Analysis of Non-Polar Low-Molecular Metabolites in Citron (Citrus medica L.) Peel Essential Oil at Different Developmental Stages and a Combined Study of Transcriptomics Revealed Genes Related to the Synthesis Regulation of the Monoterpenoid Compound Nerol
Abstract
1. Introduction
2. Results
2.1. Volatile Compound Analysis of Citron Peel Essential Oil at Different Developmental Stages
2.2. Identification of Key Differential Volatile Metabolites in the Peel Essential Oil of Citron at Different Developmental Stages
2.3. Transcriptome Sequencing and Analysis
2.3.1. Transcriptome Sequencing Data Analysis
2.3.2. Alignment and Annotation Information of Unigene Sequences
2.4. Transcription Factor Annotation
2.5. Analysis of Differentially Expressed Genes in Citrus Fruit Peel at Different Developmental Stages
2.6. Weighted Gene Co-Expression Network Analysis (WGCNA) of DEGs
2.7. Correlation Analysis of Differentially Expressed Genes and Terpenoid Metabolites at Different Developmental Stages
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Extraction and Collection of Fruit Peel Essential Oil
4.3. GC-MS Analysis of the Chemical Components of Citron Peel Essential Oil at Different Developmental Stages
4.4. Transcriptome Sequencing
4.4.1. RNA Extraction
4.4.2. cDNA Library Construction and Sequencing
4.4.3. Sequence Data Processing and Transcriptome Assembly
4.5. Functional Annotation of Unigenes and Annotation of Plant Transcription Factors
4.6. Integrated Analysis of Transcriptome and Metabolome
4.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| DEGs | Differentially expressed genes |
| QF | Green fruit |
| QHF | Green -yellow fruit |
| YF | Yellow fruit |
| WGCNA | Weighted Gene Co-expression Network Analysis |
| IPP | Isopentenyl diphosphate |
| DMAPP | Dimethylallyl diphosphate |
| MVA | Mevalonate pathway |
| MEP | Methylerythritol phosphate pathway |
| GPP | Geranyl diphosphate |
| FPP | Farnesyl diphosphate |
| GGPP | Geranylgeranyl diphosphate |
| TPS | Terpene synthases |
| GC-MS | Gas Chromatography-Mass Spectrometry |
| Q20 | Phred Quality Score of 20 |
| Q30 | Phred Quality Score of 30 |
| GC | Guanine-Cytosine Content |
| UID | Unique Identifier |
| NA | Not Available or Missing Data |
| TFs | Transcription Factors |
| GPPS | Geranyl Pyrophosphate Synthase |
| MAPK | Mitogen-activated protein kinase |
| ROS | Reactive Oxygen Species |
| cDNA | complementary Deoxyribonucleic Acid |
| FPKM | Fragments Per Kilobase of transcript per Million mapped reads |
| EMSA | Electrophoretic Mobility Shift Assay |
References
- Wu, G.A.; Terol, J.; Ibanez, V.; Lopez-Garcia, A.; Perez-Roman, E.; Borreda, C.; Domingo, C.; Tadeo, F.R.; Carbonell-Caballero, J.; Alonso, R.; et al. Genomics of the origin and evolution of Citrus. Nature 2018, 554, 311–316. [Google Scholar] [CrossRef]
- Jiang, H.; Li, Y.R.; Xu, Y.C.; Dong, C.L.; Zhai, M. Research on Cold Resistance of 10 Half-Sibling Citrus Family Progenies Seedlings. For. Sci. Technol. Dev. 2012, 26, 36–41. [Google Scholar]
- National Pharmacopoeia Commission. Pharmacopoeia of the People’s Republic of China, 11th ed.; China Medical Science and Technology Press: Beijing, China, 2020; p. 270. [Google Scholar]
- Liu, H.X.; Feng, D.; Long, C.R.; Zhou, X.Y.; Liu, H.M.; Yang, H.X.; Du, Y.X.; Guo, L.N.; Fu, X.M.; Ma, Z.C.; et al. Fruit variation and geographical distribution of citron. China J. Chin. Mater. Medica 2021, 23, 6289–6293. [Google Scholar]
- Liu, C.-Q.; Li, D.-J.; Niu, L.-Y.; Yu, M.; Song, J.-F. Research Progress in Development and Utilization of Citron. Jiangsu Agric. Sci. 2014, 42, 1–5. [Google Scholar]
- Gonzalez, M.C.; Rambla, J.L.; Lopez-Gresa, M.P.; Blazquez, M.A.; Granell, A. Volatile compounds in Citrus essential oils: A comprehensive review. Front. Plant Sci. 2019, 10, 12. [Google Scholar] [CrossRef] [PubMed]
- Hu, F.H.; Liang, L.X.; Huang, X.Y.; Yang, M.; Ma, G.Q.; Hang, F. Research Progress on the Mechanisms of Natural Plant Volatile Oils in the Intervention of Type 2 Diabetes Mellitus. Chin. J. Exp. Prescr. 2023, 29, 276–282. [Google Scholar]
- Ghani, A.; Taghvaeefard, N.; Hosseinifarahi, M.; Dakhlaoui, S.; Msaada, K. Essential oil composition and antioxidant activity of citron fruit (Citrus medica var. macrocarpa Risso) peel as relation to ripening stages. Int. J. Environ. Health Res. 2023, 33, 1278–1288. [Google Scholar]
- Liu, C.J.; Niu, L.Y.; Yu, M.; Li, D.J.; Liu, C.Q. In Vitro Antioxidant and Antimicrobial Activities of Citron Essential Oil. Food Ind. Technol. 2016, 37, 132–137. [Google Scholar]
- Keyvanara, A.H.; Yegdaneh, A.; Talebi, A.; Minaiyan, M. Evaluating anti-inflammatory effect of hydroalcoholic extracts of Citrus medica L. pulp and peel on rat model of acute colitis. Res. J. Pharmacogn. 2023, 10, 29–38. [Google Scholar]
- Entezari, M.; Majd, A.; Falahian, F.; Mehrabian, S.; Hashemi, M.; Lajimi, A.A. Antimutagenicity and anticancer effects of Citrus medica fruit juice. Acta Med. Iran. 2009, 47, 373–377. [Google Scholar]
- Zhang, Z.J. Study on the Intervention Effect and Mechanism of Herbal and Food Homologous Plant Extracts on Obesity Mice Induced by High Fat Diet. Master’s Thesis, Guangdong University of Technology, Guangdong, China, 29 May 2021. [Google Scholar]
- Mondal, M.; Saha, S.; Sarkar, C.; Hossen, M.S.; Hossain, M.S.; Khalipha, A.B.R.; Islam, M.F.; Wahed, T.B.; Islam, M.T.; Rauf, A.; et al. Role of Citrus medica L. Fruits Extract in Combatting the Hema-tological and Hepatic Toxic Effects of Carbofuran. Chem. Res. Toxicol. 2021, 34, 1890–1902. [Google Scholar] [CrossRef] [PubMed]
- Saravanan, S.; Pari, L. Protective effect of thymol on high fat diet induced diabetic nephropathy in C57BL/6J mice. Chem. Biol. Interact. 2016, 5, 1–11. [Google Scholar] [CrossRef]
- Deepa, B.; Anuradha, C.V. Effects of linalool on inflammation, matrix accumulation and podocyte loss in kidney of streptozotocin-induced diabetic rats. Toxicol. Mech. Methods 2013, 3, 223–234. [Google Scholar] [CrossRef]
- Murali, R.; Karthikeyan, A.; Saravanan, R. Protective effects of D-limonene on lipid peroxidation and antioxidant enzymes in streptozotocin-induced diabetic rats. Basic Clin. Pharmacol. Toxicol. 2013, 112, 175–181. [Google Scholar] [CrossRef]
- Santos, E.S.; Coelho, G.L.A.; Loula, Y.K.S.F.; Landim, B.L.S.; Lima, C.N.F.; Machado, S.T.D.S.; Lopes, M.J.P.; Gomes, A.D.S.; Costa, J.G.M.D.; Menezes, I.R.A.D.; et al. Hypoglycemic, Hypolipidemic, and Anti-Inflammatory Effects of Beta-Pinene in Diabetic Rats. Evid. Based Complement. Alternat Med. 2022, 17, 8173307. [Google Scholar] [CrossRef]
- Subramaniyan, S.D.; Natarajan, A.K. Citral, A Monoterpene Protect Against High Glucose Induced Oxidative Injury in HepG2 Cell In Vitro-An Experimental Study. J. Clin. Diagn. Res. 2017, 11, 10–15. [Google Scholar] [CrossRef]
- Babukumar, S.; Vinothkumar, V.; Sankaranarayanan, C.; Srinivasan, S. Geraniol, a natural monoterpene, ameliorates hyperglycemia by attenuating the key enzymes of carbohydrate metabolism in streptozotocin-induced diabetic rats. Pharm. Biol. 2017, 55, 1442–1449. [Google Scholar] [CrossRef]
- Bergman, M.E.; Davis, B.; Phillips, M.A. Medically Useful Plant Terpenoids: Biosynthesis, Occurrence, and Mechanism of Action. Molecules 2019, 24, 3961. [Google Scholar] [CrossRef]
- Vranova, E.; Coman, D.; Gruissem, W. Network analysis of the MVA and MEP pathways for isoprenoid synthesis. Annu. Rev. Plant Biol. 2013, 64, 665–700. [Google Scholar] [CrossRef] [PubMed]
- Banthorpe, D.V.; LePatourel, G.N.; Francis, M.J. Biosynthesis of geraniol and nerol and beta-D-glucosides in Pelargonium graveolens and Rosa dilecta. Biochem. J. 1972, 130, 1045–1054. [Google Scholar] [CrossRef] [PubMed]
- Kuzuyama, T. Mevalonate and nonmevalonate pathways for the biosynthesis of isoprene units. Biosci. Biotechnol. Biochem. 2002, 66, 1619–1627. [Google Scholar] [CrossRef] [PubMed]
- Degenhardt, J.; Kollner, T.G.; Gershenzon, J. Monoterpene and sesquiterpene synthases and the origin of terpene skeletal diversity in plants. Phytochemistry 2009, 70, 1621–1637. [Google Scholar] [CrossRef]
- Chen, F.; Tholl, D.; Bohlmann, J.; Pichersky, E. The family of terpene synthases in plants: A mid-size family of genes for specialized metabolism that is highly diversified throughout the kingdom. Plant J. 2011, 66, 212–229. [Google Scholar] [CrossRef]
- Bhargav, P.; Chaurasia, S.; Kumar, A.; Srivastava, G.; Pant, Y.; Chanotiya, C.S.; Ghosh, S. Unraveling the terpene synthase family and characterization of BsTPS2 contributing to (S)-(+)-linalool biosynthesis in Boswellia. Plant Mol. Biol. 2023, 113, 219–236. [Google Scholar] [CrossRef]
- Zhou, Y.; Liu, X.; Yang, Z. Characterization of Terpene Synthase from Tea Green Leafhopper Being Involved in Formation of Geraniol in Tea (Camellia sinensis) Leaves and Potential Effect of Geraniol on Insect-Derived Endobacteria. Biomolecules 2019, 9, 808. [Google Scholar] [CrossRef]
- Li, N.; Dong, Y.; Lv, M.; Li, Q.; Sun, X.; Liu, L.; Cai, Y.P.; Fan, H.H. Combined Analysis of Volatile Terpenoid Metabolism and Transcriptome Reveals Transcription Factors Related to Terpene Synthase in Two Cultivars of Dendrobium officinale Flowers. Front. Genet. 2021, 12, 661296. [Google Scholar] [CrossRef] [PubMed]
- Ji, Y.; Xiao, J.; Shen, Y.; Ma, D.M.; Li, Z.Q.; Pu, G.B.; Li, X.; Huang, L.L.; Liu, B.Y.; Ye, H.C.; et al. Cloning and characterization of AabHLH1, a bHLH transcription factor that positively regulates artemisinin biosynthesis in Artemisia annua. Plant Cell Physiol. 2014, 5, 1592–1604. [Google Scholar] [CrossRef]
- Lu, Z.; Wang, X.; Mostafa, S.; Noor, I.; Lin, X.Y.; Ren, S.X.; Cui, J.W.; Jin, B. WRKY Transcription Factors in Jasminum sambac: An Insight into the Regulation of Aroma Synthesis. Biomolecules 2023, 13, 1679. [Google Scholar] [CrossRef]
- Li, X.; Xu, Y.; Shen, S.; Yin, X.R.; Klee, H.; Zhang, B.; Chen, K.S.; Hancock, R. Transcription factor CitERF71 activates the terpene synthase gene CitTPS16 involved in the synthesis of E-geraniol in sweet orange fruit. J. Exp. Bot. 2017, 68, 4929–4938. [Google Scholar] [CrossRef] [PubMed]
- Shen, S.; Yin, X.; Zhang, B.; Xie, X.L.; Jiang, Q.; Grierson, D.; Chen, K.S. CitAP2.10 activation of the terpene synthase CsTPS1 is associated with the synthesis of (+)-valencene in ‘Newhall’ orange. J. Exp. Bot. 2016, 67, 4105–4115. [Google Scholar] [CrossRef]
- Pan, Y.; Chen, D.; Li, L. Comparative Transcriptomics Analysis of Citrus medica var. sarcodactylis at Different Developmental Stages. China J. Chin. Mater. Medica 2020, 45, 5169–5176. [Google Scholar]
- Wu, D.; Austin, R.S.; Zhou, S. The root transcriptome for North American ginseng assembled and profiled across seasonal development. BMC Genom. 2013, 14, 564. [Google Scholar] [CrossRef]
- Huang, X.; Jing, J.; Yu, J. Transcriptome sequencing of Panax pseudoginseng and identification of key enzyme genes in rriterpenoids saponin biosynthesis. Genom. Appl. Biol. 2017, 36, 2531–2538. [Google Scholar]
- Zhu, M.; Yu, N.; Wang, Q. Biosynthetic pathways of Polygonatum cyrtonemapolysaccharide and diosgenin based on its transcriptomic data. China J. Chin. Mater. Medica 2020, 45, 85–91. [Google Scholar]
- Cong, K.; Yang, S.C.; Zhang, G.H.; Liu, T.; Yan, L.L. SSR Loci Information Analysis and Polymorphism Primers Development in Two Kinds of Coptidis transcriptome. Genom. Appl. Biol. 2018, 37, 2034–2042. [Google Scholar]
- Xu, Y.; Zhu, C.; Xu, C.; Sun, J.; Grierson, D.; Zhang, B.; Chen, K.S. Integration of Metabolite Profiling and Transcriptome Analysis Reveals Genes Related to Volatile Terpenoid Metabolism in Finger Citron (C. medica var. sarcodactylis). Molecules 2019, 24, 2564. [Google Scholar] [CrossRef]
- Xu, J.; Tan, L.; Fu, H. Genetic diversity analysis of 14 citron genotypes based on molecular markers. Mol. Plant Breed. 2021, 19, 4726–4737. [Google Scholar]
- Nicolosi, E.; Deng, Z.; Gentile, A.; Malfa, S.L.; Continella, G.C.; Tribulato, E. Citrus phylogeny and genetic origin of important species as investigated by molecular markers. Theor. Appl. Genet. 2000, 100, 1155–1166. [Google Scholar] [CrossRef]
- Xian, W.; Yuan, Y.; Zhaxi, L.B.; Zhang, L.Y.; Yang, Z.; Zheng, L.X.; Zhang, W.S. Study on the Volatile Aroma Components of Citron Leaves and Fruits. J. Foshan Univ. (Nat. Sci. Ed.) 2022, 4, 74–80. [Google Scholar]
- Niu, L.; Yu, M.; Liu, F.; Li, D.; Liu, C. Composition and aroma-active components determination of Xiangyuan (Citrus wilsonii Tanaka) essential oil by GC-MS-O. Food Ferment. Ind. 2013, 4, 186–191. [Google Scholar]
- Li, Z.; Cai, M.; Liu, Y.; Sun, P.L.; Luo, S.L. Antibacterial Activity and Mechanisms of Essential Oil from Citrus medica L. var. sarcodactylis. Molecules 2019, 24, 1577. [Google Scholar] [CrossRef]
- Wu, Z.; Li, H.; Yang, Y.; Zhang, Y.; Tu, D.W. Variation in the components and antioxidant activity of Citrus medica L. var. sarcodactylis essential oils at different stages of maturity. Ind. Crop Prod. 2013, 46, 311–315. [Google Scholar]
- Gao, J.; Wu, B.P.; Gao, L.X.; Liu, H.R.; Zhang, B.; Sun, C.D.; Chen, K.S. Glycosidically bound volatiles as a ected by ripening stages of Satsuma mandarin fruit. Food Chem. 2018, 240, 1097–1105. [Google Scholar] [CrossRef]
- Ma, L.; Xue, N.; Fu, X.; Zhang, H.S.; Li, G. Arabidopsis thaliana FAR-RED ELONGATED HYPOCOTYLS3 (FHY3) and FAR-RED-IMPAIRED RESPONSE1 (FAR1) modulate starch synthesis in response to light and sugar. New Phytol. 2017, 213, 1682–1696. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Tian, T.; Lin, R.; Deng, X.W.; Wang, H.Y.; Li, G. Arabidopsis FHY3 and FAR1 regulate light-induced myo-inositol biosynthesis and oxidative stress responses by transcriptional activation of MIPS1. Mol. Plant 2016, 9, 541–557. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Zhang, S.; Li, J.; Pei, R.; Tian, L.; Qi, J.; Azam, M.; Agyenim-Boateng, K.G.; Shaibu, A.S.; Liu, Y.T.; et al. Dual-function C2H2-type zinc-finger transcription factor GmZFP7 contributes to isoflavone accumulation in soybean. New Phytol. 2023, 237, 1794–1809. [Google Scholar] [CrossRef]
- Liu, Y.; Liu, D.; Hu, R.; Hua, C.M.; Ali, I.; Zhang, A.D.; Liu, B.H.; Wu, M.J.; Huang, L.L.; Gan, Y.B. AtGIS, a C2H2 zinc-finger transcription factor from Arabidopsis regulates glandular trichome development through GA signaling in tobacco. Biochem. Biophys. Res. Commun. 2017, 483, 209–215. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Liu, G.; Yang, F.; Liang, Y.L.; Gao, Q.Q.; Xian, C.F.; Li, X.; Yang, R.; Zhang, G.H.; Jiang, H.F.; et al. Multilayered regulation of secondary metabolism in medicinal plants. Mol. Hortic. 2023, 3, 11. [Google Scholar] [CrossRef]
- Shang, Y.; Ma, Y.; Zhou, Y.; Zhang, H.M.; Duan, L.X.; Chen, H.M.; Zeng, J.G.; Zhou, Q.; Wang, S.H.; Gu, W.J.; et al. Plant science. Biosynthesis, regulation, and domestication of bitterness in cucumber. Science 2014, 346, 1084–1088. [Google Scholar] [CrossRef]
- Tamura, K.; Yoshida, K.; Hiraoka, Y.; Sakaguchi, D.; Chikugo, A.; Mochida, K.; Kojoma, M.; Mitsuda, N.; Saito, K.; Muranaka, T.; et al. The Basic Helix-Loop-Helix transcription factor GubHLH3 positively regulates soyasaponin biosynthetic genes in Glycyrrhiza uralensis. Plant Cell Physiol. 2018, 59, 778–791, Erratum in Plant Cell Physiol. 2018, 59, 783–796. [Google Scholar] [CrossRef]
- Wu, Z.; Li, L.; Liu, H.; Yan, X.; Ma, Y.; Li, Y.; Chen, T.; Wang, C.; Xie, L.; Hao, X.; et al. AaMYB15, an R2R3-MYB TF in Artemisia annua, acts as a negative regulator of artemisinin biosynthesis. Plant Sci. 2021, 308, 110920. [Google Scholar] [CrossRef]
- Xia, P.G.; Hu, W.Y.; Zheng, Y.J.; Wang, Y.; Yan, K.J.; Liang, Z.S. Structural and interactions analysis of a transcription factor PnMYB2 in Panax notoginseng. J. Plant Physiol. 2022, 275, 153756. [Google Scholar] [CrossRef]
- Eulgem, T.; Somssich, I.E. Networks of WRKY transcription factors in defense signaling. Curr. Opin. Plant Biol. 2007, 10, 366–371. [Google Scholar] [CrossRef]
- Gao, J.; Chen, Y.; Gao, M.; Wu, L.W.; Zhao, Y.X.; Wang, Y.D. LcWRKY17, a WRKY Transcription Factor from Litsea cubeba, Effectively Promotes Monoterpene Synthesis. Int. J. Mol. Sci. 2023, 24, 7210. [Google Scholar] [CrossRef] [PubMed]
- Sharma, T.; Kawoosa, T.; Gahlan, P.; Sharma, D.; Kaachra, A.; Hallan, V.; Kumar, S. Two light responsive WRKY genes exhibit positive and negative correlation with picroside content in Picrorhiza kurrooa Royle ex Benth, an endangered medicinal herb. 3 Biotech 2020, 10, 255. [Google Scholar] [CrossRef] [PubMed]
- Jin, Z.; Kwon, M.; Lee, A.R.; Ro, D.K.; Wungsintaweekul, J.; Kim, S.U. Molecular cloning and functional characterization of three terpene synthases from unripe fruit of black pepper (Piper nigrum). Arch. Biochem. Biophys. 2018, 638, 35–40. [Google Scholar] [CrossRef] [PubMed]
- Anand, A.; Jayaramaiah, R.H.; Beedkar, S.D.; Dholakia, B.B.; Lavhale, S.G.; Punekar, S.A.; Gade, W.N.; Thulasiram, H.V.; Giri, A.P. Terpene profiling, transcriptome analysis and characterization of cis-β-terpineol synthase from Ocimum. Physiol. Mol. Biol. Plants 2019, 25, 47–57. [Google Scholar] [CrossRef]
- Li, M.; Shao, Y.; Pan, B.; Liu, C.; Tan, H.X. Regulation of important natural products biosynthesis by WRKY transcription factors in plants. J. Adv. Res. 2025; in press. [Google Scholar] [CrossRef]
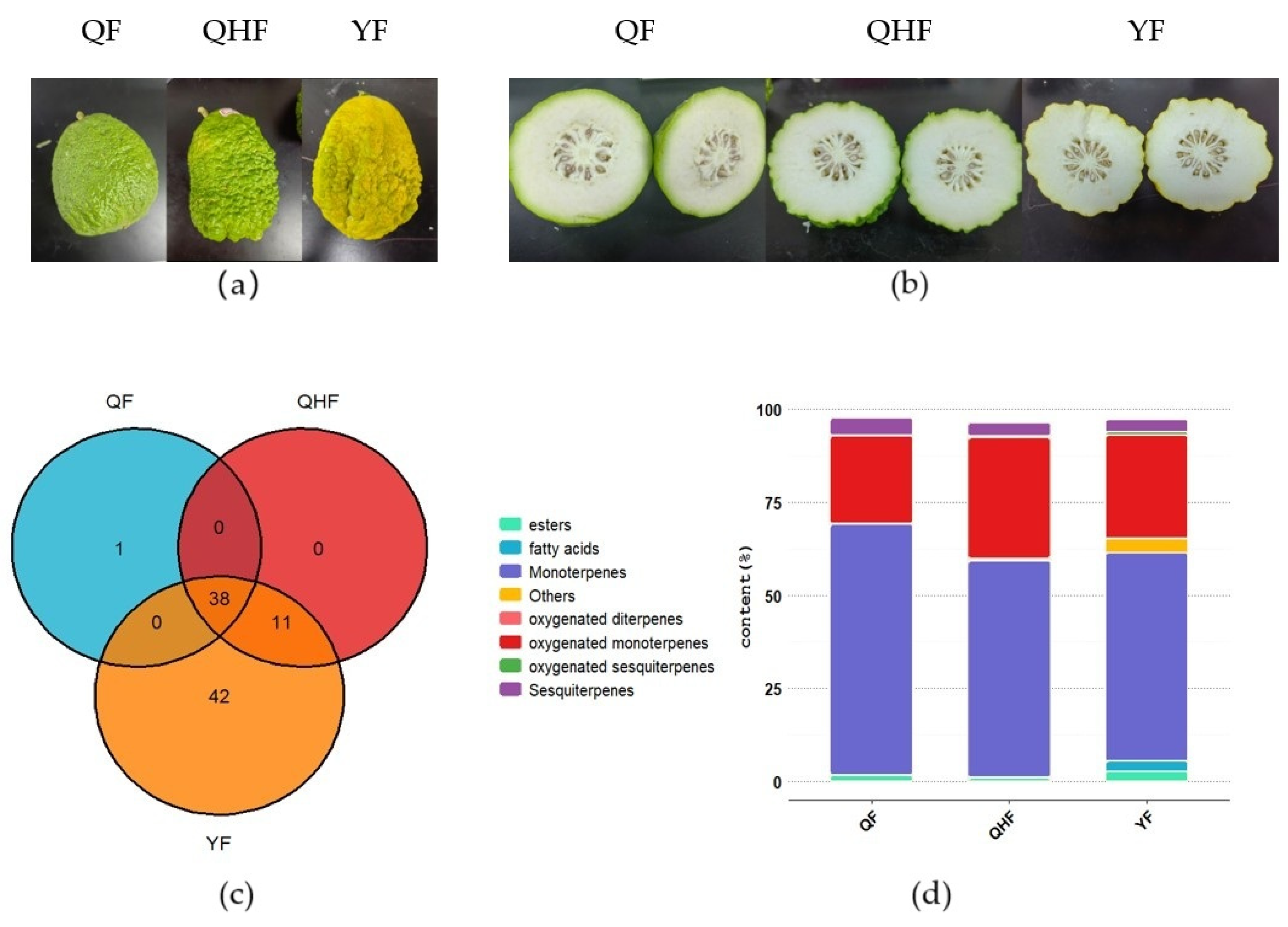
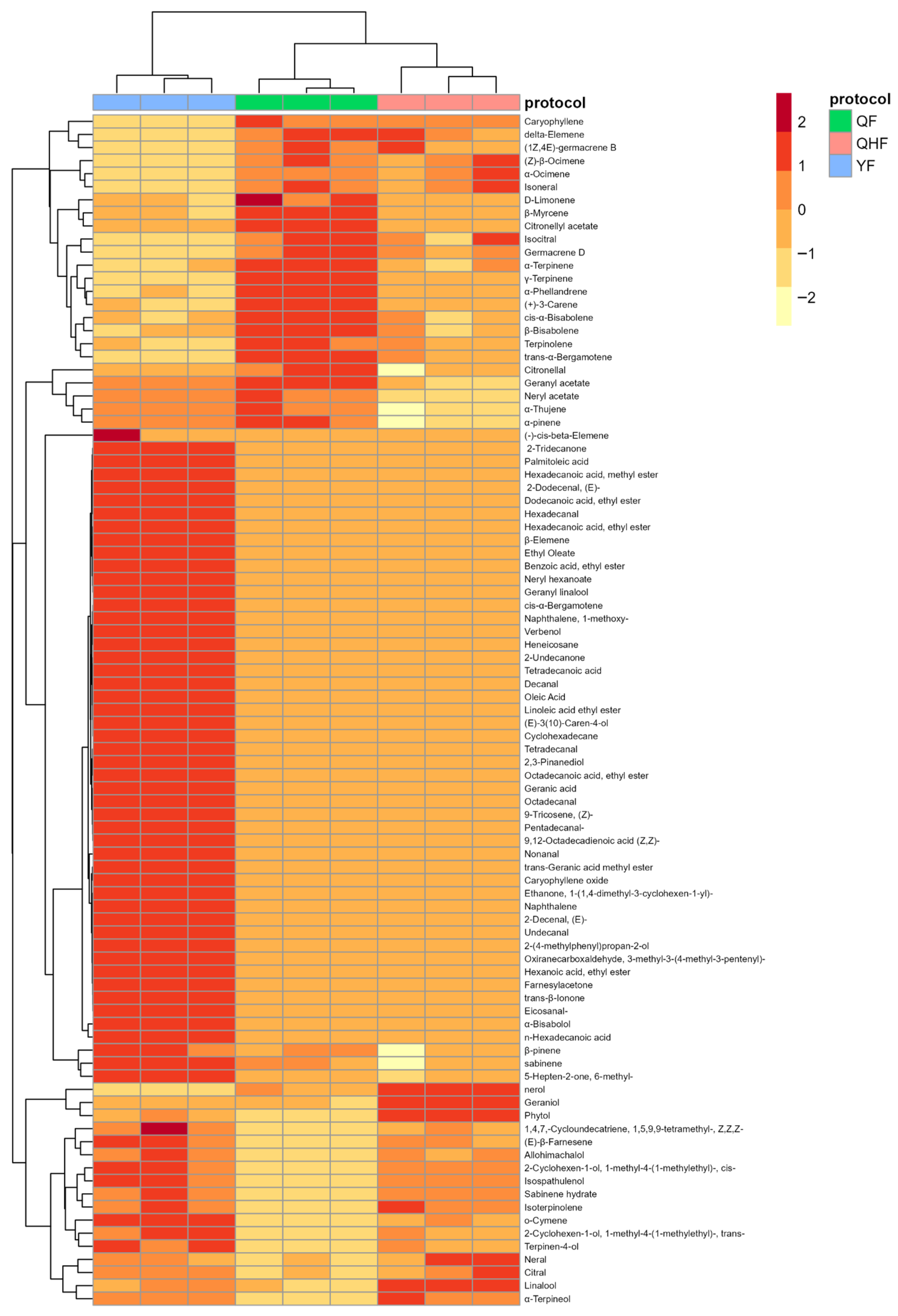
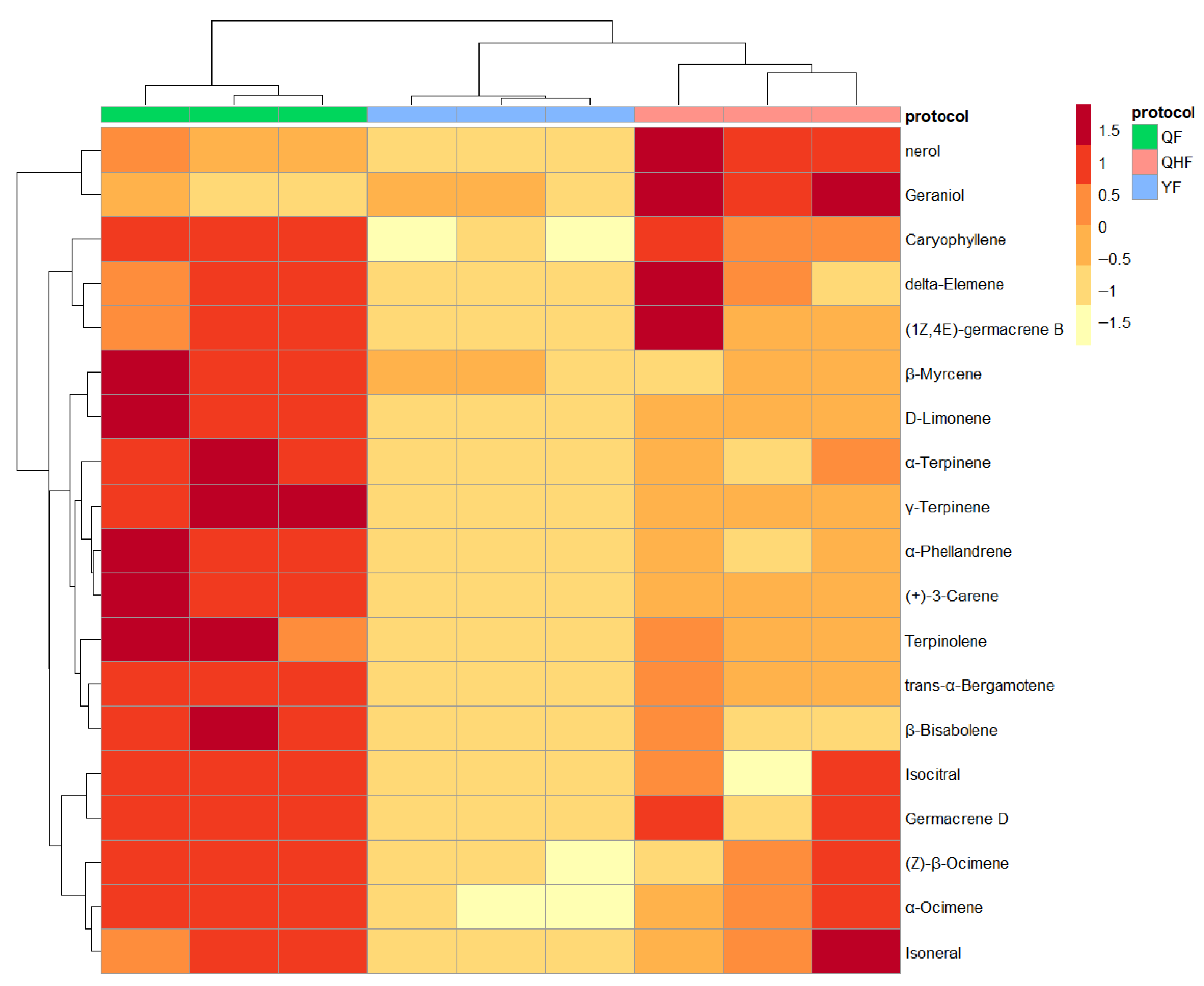
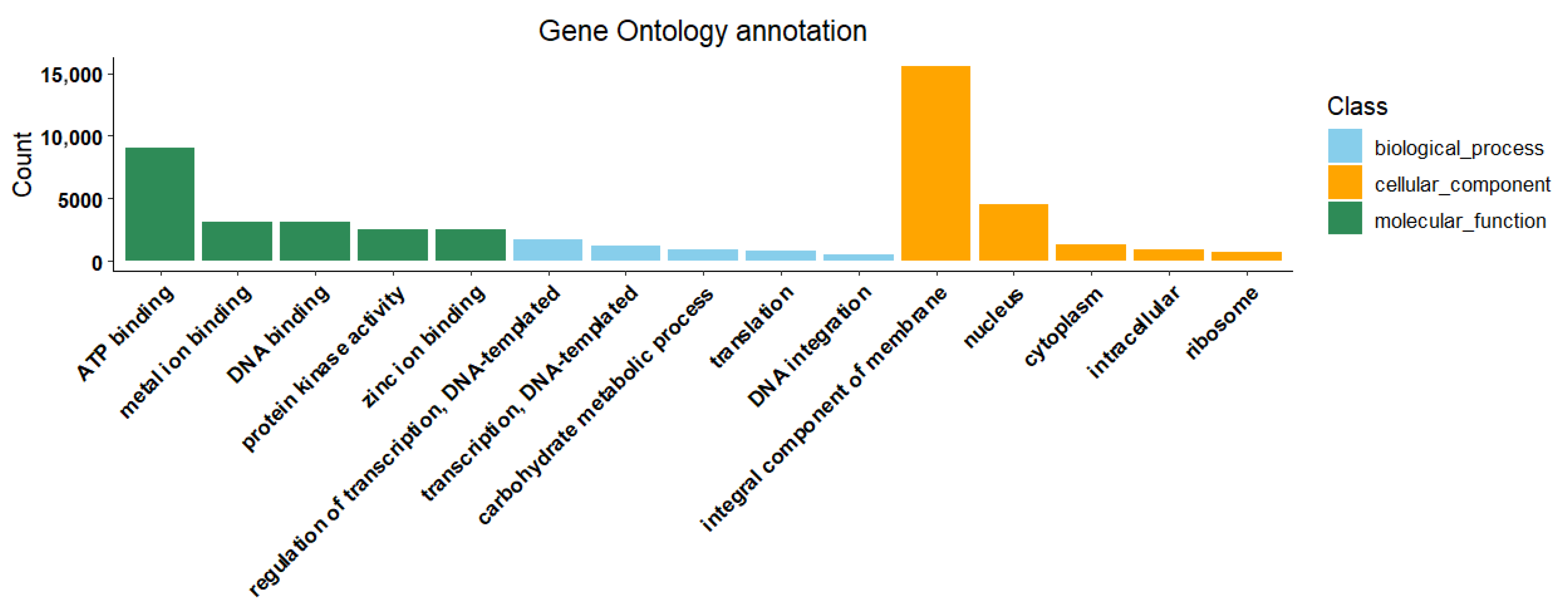
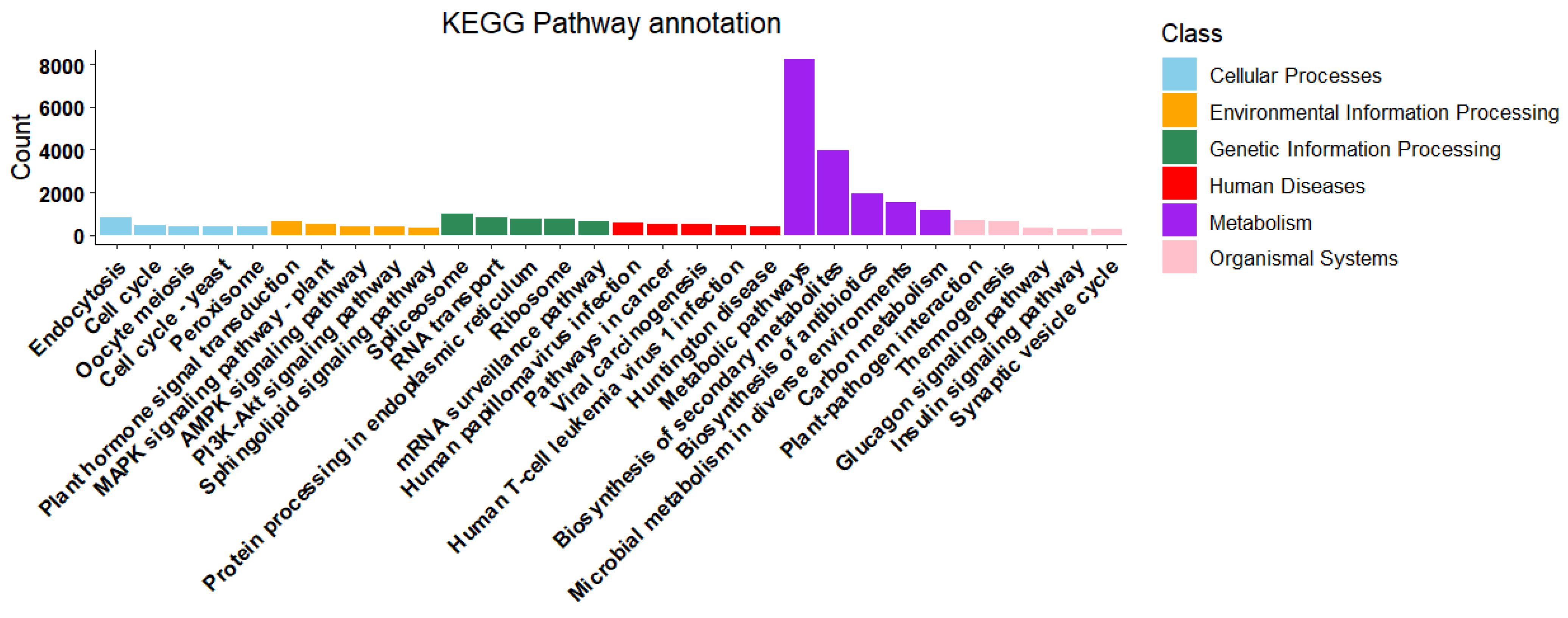

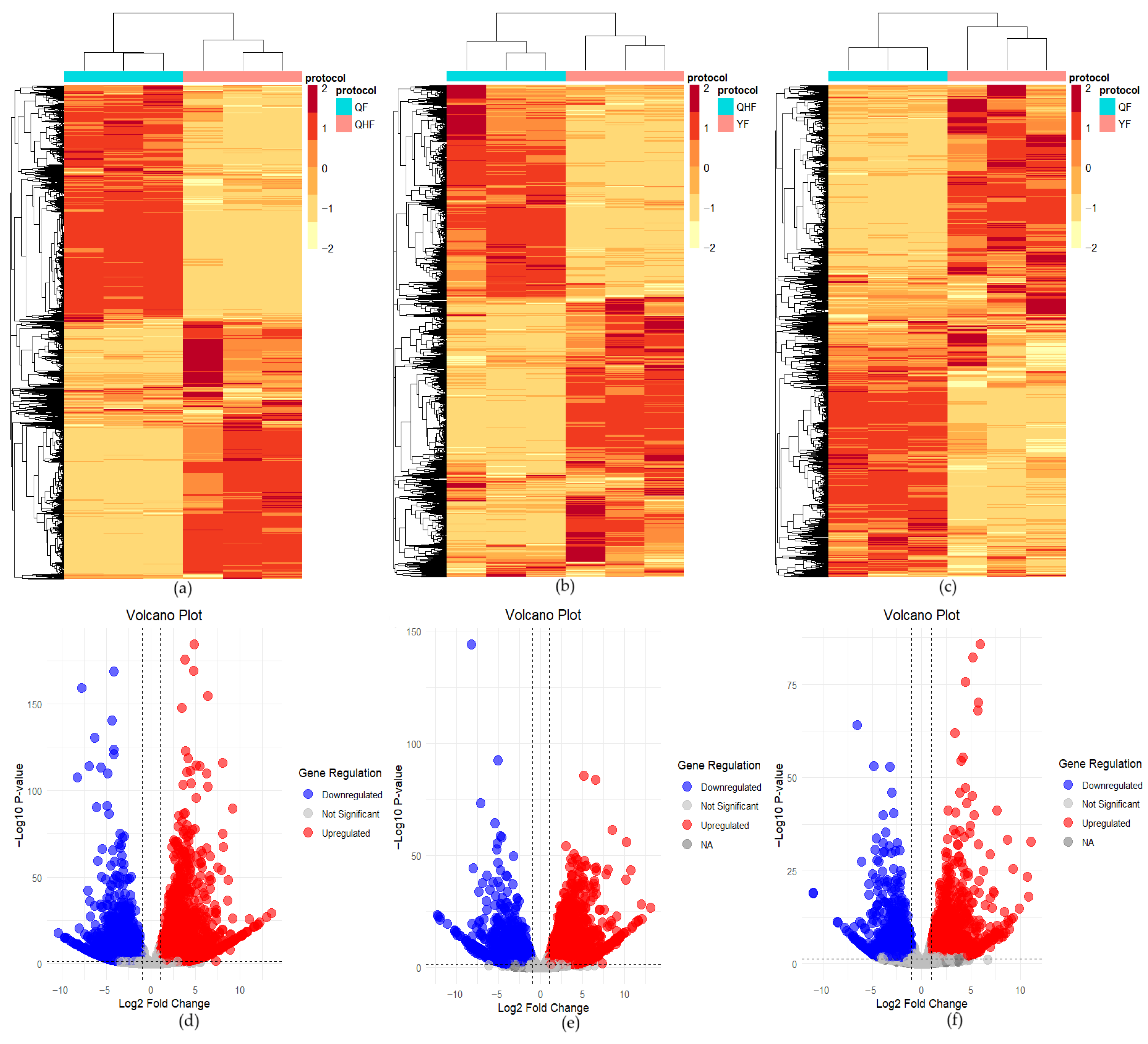
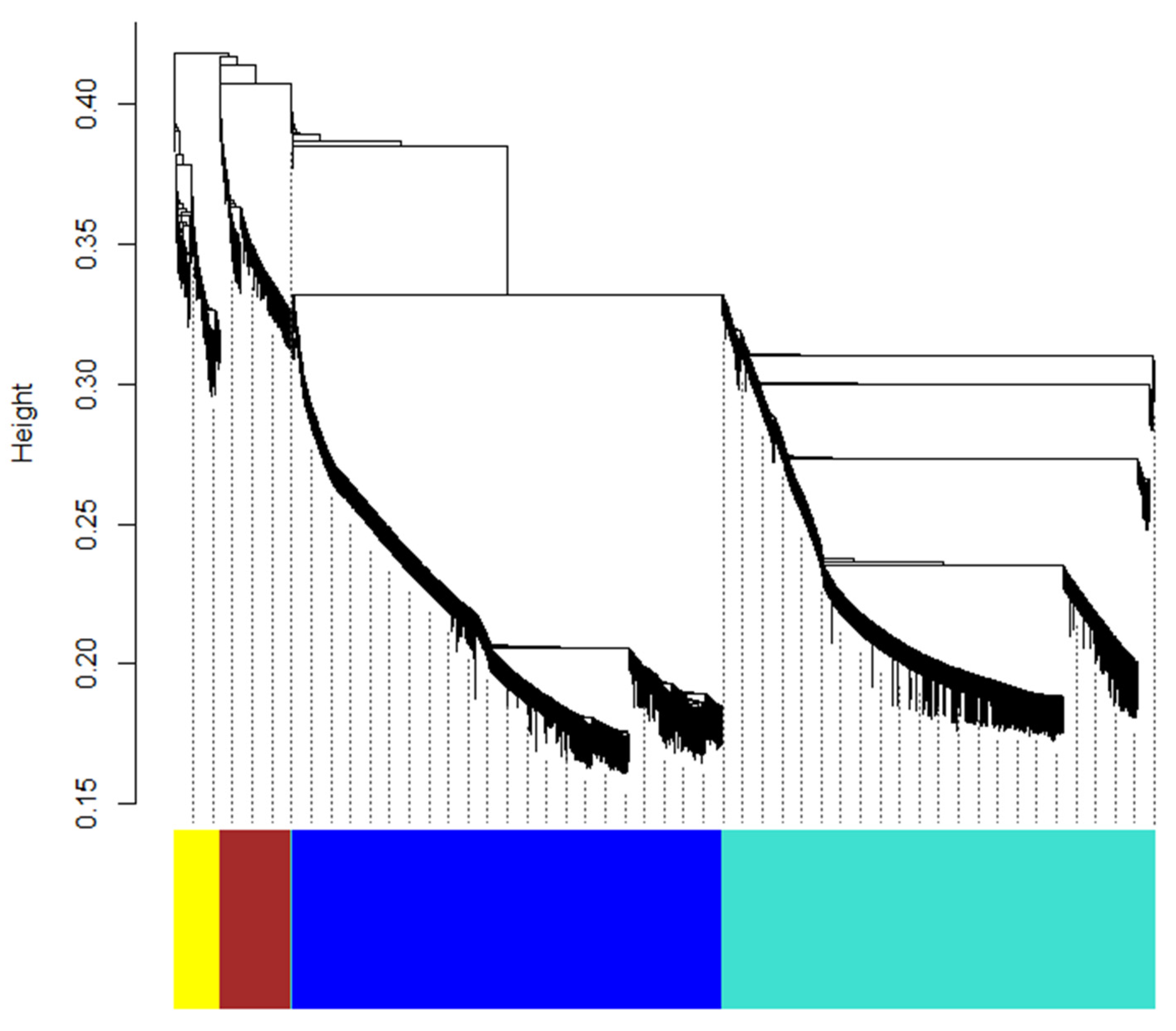


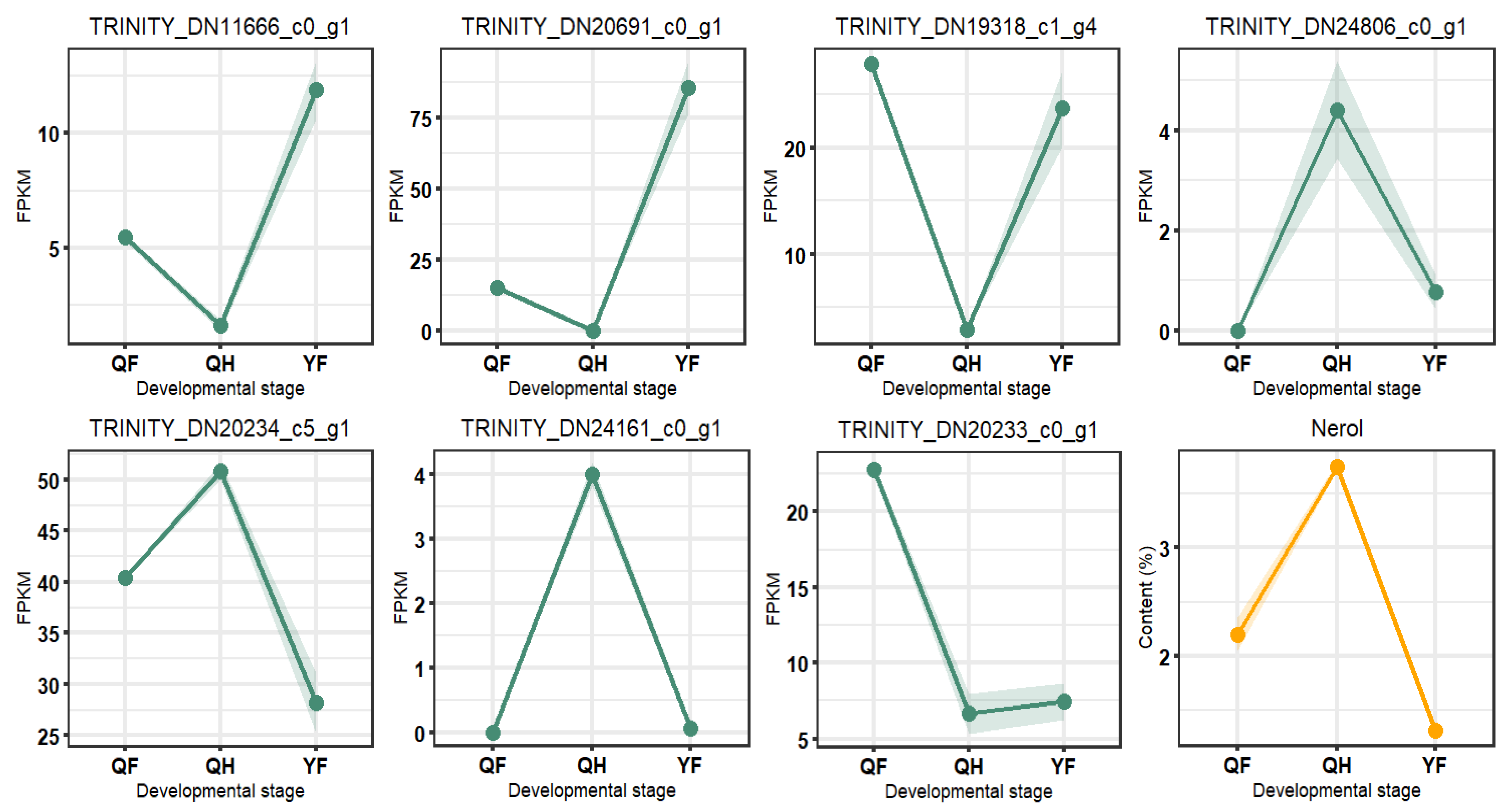
| Compound | Log2FC | p Value | p.adj | B | sig |
|---|---|---|---|---|---|
| γ-Terpinene | −1.45 | 2.49 × 10−43 | 2.27 × 10−41 | 106.16 | Down |
| (+)-3-Carene | −1.45 | 1.66 × 10−42 | 7.57 × 10−41 | 103.50 | Down |
| α-Phellandrene | −1.42 | 7.29 × 10−41 | 1.66 × 10−39 | 98.25 | Down |
| Caryophyllene | −1.42 | 2.42 × 10−41 | 7.33 × 10−40 | 99.77 | Down |
| nerol | −1.39 | 6.45 × 10−40 | 9.78 × 10−39 | 95.25 | Down |
| trans-α-Bergamotene | −1.35 | 2.06 × 10−40 | 3.75 × 10−39 | 96.80 | Down |
| D-Limonene | −1.27 | 2.95 × 10−34 | 2.99 × 10−33 | 77.84 | Down |
| Isocitral | −1.25 | 3.83 × 10−39 | 4.98 × 10−38 | 92.76 | Down |
| (1Z,4E)-germacrene B | −1.19 | 2.67 × 10−31 | 2.43 × 10−30 | 69.07 | Down |
| β-Bisabolene | −1.17 | 7.73 × 10−31 | 5.41 × 10−30 | 67.72 | Down |
| α-Terpinene | −1.14 | 1.54 × 10−29 | 9.99 × 10−29 | 63.95 | Down |
| Germacrene D | −1.13 | 5.97 × 10−31 | 4.53 × 10−30 | 68.01 | Down |
| Terpinolene | −1.12 | 1.56 × 10−27 | 9.46 × 10−27 | 58.24 | Down |
| n-Hexadecanoic acid | 1.11 | 3.01 × 10−26 | 1.61 × 10−25 | 54.63 | Up |
| delta-Elemene | −1.11 | 5.94 × 10−31 | 4.53 × 10−30 | 68.01 | Down |
| α-Ocimene | −1.07 | 4.78 × 10−25 | 2.29 × 10−24 | 51.27 | Down |
| Citronellyl acetate | −1.07 | 1.97 × 10−38 | 2.24 × 10−37 | 90.43 | Down |
| Geranyl acetate | −1.05 | 2.07 × 10−26 | 1.18 × 10−25 | 55.03 | Down |
| Isoneral | −1.01 | 2.78 × 10−25 | 1.40 × 10−24 | 51.88 | Down |
| (Z)-β-Ocimene | −1.01 | 3.19 × 10−23 | 1.38 × 10−22 | 46.24 | Down |
| β-Myrcene | −1.00 | 1.76 × 10−22 | 7.29 × 10−22 | 44.23 | Down |
| Sample | R.Reads | R.Bases (G) | R.Q20 (%) | R.Q30 (%) | R.GC (%) | H.Reads | H.Bases (G) | H.Q20 (%) | H.Q30 (%) | H.GC (%) | E.Rate (%) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| QF_1 | 40,722,928 | 6.15 | 97.24 | 93.29 | 47.1 | 36,309,012 | 5.39 | 98.67 | 95.35 | 45.91 | 89.16 |
| QF_2 | 47,874,334 | 7.23 | 97.51 | 93.69 | 47 | 42,815,356 | 6.35 | 98.72 | 95.52 | 45.89 | 89.43 |
| QF_3 | 39,764,314 | 6.00 | 97.13 | 93.09 | 47.13 | 35,436,944 | 5.25 | 98.63 | 95.23 | 45.91 | 89.12 |
| QH_1 | 53,984,240 | 8.15 | 98.04 | 94.37 | 46.94 | 51,947,994 | 7.72 | 98.73 | 95.39 | 46.66 | 96.23 |
| QH_2 | 61,495,622 | 9.29 | 98.15 | 94.67 | 47 | 59,437,138 | 8.84 | 98.82 | 95.64 | 46.72 | 96.65 |
| QH_3 | 61,405,466 | 9.27 | 98.07 | 94.4 | 46.97 | 58,942,878 | 8.76 | 98.73 | 95.39 | 46.64 | 95.99 |
| YF_1 | 43,738,378 | 6.59 | 95.6 | 89.21 | 45.95 | 40,036,546 | 5.83 | 97.56 | 92.39 | 45.55 | 91.54 |
| YF_2 | 42,906,236 | 6.48 | 94.81 | 87.74 | 45.37 | 38,117,590 | 5.56 | 96.98 | 90.96 | 45.03 | 88.84 |
| YF_3 | 43,824,350 | 6.62 | 94.76 | 87.7 | 45.3 | 38,996,566 | 5.71 | 96.95 | 90.89 | 44.95 | 88.98 |
| Sample | UID Reads | Filt.rRNA Reads | Re.Overrep Reads | Correct Reads | Dedup Reads |
|---|---|---|---|---|---|
| QF_1 | 21,048,354 | 20,741,018 (98.54%) | 20,702,696 (98.36%) | 19,759,764 (93.88%) | 16,962,776 (80.59%) |
| QF_2 | 23,886,132 | 23,544,430 (98.57%) | 23,499,452 (98.38%) | 22,445,078 (93.97%) | 19,049,770 (79.75%) |
| QF_3 | 21,215,766 | 20,910,724 (98.56%) | 20,870,194 (98.37%) | 19,916,106 (93.87%) | 17,075,094 (80.48%) |
| QH_1 | 31,546,066 | 31,403,970 (99.55%) | 30,848,100 (97.79%) | 29,386,708 (93.15%) | 23,097,250 (73.22%) |
| QH_2 | 35,541,622 | 35,329,910 (99.40%) | 34,324,848 (96.58%) | 32,657,494 (91.89%) | 25,371,004 (71.38%) |
| QH_3 | 35,171,062 | 35,015,082 (99.56%) | 34,415,500 (97.85%) | 32,806,746 (93.28%) | 25,344,526 (72.06%) |
| YF_1 | 32,750,846 | 32,662,144 (99.73%) | 32,127,036 (98.10%) | 30,376,082 (92.75%) | 25,751,970 (78.63%) |
| YF_2 | 29,815,338 | 29,756,896 (99.80%) | 29,756,896 (99.80%) | 28,109,788 (94.28%) | 23,585,060 (79.10%) |
| YF_3 | 30,465,698 | 30,401,254 (99.79%) | 30,401,254 (99.79%) | 28,688,804 (94.17%) | 24,406,710 (80.11%) |
| Type | Trinity | Unigene |
|---|---|---|
| N50 | 2137.0 | 1591.0 |
| N90 | 594.0 | 331.0 |
| avaragelength | 1320.1 | 872.5 |
| maxlength | 13,377.0 | 13,377.0 |
| minlength | 201.0 | 201.0 |
| totalbase | 134,384,342.0 | 43,271,578.0 |
| totalcontigs | 101,796.0 | 49,596.0 |
| GC_content | 40.4 | 40.4 |
| GC_content_max | 84.2 | 84.2 |
| GC_content_min | 7.3 | 7.3 |
| Data Base | Annotated Number |
|---|---|
| Uniprot | 80,800 |
| NR | 25,044 |
| Pfam | 75,201 |
| Rfam | 95,979 |
| eggNog | 72,146 |
| GO | 57,106 |
| KEGG | 22,160 |
| Total | 100,765 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Luo, J.; Zhong, W.; Xiong, Y.; Wang, J. Analysis of Non-Polar Low-Molecular Metabolites in Citron (Citrus medica L.) Peel Essential Oil at Different Developmental Stages and a Combined Study of Transcriptomics Revealed Genes Related to the Synthesis Regulation of the Monoterpenoid Compound Nerol. Int. J. Mol. Sci. 2025, 26, 9034. https://doi.org/10.3390/ijms26189034
Luo J, Zhong W, Xiong Y, Wang J. Analysis of Non-Polar Low-Molecular Metabolites in Citron (Citrus medica L.) Peel Essential Oil at Different Developmental Stages and a Combined Study of Transcriptomics Revealed Genes Related to the Synthesis Regulation of the Monoterpenoid Compound Nerol. International Journal of Molecular Sciences. 2025; 26(18):9034. https://doi.org/10.3390/ijms26189034
Chicago/Turabian StyleLuo, Jie, Wanting Zhong, Yayi Xiong, and Jian Wang. 2025. "Analysis of Non-Polar Low-Molecular Metabolites in Citron (Citrus medica L.) Peel Essential Oil at Different Developmental Stages and a Combined Study of Transcriptomics Revealed Genes Related to the Synthesis Regulation of the Monoterpenoid Compound Nerol" International Journal of Molecular Sciences 26, no. 18: 9034. https://doi.org/10.3390/ijms26189034
APA StyleLuo, J., Zhong, W., Xiong, Y., & Wang, J. (2025). Analysis of Non-Polar Low-Molecular Metabolites in Citron (Citrus medica L.) Peel Essential Oil at Different Developmental Stages and a Combined Study of Transcriptomics Revealed Genes Related to the Synthesis Regulation of the Monoterpenoid Compound Nerol. International Journal of Molecular Sciences, 26(18), 9034. https://doi.org/10.3390/ijms26189034







