran, B.; Zhang, S.; Jiang, Z.; Yew, P.R.; Peng, J.; Ye, Q.; Hu, Y. The F-box protein FBXO44 mediates BRCA1 ubiquitination and degradation. J. Biol. Chem. 2012, 287, 41014–41022. [Google Scholar] [CrossRef]
- Deng, J.; Zhang, T.; Liu, F.; Han, Q.; Li, Q.; Guo, X.; Ma, Y.; Li, L.; Shao, G. CRL4-DCAF8L2 E3 ligase promotes ubiquitination and degradation of BARD1. Biochem. Biophys. Res. Commun. 2022, 611, 107–113. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Han, Q.; Zhang, T.; Chang, F.; Deng, J.; Huang, X.; Wang, W.; Xu, Y.; Li, Q.; Xu, L.; et al. CRL4-DCAF8L1 Regulates BRCA1 and BARD1 Protein Stability. Int. J. Biol. Sci. 2022, 18, 1434–1450. [Google Scholar] [CrossRef]
- Song, L.; Rape, M. Regulated degradation of spindle assembly factors by the anaphase-promoting complex. Mol. Cell 2010, 38, 369–382. [Google Scholar] [CrossRef]
- Minten, E.V.; Kapoor-Vazirani, P.; Li, C.; Zhang, H.; Balakrishnan, K.; Yu, D.S. SIRT2 promotes BRCA1-BARD1 heterodimerization through deacetylation. Cell. Rep. 2021, 34, 108921. [Google Scholar] [CrossRef]
- Lin, Z.; Fang, D. The Roles of SIRT1 in Cancer. Genes Cancer 2013, 4, 97–104. [Google Scholar] [CrossRef] [PubMed]
- Fiorentino, F.; Carafa, V.; Favale, G.; Altucci, L.; Mai, A.; Rotili, D. The Two-Faced Role of SIRT6 in Cancer. Cancers 2021, 13, 1156. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Chen, W. Emerging Roles of SIRT1 in Cancer Drug Resistance. Genes Cancer 2013, 4, 82–90. [Google Scholar] [CrossRef] [PubMed]
- Irminger-Finger, I.; Ratajska, M.; Pilyugin, M. New concepts on BARD1: Regulator of BRCA pathways and beyond. Int. J. Biochem. Cell Biol. 2016, 72, 1–17. [Google Scholar] [CrossRef]
- Wang, Y.Q.; Yan, Q.; Zhang, J.R.; Li, S.D.; Yang, Y.X.; Wan, X.P. Epigenetic inactivation of BRCA1 through promoter hypermethylation in ovarian cancer progression. J. Obstet. Gynaecol. Res. 2013, 39, 549–554. [Google Scholar] [CrossRef]
- Zhang, L.; Long, X. Association of BRCA1 promoter methylation with sporadic breast cancers: Evidence from 40 studies. Sci. Rep. 2015, 5, 17869. [Google Scholar] [CrossRef]
- Li, L.; Cohen, M.; Wu, J.; Sow, M.H.; Nikolic, B.; Bischof, P.; Irminger-Finger, I. Identification of BARD1 splice-isoforms involved in human trophoblast invasion. Int. J. Biochem. Cell Biol. 2007, 39, 1659–1672. [Google Scholar] [CrossRef]
- Bayraktar, E.; Rodriguez, C.; Kumar, S.; Stur, E.; Mangala, L.S.; Baylin, S.; Lopez-Berestein, G.; Pradeep, S.; Sood, A.K. Abstract 6001: Epigenetic regulation of BARD1 confers resistance to anti-VEGF therapy in ovarian cancer. In Proceedings of the American Association for Cancer Research Annual Meeting 2023, Orlando, FL, USA, 14–19 April 2023. [Google Scholar]
- Lubecka, K.; Flower, K.; Beetch, M.; Qiu, J.; Kurzava, L.; Buvala, H.; Ruhayel, A.; Gawrieh, S.; Liangpunsakul, S.; Gonzalez, T.; et al. Loci-specific differences in blood DNA methylation in HBV-negative populations at risk for hepatocellular carcinoma development. Epigenetics 2018, 13, 605–626. [Google Scholar] [CrossRef]
- Murfuni, I.; Basile, G.; Subramanyam, S.; Malacaria, E.; Bignami, M.; Spies, M.; Franchitto, A.; Pichierri, P. Survival of the replication checkpoint deficient cells requires MUS81-RAD52 function. PLoS Genet. 2013, 9, e1003910. [Google Scholar] [CrossRef] [PubMed]
- Sporn, J.C.; Hothorn, T.; Jung, B. BARD1 expression predicts outcome in colon cancer. Clin. Cancer Res. 2011, 17, 5451–5462. [Google Scholar] [CrossRef]
- McDougall, L.I.; Powell, R.M.; Ratajska, M.; Lynch-Sutherland, C.F.; Hossain, S.M.; Wiggins, G.A.R.; Harazin-Lechowska, A.; Cybulska-Stopa, B.; Motwani, J.; Macaulay, E.C.; et al. Differential Expression of BARD1 Isoforms in Melanoma. Genes 2021, 12, 320. [Google Scholar] [CrossRef] [PubMed]
- Ozden, O.; Bishehsari, F.; Bauer, J.; Park, S.H.; Jana, A.; Baik, S.H.; Sporn, J.C.; Staudacher, J.J.; Yazici, C.; Krett, N.; et al. Expression of an Oncogenic BARD1 Splice Variant Impairs Homologous Recombination and Predicts Response to PARP-1 Inhibitor Therapy in Colon Cancer. Sci. Rep. 2016, 6, 26273. [Google Scholar] [CrossRef] [PubMed]
- Dizin, E.; Irminger-Finger, I. Negative feedback loop of BRCA1-BARD1 ubiquitin ligase on estrogen receptor alpha stability and activity antagonized by cancer-associated isoform of BARD1. Int. J. Biochem. Cell Biol. 2010, 42, 693–700. [Google Scholar] [CrossRef]
- Jasiak, A.; Krawczynska, N.; Iliszko, M.; Czarnota, K.; Buczkowski, K.; Stefanowicz, J.; Adamkiewicz-Drozynska, E.; Cichosz, G.; Izycka-Swieszewska, E. Expression of BARD1 beta Isoform in Selected Pediatric Tumors. Genes 2021, 12, 168. [Google Scholar] [CrossRef]
- Wiener, D.; Gajardo-Meneses, P.; Ortega-Hernandez, V.; Herrera-Cares, C.; Diaz, S.; Fernandez, W.; Cornejo, V.; Gamboa, J.; Tapia, T.; Alvarez, C.; et al. BRCA1 and BARD1 colocalize mainly in the cytoplasm of breast cancer tumors, and their isoforms show differential expression. Breast Cancer Res. Treat. 2015, 153, 669–678. [Google Scholar] [CrossRef]
- Wu, J.Y.; Vlastos, A.T.; Pelte, M.F.; Caligo, M.A.; Bianco, A.; Krause, K.H.; Laurent, G.J.; Irminger-Finger, I. Aberrant expression of BARD1 in breast and ovarian cancers with poor prognosis. Int. J. Cancer 2006, 118, 1215–1226. [Google Scholar]
- Zhang, Y.Q.; Bianco, A.; Malkinson, A.M.; Leoni, V.P.; Frau, G.; De Rosa, N.; Andre, P.A.; Versace, R.; Boulvain, M.; Laurent, G.J.; et al. BARD1: An independent predictor of survival in non-small cell lung cancer. Int. J. Cancer 2012, 131, 83–94. [Google Scholar]
- Li, L.; Ryser, S.; Dizin, E.; Pils, D.; Krainer, M.; Jefford, C.E.; Bertoni, F.; Zeillinger, R.; Irminger-Finger, I. Oncogenic BARD1 isoforms expressed in gynecological cancers. Cancer Res. 2007, 67, 11876–11885. [Google Scholar] [CrossRef] [PubMed]
- Lepore, I.; Dell’Aversana, C.; Pilyugin, M.; Conte, M.; Nebbioso, A.; De Bellis, F.; Tambaro, F.P.; Izzo, T.; Garcia-Manero, G.; Ferrara, F.; et al. HDAC inhibitors repress BARD1 isoform expression in acute myeloid leukemia cells via activation of miR-19a and/or b. PLoS ONE 2013, 8, e83018. [Google Scholar] [CrossRef] [PubMed]
- Bosse, K.R.; Diskin, S.J.; Cole, K.A.; Wood, A.C.; Schnepp, R.W.; Norris, G.; Nguyen le, B.; Jagannathan, J.; Laquaglia, M.; Winter, C.; et al. Common variation at BARD1 results in the expression of an oncogenic isoform that influences neuroblastoma susceptibility and oncogenicity. Cancer Res. 2012, 72, 2068–2078. [Google Scholar] [CrossRef]
- Zhang, Y.Q.; Pilyugin, M.; Kuester, D.; Leoni, V.P.; Li, L.; Casula, G.; Zorcolo, L.; Schneider-Stock, R.; Atzori, L.; Irminger-Finger, I. Expression of oncogenic BARD1 isoforms affects colon cancer progression and correlates with clinical outcome. Br. J. Cancer 2012, 107, 675–683. [Google Scholar] [CrossRef]
- Pilyugin, M.; André, P.; Ratajska, M.; Kuzniacka, A.; Limon, J.; Tournier, B.B.; Colas, J.; Laurent, G.J.; Irminger-Finger, I. Antagonizing functions of BARD1 and its alternatively spliced variant BARD1δ in telomere stability. Oncotarget 2017, 8, 9339–9353. [Google Scholar] [CrossRef]
- Marzec, K.A.; Martino-Echarri, E.; Irminger-Finger, I.; Henderson, B.R. BARD1 splice variants display mislocalization in breast cancer cells and can alter the apoptotic response to cisplatin. Cancer Lett. 2016, 381, 149–155. [Google Scholar] [CrossRef] [PubMed]
- Herrera, C.P.; Pérez, E.; Blanco, E.; Gajardo, P.; Carvallo, P. Abstract 2455: BRCA1 and BARD1 expression and localization in breast cell lines. In Proceedings of the 105th Annual Meeting of the American Association for Cancer Research, San Diego, CA, USA, 5–9 April 2014. [Google Scholar]
- Pilyugin, M.; Irminger-Finger, I. Long non-coding RNA and microRNAs might act in regulating the expression of BARD1 mRNAs. Int. J. Biochem. Cell Biol. 2014, 54, 356–367. [Google Scholar] [CrossRef]
- Finan, J.M.; Sutton, T.L.; Dixon, D.A.; Brody, J.R. Targeting the RNA-Binding Protein HuR in Cancer. Cancer Res. 2023, 83, 3507–3516. [Google Scholar] [CrossRef]
- Jain, A.; McCoy, M.; Coats, C.; Brown, S.Z.; Addya, S.; Pelz, C.; Sears, R.C.; Yeo, C.J.; Brody, J.R. HuR Plays a Role in Double-Strand Break Repair in Pancreatic Cancer Cells and Regulates Functional BRCA1-Associated-Ring-Domain-1(BARD1) Isoforms. Cancers 2022, 14, 1848. [Google Scholar] [CrossRef]
- Blanco, F.F.; Jimbo, M.; Wulfkuhle, J.; Gallagher, I.; Deng, J.; Enyenihi, L.; Meisner-Kober, N.; Londin, E.; Rigoutsos, I.; Sawicki, J.A.; et al. The mRNA-binding protein HuR promotes hypoxia-induced chemoresistance through posttranscriptional regulation of the proto-oncogene PIM1 in pancreatic cancer cells. Oncogene 2016, 35, 2529–2541. [Google Scholar] [CrossRef]
- Brody, J.R.; Dixon, D.A. Complex HuR function in pancreatic cancer cells. Wiley Interdiscip. Rev. RNA 2018, 9, e1469. [Google Scholar] [CrossRef]
- Dhanasekaran, R.; Deutzmann, A.; Mahauad-Fernandez, W.D.; Hansen, A.S.; Gouw, A.M.; Felsher, D.W. The MYC oncogene—the grand orchestrator of cancer growth and immune evasion. Nat. Rev. Clin. Oncol. 2022, 19, 23–36. [Google Scholar] [CrossRef] [PubMed]
- Patel, S.; Jenkins, E.; Kusurkar, R.P.; Lee, S.; Jiang, W.; Nevler, A.; McCoy, M.; Pishvaian, M.J.; Sears, R.C.; Brody, J.R.; et al. Targeting BARD1 suppresses a Myc-dependent transcriptional program and tumor growth in pancreatic ductal adenocarcinoma. Neoplasia 2025, 63, 101152. [Google Scholar] [CrossRef] [PubMed]
- Oliver, T.G.; Mercer, K.L.; Sayles, L.C.; Burke, J.R.; Mendus, D.; Lovejoy, K.S.; Cheng, M.H.; Subramanian, A.; Mu, D.; Powers, S.; et al. Chronic cisplatin treatment promotes enhanced damage repair and tumor progression in a mouse model of lung cancer. Genes Dev. 2010, 24, 837–852. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.E.; Yin, M.; Dong, Q.; Stewart, D.J.; Merriman, K.W.; Amos, C.I.; Spitz, M.R.; Wei, Q. DNA repair capacity in peripheral lymphocytes predicts survival of patients with non-small-cell lung cancer treated with first-line platinum-based chemotherapy. J. Clin. Oncol. 2011, 29, 4121–4128. [Google Scholar] [CrossRef]
- Parsels, L.A.; Engelke, C.G.; Parsels, J.; Flanagan, S.A.; Zhang, Q.; Tanska, D.; Wahl, D.R.; Canman, C.E.; Lawrence, T.S.; Morgan, M.A. Combinatorial Efficacy of Olaparib with Radiation and ATR Inhibitor Requires PARP1 Protein in Homologous Recombination-Proficient Pancreatic Cancer. Mol. Cancer Ther. 2021, 20, 263–273. [Google Scholar] [CrossRef]
- Dong, W.; Li, L.; Teng, X.; Yang, X.; Si, S.; Chai, J. End Processing Factor APLF Promotes NHEJ Efficiency and Contributes to TMZ- and Ionizing Radiation-Resistance in Glioblastoma Cells. Onco Targets Ther. 2020, 13, 10593–10605. [Google Scholar] [CrossRef]
- Oyama, S.; Matsuda, A.; Murakami, R.; Kakizaki, Y.; Ishizawa, T.; Kobayashi, T.; Nakamura, H.; Nawa, Y.; Otaki, Y.; Nagata, Y.; et al. Pancreatic cancer organoids derived from EUS-guided fine needle aspiration specimens can be used to predict chemotherapy resistance. Sci. Rep. 2025, 15, 23818. [Google Scholar] [CrossRef]
- Mathews, L.A.; Cabarcas, S.M.; Hurt, E.M.; Zhang, X.; Jaffee, E.M.; Farrar, W.L. Increased expression of DNA repair genes in invasive human pancreatic cancer cells. Pancreas 2011, 40, 730–739. [Google Scholar] [CrossRef]
- Zhu, Y.; Liu, Y.; Zhang, C.; Chu, J.; Wu, Y.; Li, Y.; Liu, J.; Li, Q.; Li, S.; Shi, Q.; et al. Tamoxifen-resistant breast cancer cells are resistant to DNA-damaging chemotherapy because of upregulated BARD1 and BRCA1. Nat. Commun. 2018, 9, 1595. [Google Scholar] [CrossRef]
- Pantelidou, C.; Sonzogni, O.; De Oliveria Taveira, M.; Mehta, A.K.; Kothari, A.; Wang, D.; Visal, T.; Li, M.K.; Pinto, J.; Castrillon, J.A.; et al. PARP Inhibitor Efficacy Depends on CD8(+) T-cell Recruitment via Intratumoral STING Pathway Activation in BRCA-Deficient Models of Triple-Negative Breast Cancer. Cancer Discov. 2019, 9, 722–737. [Google Scholar] [CrossRef]
- Gurjao, C.; Liu, D.; Hofree, M.; AlDubayan, S.H.; Wakiro, I.; Su, M.J.; Felt, K.; Gjini, E.; Brais, L.K.; Rotem, A.; et al. Intrinsic Resistance to Immune Checkpoint Blockade in a Mismatch Repair-Deficient Colorectal Cancer. Cancer Immunol. Res. 2019, 7, 1230–1236. [Google Scholar] [CrossRef] [PubMed]
- Gu, P.; Xue, L.; Zhao, C.; Li, W.; Jiang, Z.; Liu, A.; Li, T.; Liu, L.; Decker, M.; Cheng, X.; et al. Targeting the Homologous Recombination Pathway in Cancer With a Novel Class of RAD51 Inhibitors. Front. Oncol. 2022, 12, 885186. [Google Scholar] [CrossRef] [PubMed]
- U.S. Food and Drug Administration. FDA Approves Lynparza for Treatment of Germline BRCA-Mutated Metastatic Pancreatic Cancer. Available online: https://www.fda.gov/drugs/resources-information-approved-drugs/fda-approves-olaparib-gbrcam-metastatic-pancreatic-adenocarcinoma (accessed on 10 September 2025).
- Park, W.; Connor, C.O.’; Chou, J.; Schwartz, C.; Larsen, M.; Varghese, A.; Yu, K.; Balogun, F.; Yang, J.; Katz, S.; et al. 1504MO Phase II trial of Pembrolizumab and OLApaRib (POLAR) maintenance for select patients (pts) with metastatic pancreatic cancer (mPC) with (A) homologous recombination deficiency (HRD), (B) non-core HRD (ncHRD) and (C) exceptional response to platinum. Ann. Oncol. 2024, 35, S922. [Google Scholar] [CrossRef]
- Phase II Trial of Ceralasertib (AZD6738) Alone and in Combination with Olaparib or Durvalumab in Patients with Selected Solid Tumor Malignancies; AstraZeneca: Singapore, 2018.
- Olaparib for BRCAness Phenotype in Pancreatic Cancer: Phase II Study; National Cancer Institute and AstraZeneca: Singapore, 2016.
- A Phase 2 Study of Cediranib in Combination with Olaparib in Advanced Solid Tumors; AstraZeneca: Singapore, 2015.
- A Window of Opportunity Strategy for Targeted Pathway Inhibition in Patients with Pancreatic Ductal Adenocarcinoma; American Association for Cancer: Philadelphia, PA, USA, 2019.
- Ko, B.; Coyne, G.H.; Naqash, A.R.; George, T.T.; DeRemer, D.; Rubinstein, L.; Zlott, J.; Wilsker, D.; Ketchum, D.; Runkle, S.; et al. Abstract B138: Pharmacodynamics-driven phase 2 trial of talazoparib in patients with advanced solid tumors and aberrations in genes involved in DNA damage response. Mol. Cancer Ther. 2023, 22 (Suppl. 12), B138. [Google Scholar] [CrossRef]
- Rodon Ahnert, J.; Tan, D.S.; Garrido-Laguna, I.; Harb, W.; Bessudo, A.; Beck, J.T.; Rottey, S.; Bahary, N.; Kotecki, N.; Zhu, Z.; et al. Avelumab or talazoparib in combination with binimetinib in metastatic pancreatic ductal adenocarcinoma: Dose-finding results from phase Ib of the JAVELIN PARP MEKi trial. ESMO Open 2023, 8, 101584. [Google Scholar] [CrossRef] [PubMed]
- Phase 2 Proof-of-Concept Trial Testing the PARP Inhibitor Niraparib in Patients with Pancreatic Cancer Harboring Deficiencies in Homologous Recombination DNA Repair; Tesaro, Inc.: Waltham, MA, USA, 2018.
- Niraparib in Metastatic Pancreatic Cancer After Previous Chemotherapy (NIRA-PANC): A Phase 2 Trial; Tesaro, Inc.: Waltham, MA, USA, 2018.
- Domchek, S.M.; McWilliams, R.R.; Hendifar, A.H.; Shroff, R.T.; Leichman, L.P.; Epelbaum, R.; Geva, R.; Kim, G.P.; Alberts, S.R.; Wolff, R.A.; et al. A phase 2, open-label study of rucaparib in patients with pancreatic cancer and a known deleterious BRCA mutation. J. Clin. Oncol. 2014, 32, TPS4161. [Google Scholar] [CrossRef]
- O’Reilly, E.M.; Lee, J.W.; Zalupski, M.; Capanu, M.; Park, J.; Golan, T.; Tahover, E.; Lowery, M.A.; Chou, J.F.; Sahai, V.; et al. Randomized, Multicenter, Phase II Trial of Gemcitabine and Cisplatin With or Without Veliparib in Patients With Pancreas Adenocarcinoma and a Germline BRCA/PALB2 Mutation. J. Clin. Oncol. 2020, 38, 1378–1388. [Google Scholar] [CrossRef]
- AbbVie. AbbVie Receives FDA Orphan Drug Designation for Investigational Medicine Veliparib for the Treatment of Advanced Squamous Non-Small Cell Lung Cancer; PRNewswire: North Chicago, IL, USA, 2016. [Google Scholar]
- Jain, A.; Agostini, L.C.; McCarthy, G.A.; Chand, S.N.; Ramirez, A.; Nevler, A.; Cozzitorto, J.; Schultz, C.W.; Lowder, C.Y.; Smith, K.M.; et al. Poly (ADP) Ribose Glycohydrolase Can Be Effectively Targeted in Pancreatic Cancer. Cancer Res. 2019, 79, 4491–4502. [Google Scholar] [CrossRef]
- A Study of PARG Inhibitor IDE161 in Participants with Advanced Solid Tumors; IDEAYA Biosciences: San Diego, CA, USA, 2023.
- Ravindranathan, R.; Somuncu, O.; da Costa, A.; Mukkavalli, S.; Lamarre, B.P.; Nguyen, H.; Grochala, C.; Jiao, Y.; Liu, J.; Kochupurakkal, B.; et al. PARG inhibitor sensitivity correlates with accumulation of single-stranded DNA gaps in preclinical models of ovarian cancer. Proc. Natl. Acad. Sci. USA 2024, 121, e2413954121. [Google Scholar] [CrossRef]
- Villaruz, L.C.; Kelly, K.; Wagar, S.N.; Davis, E.J.; Shapiro, G.; LoRusso, P.; Dees, E.C.; Normolle, D.P.; Rhee, J.C.; Chu, E.; et al. NCI 9938: Phase I clinical trial of ATR inhibitor berzosertib (M6620, VX-970) in combination with irinotecan in patients with advanced solid tumors. J. Clin. Oncol. 2022, 40, 3012. [Google Scholar] [CrossRef]
- Phase 1 Trial of Gemcitabine Combined with the Elimusertib (BAY 1895344) ATR Inhibitor with Expansion Cohorts in Advanced Pancreatic and Ovarian Cancer; NCI: Bethesda, MD, USA, 2020.
- Phase 2 Single Arm Trial Testing the ZN-c3 WEE1 Inhibitor in Combination with Gemcitabine in Second-Line Advanced Pancreatic Adenocarcinoma; Lustgarten, F., K-Group, Beta, Inc., Stand Up To Cancer: Boston, MA, USA, 2023.
- A Phase 1/2 Dose Escalation and Cohort Expansion Study of LP-184 in Patients with Advanced or Metastatic Solid Tumors; Lantern Pharma, Inc.: Dallas, TX, USA, 2023.
- A Phase 1 Study of MOMA-313 Given as Monotherapy or in Combination with a PARP Inhibitor in Participants with Advanced or Metastatic Solid Tumors; MOMA therap.: Cambridge, MA, USA, 2024.
- Calheiros, J.; Silva, R.; Barbosa, F.; Morais, J.; Moura, S.R.; Almeida, S.; Fiorini, E.; Mulhovo, S.; Aguiar, T.Q.; Wang, T.; et al. A first-in-class inhibitor of homologous recombination DNA repair counteracts tumour growth, metastasis and therapeutic resistance in pancreatic cancer. J. Exp. Clin. Cancer Res. 2025, 44, 129. [Google Scholar] [PubMed]
- Li, M.; Yu, X. Function of BRCA1 in the DNA damage response is mediated by ADP-ribosylation. Cancer Cell 2013, 23, 693–704. [Google Scholar] [CrossRef] [PubMed]
- Pilyugin, M.; Ratajska, M.; Stukan, M.; Concin, N.; Zeillinger, R.; Irminger-Finger, I. BARD1 Autoantibody Blood Test for Early Detection of Ovarian Cancer. Genes 2021, 12, 969. [Google Scholar] [CrossRef] [PubMed]
- Pilyugin, M.; Descloux, P.; André, P.A.; Laszlo, V.; Dome, B.; Hegedus, B.; Sardy, S.; Janes, S.; Bianco, A.; Laurent, G.J.; et al. BARD1 serum autoantibodies for the detection of lung cancer. PLoS ONE 2017, 12, e0182356. [Google Scholar]
| Variant Type | dbSNP ID | Coding Change | Amino Acid Change | Exon | Protein Domain | ClinVar Classification | Reference |
|---|---|---|---|---|---|---|---|
| Nonsense | rs762171436 | c.632T > A | p.Leu211Ter | 4 | None | Pathogenic/Likely Pathogenic | Chaffee et al., 2018 [47] |
| Frameshift | rs28997575 | c.1075_1095del | p.Leu359_Pro365del | 4 | None | Benign/Likely Benign | Amemiya et al., 2015 [65]; Hu et al., 2016 [46] |
| Missense | rs2229571 | c.1134G > C | p.Arg378Ser | 4 | None | Benign | Hu et al., 2016 [46]; Zhou et al., 2009 [66] |
| Missense | rs2070094 | c.1519G > A | p.Val507Met | 6 | ANK | Benign/Likely Benign | Sauer et al., 2005 [67]; Hu et al., 2016 [46] |
| Missense | rs28997576 | c.1670G > C | p.Cys557Ser | 7 | None | Benign | Stacey et al., 2006 [68]; Hu et al., 2016 [46] |
| Missense | rs730881420 | c.1685C > T | p.Thr562Ile | 8 | None | Uncertain Significance | Hu et al., 2016 [46] |
| Missense | rs587782279 | c.1693C > T | p.Arg565Cys | 7 | None | Uncertain Significance | Chaffee et al., 2018 [47] |
| Missense | rs35306212 | c.1738G > A | p.Glu580Lys | 8 | BRCT | Benign/Likely Benign | Hu et al., 2016 [46] |
| Nonsense | rs587781948 | c.1921C > T | p.Arg641Ter | 10 | BRCT | Pathogenic | Ramus et al., 2015 [69]; De Brakeleer et al., 2016 [70], Hu et al., 2016 [46] |
| Missense | rs3738888 | c.1972C > T | p.Arg658Cys | 10 | None | Benign/Likely Benign | De Brakeleer et al., 2010 [71]; Klonowska et al., 2015 [72]; Hu et al., 2016 [46] |
| Missense | rs61754118 | c.2212A > G | p.Ile738Val | 11 | BRCT | Benign/Likely Benign | Sauer et al., 2005 [67]; Gorringe et al., 2008 [73]; Hu et al., 2016 [46] |
| NCT Number | Phase | Interventions (Mechanism of Action) | Patient Population | Primary Endpoint |
|---|---|---|---|---|
| NCT03601923 | II | Niraparib (PARP inhibitor) | Patients with advanced pancreatic adenocarcinoma with mutations in BRCA1, BRCA2, PALB2, CHEK2, or ATM | PFS at 6 months |
| NCT03553004 | II | Niraparib (PARP inhibitor) | Patients with metastatic PDAC exposed to prior chemotherapy with genes involved in DNA repair | ORR at 8 weeks |
| NCT04550494 | II | Talazoparib (PARP inhibitor) | Patients with solid tumors and documented aberrations in DDR-related genes, including BRCA1/2 and BARD1 | Percent of patients who demonstrate simultaneous RAD51 activation and lack of γ-H2AX activation, defined as ≥5% of cells with ≥5 RAD51 foci and <4% nuclear area positive for γ-H2AX at the cycle two, day one biopsy |
| NCT03140670 | II | Rucaparib (PARP inhibitor) | Patients with pancreatic adenocarcinoma on platinum-based treatment and documented BRCA1/2 or PALB2 mutation | PFS at 6 months |
| NCT02677038 | II | Olaparib (PARP inhibitor) | Patients with stage IV PDAC with any genetic alterations conferring HRD, with the exception of germline BRCA1/2 | ORR at 5 years and 8 months |
| NCT04666740 | II | Pembrolizumab (PD-L1 inhibitor) + Olaparib (PARP inhibitor) in maintenance setting | Patients with metastatic pancreatic adenocarcinoma or acinar cell carcinoma with genetic alterations conferring HRD with stable disease on platinum treatment Note: Patients are stratified into core HR genes (BRCA1/2, PALB2) and non-core HR genes (including BARD1) | PFS at 6 months |
| NCT02498613 | II | Cediranib (VEGF inhibitor) + Olaparib (PARP inhibitor) | Metastatic or unresectable NSCLC, TNBC, PDAC, or SCLC with at least 1 prior line of systemic treatment | ORR at 43 months |
| NCT01585805 | II | Veliparib (PARP inhibitor) vs. Veliparib + Gemcitabine hydrochloride (nucleoside analog) + cisplatin (alkylating agent) vs. Gemcitabine + cisplatin | Metastatic pancreatic adenocarcinoma with BRCA1/2 or PALB2 mutation | OD at 21 days and RR at 5 years |
| NCT05933265 | I/II | LP-184 (alkylating prodrug) vs. LP-184 + spironolactone (mineralocorticoid receptor antagonist) vs. LP-184 + Olaparib (PARP inhibitor), in TNBC subset only | Patients with advanced solid tumors, with a preference for tumor types with high prevalence of DDR gene mutations (TNBC, lung, prostate, ovarian, pancreatic, bladder, and glioblastoma) | Incidence and severity of all AEs, MTD, and recommended Phase II dose at 12 months |
| NCT06545942 | I | MOMA-313 (DNA polymerase θ helicase inhibitor) vs. MOMA-313 + Olaparib (PARP inhibitor) | Patients with advanced or metastatic solid tumors that are not eligible for curative therapy, with any genetic alterations conferring HRD | Number of participants with AEs, DLTs, SAEs, and/or AEs leading to discontinuation |
| NCT03682289 | II | Ceralasertib (ATR kinase inhibitor) vs. ceralasertib + Olaparib (PARP inhibitor) vs. ceralasertib + durvalumab (immunotherapy) | Patients with a solid tumor malignancy with progression on at least one prior systemic therapy, including all pancreatic cancers | ORR at 3 years |
| NCT06015659 | II | ZN-c3 (Wee1 inhibitor) + gemcitabine (nucleoside analog) | Advanced pancreatic adenocarcinoma in the second line | PFS at 6 months |
| NCT04616534 | I | Elimusertib (ATR kinase inhibitor) + gemcitabine (nucleoside analog) | Patients with advanced pancreatic adenocarcinoma or ovarian cancer | MTD, incidence of AEs, ORR, DoR, PFS, and OS at 1 year |
| NCT02595931 | I | Berzosertib/M6620 (ATR kinase inhibitor) + irinotecan (topoisomerase I inhibitor) | Patients with solid tumors that are metastatic or unresectable, with known deficiencies in DDR genes | MTD |
| NCT05787587 | I | IDE-161 (PARG inhibitor) vs. IDE-161 + pembrolizumab (immunotherapy) | Patients with metastatic solid tumors with genetic alterations conferring HRD, including BRCA1/2 and BARD1 | Incidence of DLTs, AEs, TEAEs |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zekavat, L.; Jain, A. BARD1: A Friend or Foe in Pancreatic Ductal Adenocarcinoma? Int. J. Mol. Sci. 2025, 26, 9041. https://doi.org/10.3390/ijms26189041
Zekavat L, Jain A. BARD1: A Friend or Foe in Pancreatic Ductal Adenocarcinoma? International Journal of Molecular Sciences. 2025; 26(18):9041. https://doi.org/10.3390/ijms26189041
Chicago/Turabian StyleZekavat, Lily, and Aditi Jain. 2025. "BARD1: A Friend or Foe in Pancreatic Ductal Adenocarcinoma?" International Journal of Molecular Sciences 26, no. 18: 9041. https://doi.org/10.3390/ijms26189041
APA StyleZekavat, L., & Jain, A. (2025). BARD1: A Friend or Foe in Pancreatic Ductal Adenocarcinoma? International Journal of Molecular Sciences, 26(18), 9041. https://doi.org/10.3390/ijms26189041






