Lipidomic Profiling of Hyperacute Ischemic Heart Disease and Toxic Deaths: A Forensic Investigation into Metabolic Biomarkers
Abstract
1. Introduction
2. Results
2.1. Distinct Lipidomic Signatures Differentiate Hyperacute Ischemic Heart Disease from Toxic/Drug Abuse Deaths
2.2. Differential Lipid Abundance and Pathway Analysis
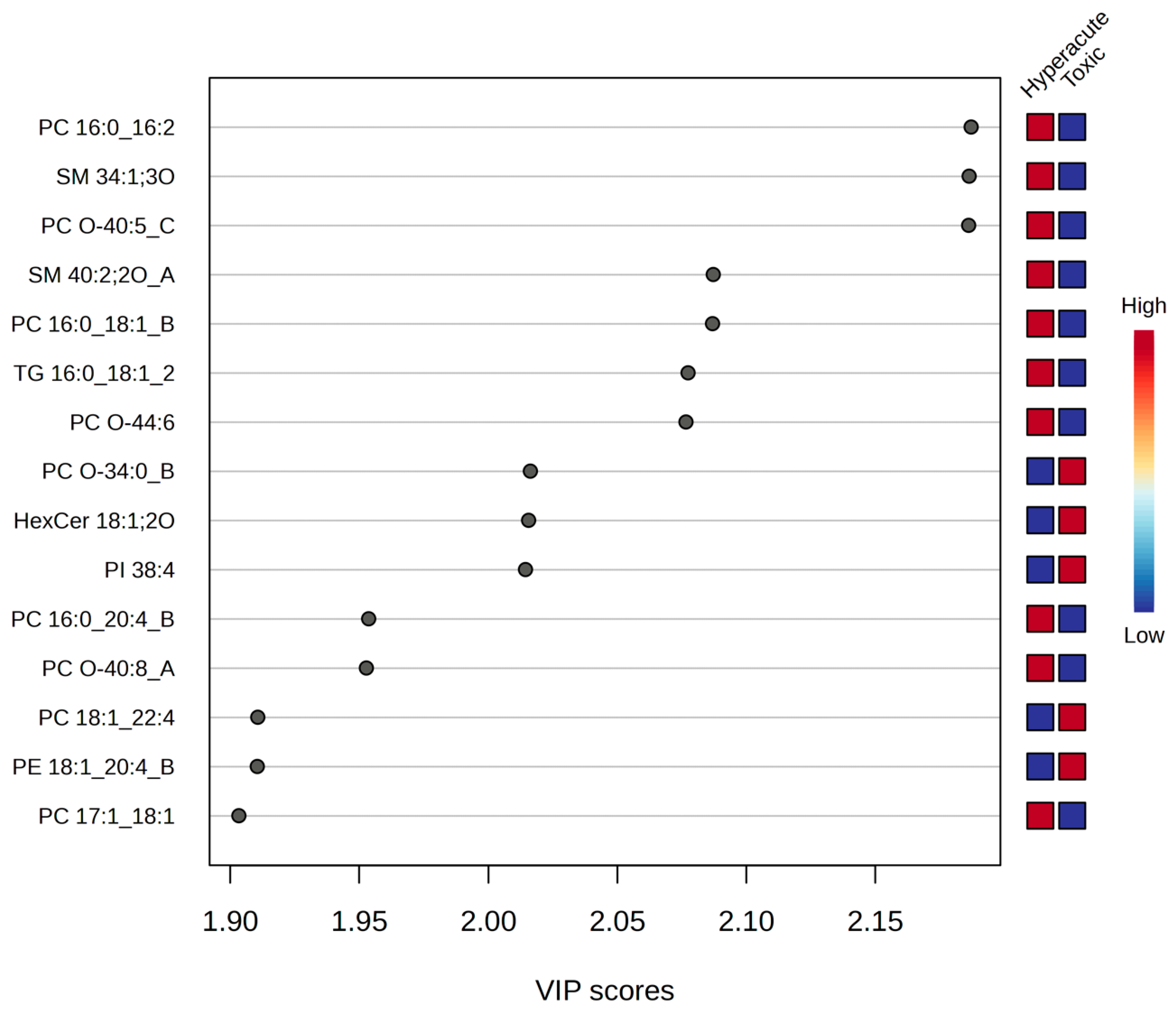
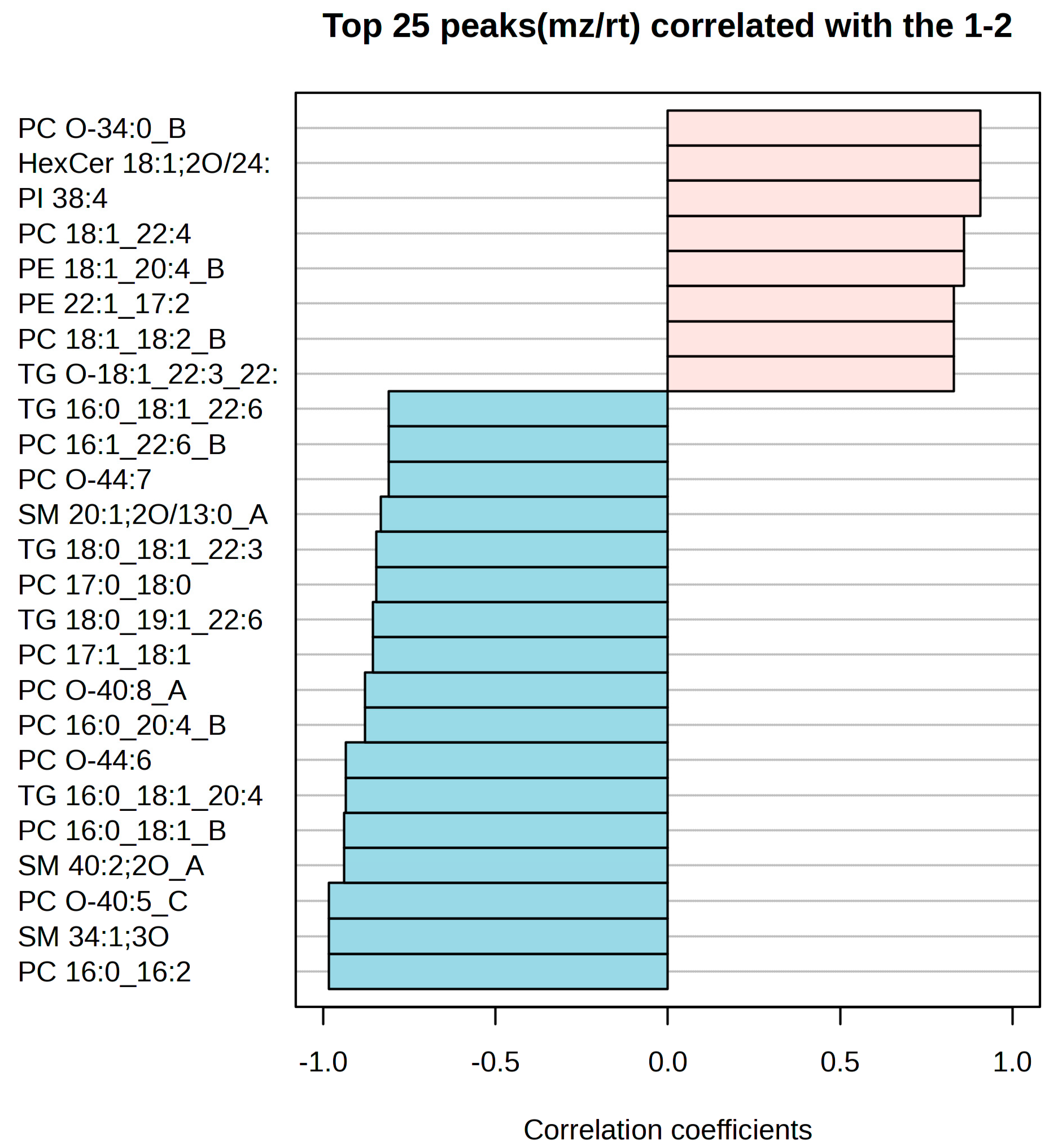
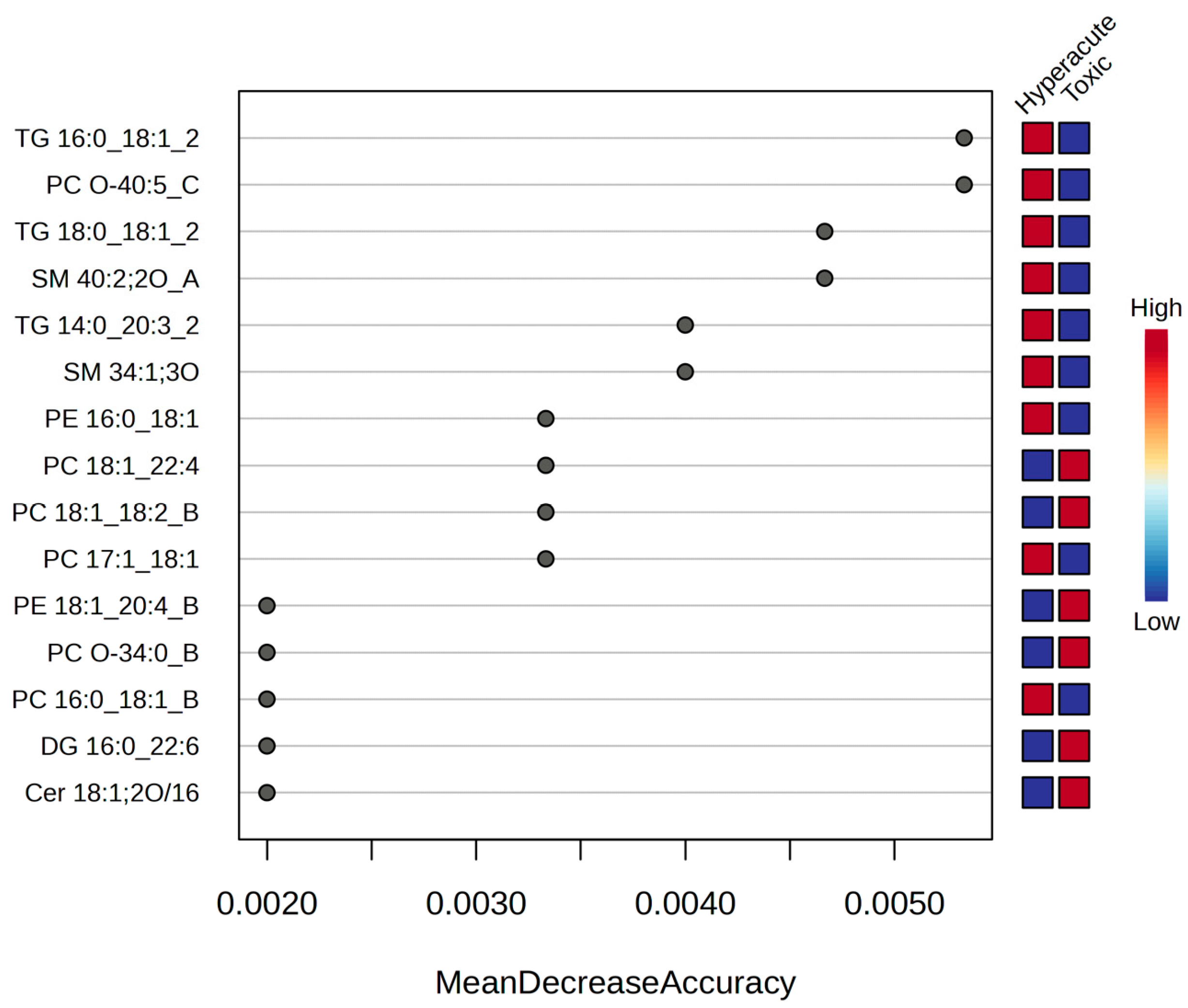
2.3. Potential Biomarkers for Hyperacute Ischemic Heart Disease and Toxic/Drug Abuse Deaths
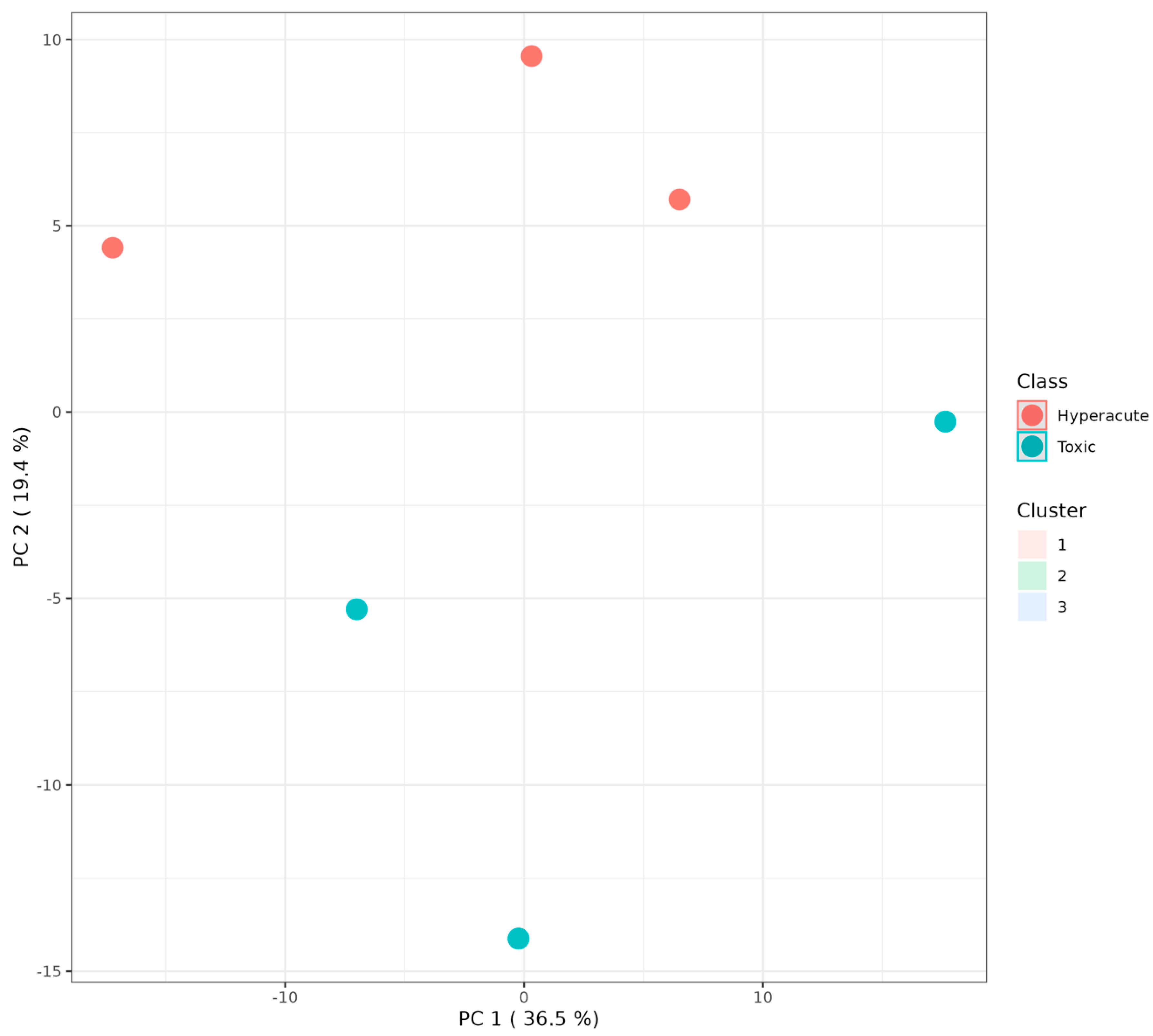
2.4. Multivariate Statistical Analyses
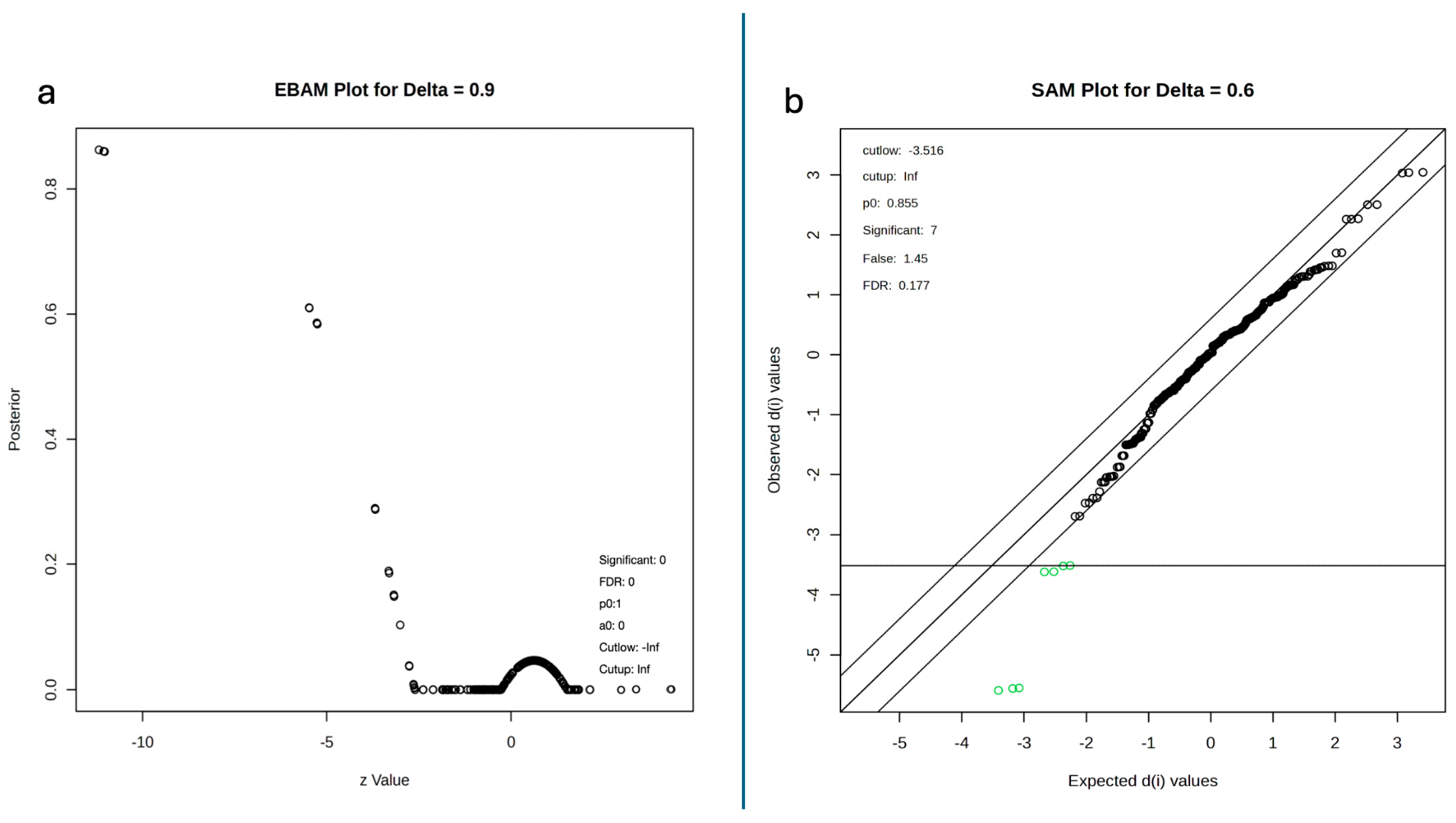
2.5. Empirical Bayes, Significance Analysis of Microarrays and Kmeans
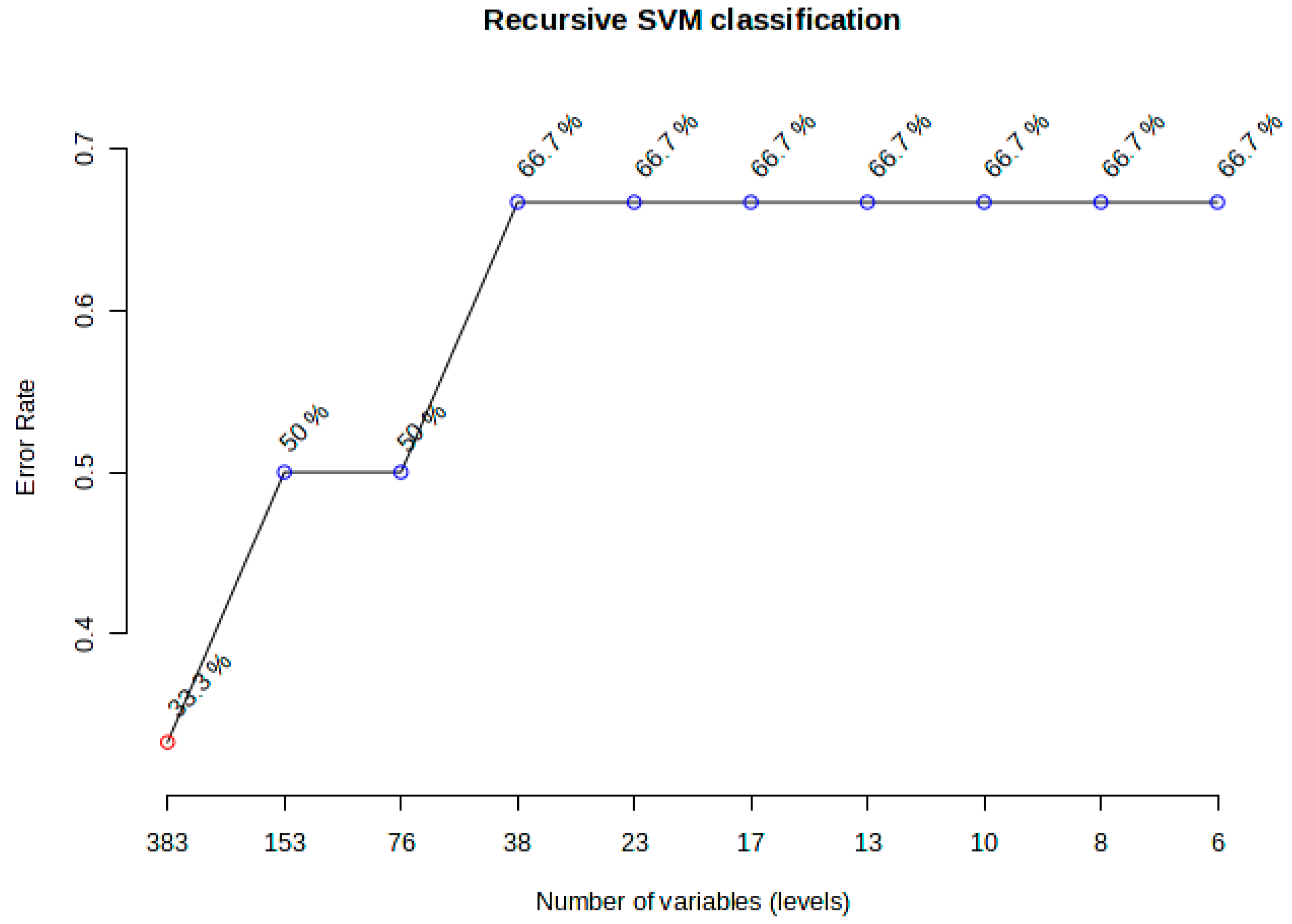
2.6. Machine Learning and Clustering Analyses
2.7. Pathway Enrichment Analysis
3. Discussion
4. Materials and Methods
4.1. Human Sample Collection During Forensic Autopsy
4.2. Reagents
4.3. Instrumentation
4.4. Sample Preparation
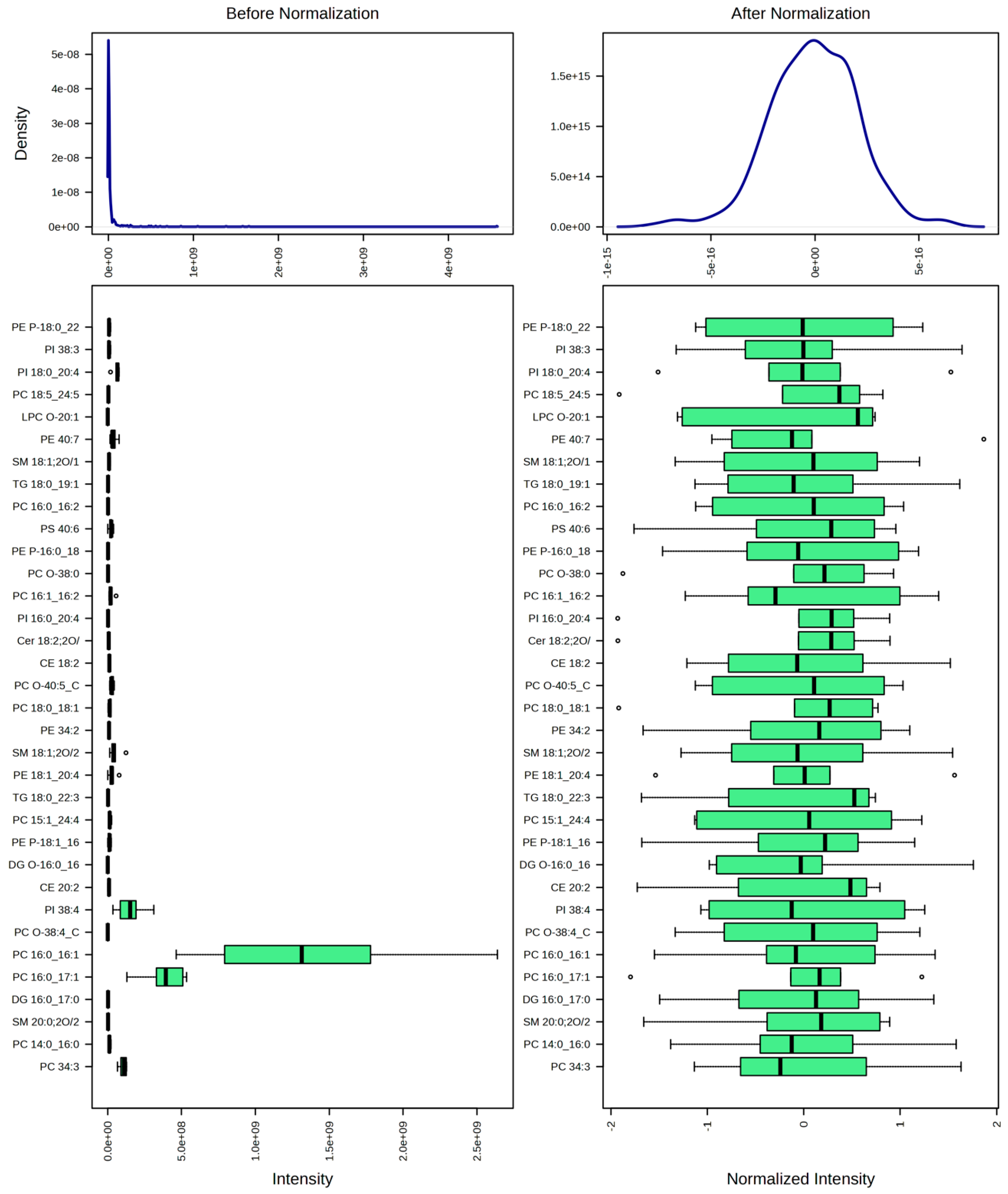
4.5. Data Processing and Statistical Analyses
- Data filtering: A 25% filter based on standard deviation was applied to remove variables with low variation across samples, which are less likely to be informative; a total of 128 lipids were removed;
- Normalization: Data were normalized by mean to correct for systematic differences between samples;
- Data transformation: A square root transformation was applied to reduce the impact of heteroscedasticity and to make the data distribution more symmetric;
- Scaling: Data were autoscaled (mean-centered and divided by the standard deviation of each variable) to make features more comparable.
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| FDR | False Discovery Rate |
| HexCer | Hexosylceramide |
| IHD | Ischemic Heart Disease |
| PC | Phosphatidylcholine |
| PCA | Principal Component Analysis |
| PE | Phosphatidylethanolamine |
| PI | Phosphatidylinositol |
| PLS-DA | Partial Least Squares Discriminant Analysis |
| SCD | Sudden Cardiac Death |
| SVM | Support Vector Machine |
| SM | Sphingomyelin |
| UHPLC-Q-TOF | Ultra-High-Performance Liquid Chromatography Quadrupole Time-of-Flight |
| TG | Triacylglycerol (Triglyceride) |
References
- Aquaro, G.D.; Guidi, B.; Emdin, M.; Pucci, A.; Chiti, E.; Santurro, A.; Scopetti, M.; Biondi, F.; Maiese, A.; Turillazzi, E.; et al. Post-Mortem Cardiac Magnetic Resonance in Explanted Heart of Patients with Sudden Death. Int. J. Environ. Res. Public Health 2022, 19, 13395. [Google Scholar] [CrossRef]
- Michaud, K.; Basso, C.; D’aMati, G.; Giordano, C.; Kholová, I.; Preston, S.D.; Rizzo, S.; Sabatasso, S.; Sheppard, M.N.; Vink, A.; et al. Diagnosis of myocardial infarction at autopsy: AECVP reappraisal in the light of the current clinical classification. Virchows Arch. 2020, 476, 179–194. [Google Scholar] [CrossRef]
- Szeremeta, M.; Pietrowska, K.; Niemcunowicz-Janica, A.; Kretowski, A.; Ciborowski, M. Applications of Metabolomics in Forensic Toxicology and Forensic Medicine. Int. J. Mol. Sci. 2021, 22, 3010. [Google Scholar] [CrossRef]
- Roberts, L.D.; Souza, A.L.; Gerszten, R.E.; Clish, C.B. Targeted metabolomics. Curr. Protoc. Mol. Biol. 2012, 98, 30.2.1–30.2.24. [Google Scholar] [CrossRef]
- Schrimpe-Rutledge, A.C.; Codreanu, S.G.; Sherrod, S.D.; McLean, J.A. Untargeted Metabolomics Strategies-Challenges and Emerging Directions. J. Am. Soc. Mass Spectrom. 2016, 27, 1897–1905. [Google Scholar] [CrossRef]
- Zhang, K.; Yan, H.; Liu, R.; Xiang, P.; Deng, K.; Zhang, J.; Tuo, Y.; Wang, Z.; Huang, P. Exploring metabolic alterations associated with death from asphyxia and the differentiation of asphyxia from sudden cardiac death by GC-HRMS-based untargeted metabolomics. J. Chromatogr. B 2021, 1171, 122638. [Google Scholar] [CrossRef] [PubMed]
- Procopio, N.; Bonicelli, A. From flesh to bones: Multi-omics approaches in forensic science. Proteomics 2024, 24, e2200335. [Google Scholar] [CrossRef]
- Cao, J.; Wei, X.; Liu, M.-F.; An, G.-S.; Li, J.; Du, Q.-X.; Sun, J.-H. Forensic identification of sudden cardiac death: A new approach combining metabolomics and machine learning. Anal. Bioanal. Chem. 2023, 415, 2291–2305. [Google Scholar] [CrossRef] [PubMed]
- Bockus, L.B.; Jensen, P.N.; Fretts, A.M.; Hoofnagle, A.N.; McKnight, B.; Sitlani, C.M.; Siscovick, D.S.; King, I.B.; Psaty, B.M.; Sotoodehnia, N.; et al. Plasma Ceramides and Sphingomyelins and SCD in the Cardiovascular Health Study. JAMA Netw. Open 2023, 6, e2343854. [Google Scholar] [CrossRef] [PubMed]
- Lv, J.; Pan, C.; Cai, Y.; Han, X.; Wang, C.; Ma, J.; Pang, J.; Xu, F.; Wu, S.; Kou, T.; et al. Plasma metabolomics reveals the shared and distinct metabolic disturbances associated with cardiovascular events in coronary artery disease. Nat. Commun. 2024, 15, 5729. [Google Scholar] [CrossRef]
- Chighine, A.; Locci, E.; Nioi, M.; D’aLoja, E. Looking for Post-Mortem Metabolomic Standardization: Waiting for Godot—The Importance of Post-Mortem Interval in Forensic Metabolomics. Chem. Res. Toxicol. 2021, 34, 1946–1947. [Google Scholar] [CrossRef]
- Ding, M.; Rexrode, K.M. A Review of Lipidomics of Cardiovascular Disease Highlights the Importance of Isolating Lipoproteins. Metabolites 2020, 10, 163. [Google Scholar] [CrossRef] [PubMed]
- Pistritu, D.-V.; Vasiliniuc, A.-C.; Vasiliu, A.; Visinescu, E.-F.; Visoiu, I.-E.; Vizdei, S.; Anghel, P.M.; Tanca, A.; Bucur, O.; Liehn, E.A. Phospholipids, the Masters in the Shadows during Healing after Acute Myocardial Infarction. Int. J. Mol. Sci. 2023, 24, 8360. [Google Scholar] [CrossRef]
- O’LEary, E.I.; Jiang, Z.; Strub, M.-P.; Lee, J.C. Effects of phosphatidylcholine membrane fluidity on the conformation and aggregation of N-terminally acetylated α-synuclein. J. Biol. Chem. 2018, 293, 11195–11205. [Google Scholar] [CrossRef]
- Patwardhan, G.A.; Beverly, L.J.; Siskind, L.J. Sphingolipids and mitochondrial apoptosis. J. Bioenerg. Biomembr. 2016, 48, 153–168. [Google Scholar] [CrossRef]
- Peters, L.; Kuebler, W.M.; Simmons, S. Sphingolipids in Atherosclerosis: Chimeras in Structure and Function. Int. J. Mol. Sci. 2022, 23, 11948. [Google Scholar] [CrossRef] [PubMed]
- Burrello, J.; Biemmi, V.; Dei Cas, M.; Amongero, M.; Bolis, S.; Lazzarini, E.; Bollini, S.; Vassalli, G.; Paroni, R.; Barile, L. Sphingolipid composition of circulating extracellular vesicles after myocardial ischemia. Sci. Rep. 2020, 10, 16182. [Google Scholar] [CrossRef]
- Bar, S.Y.; Pintel, N.; Alghne, H.A.; Khattib, H.; Avni, D. The therapeutic potential of sphingolipids for cardiovascular diseases. Front. Cardiovasc. Med. 2023, 10, 1224743. [Google Scholar] [CrossRef] [PubMed]
- Shu, H.; Peng, Y.; Hang, W.; Li, N.; Zhou, N.; Wang, D.W. Emerging Roles of Ceramide in Cardiovascular Diseases. Aging Dis. 2022, 13, 232–245. [Google Scholar] [CrossRef]
- Borodzicz-Jażdżyk, S.; Jażdżyk, P.; Łysik, W.; Cudnoch-Jȩdrzejewska, A.; Czarzasta, K. Sphingolipid metabolism and signaling in cardiovascular diseases. Front. Cardiovasc. Med. 2022, 9, 915961. [Google Scholar] [CrossRef]
- Tomczyk, M.M.; Dolinsky, V.W. The Cardiac Lipidome in Models of Cardiovascular Disease. Metabolites 2020, 10, 254. [Google Scholar] [CrossRef] [PubMed]
- Esan, O.; Wierzbicki, A.S. Triglycerides and cardiovascular disease. Curr. Opin. Cardiol. 2021, 36, 469–477. [Google Scholar] [CrossRef]
- Schulze, P.C.; Drosatos, K.; Goldberg, I.J. Lipid Use and Misuse by the Heart. Circ. Res. 2016, 118, 1736–1751. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Wu, Z.; Li, H.; Wang, Z.; Chen, Y.; Bao, J.; Chen, B.; Xu, S.; Xia, E.; Ye, D.; et al. Glycosylphosphatidylinositol anchor biosynthesis pathway-based biomarker identification with machine learning for prognosis and T cell exhaustion status prediction in breast cancer. Front. Immunol. 2024, 15, 1392940. [Google Scholar] [CrossRef]
- Guo, X.; Chen, Y.; Liu, Q. Necroptosis in heart disease: Molecular mechanisms and therapeutic implications. J. Mol. Cell. Cardiol. 2022, 169, 74–83. [Google Scholar] [CrossRef]
- Mujtaba, S.; Romero, J.; Taub, C.C. Methadone, QTc prolongation and torsades de pointes: Current concepts, management and a hidden twist in the tale? J. Cardiovasc. Dis. Res. 2013, 4, 229–235. [Google Scholar] [CrossRef]
- Fischbach, P. The role of illicit drug use in sudden death in the young. Cardiol. Young 2017, 27 (Suppl. S1), S75–S79. [Google Scholar] [CrossRef]
- Pang, Z.; Lu, Y.; Zhou, G.; Hui, F.; Xu, L.; Viau, C.; Spigelman, A.F.; E MacDonald, P.; Wishart, D.S.; Li, S.; et al. MetaboAnalyst 6.0: Towards a unified platform for metabolomics data processing, analysis and interpretation. Nucleic Acids Res. 2024, 52, W398–W406. [Google Scholar] [CrossRef]
- Leegwater, H.; Zhang, Z.; Zhang, X.; Hankemeier, T.; Harms, A.C.; Zweemer, A.J.M.; Le Dévédec, S.E.; Kindt, A. Normalization strategies for lipidome data in cell line panels. bioRxiv 2024. [Google Scholar] [CrossRef]
- Acevedo, A.; Durán, C.; Ciucci, S.; Gerl, M.; Cannistraci, C.V. LIPEA: Lipid Pathway Enrichment Analysis. bioRxiv 2018. [Google Scholar] [CrossRef]
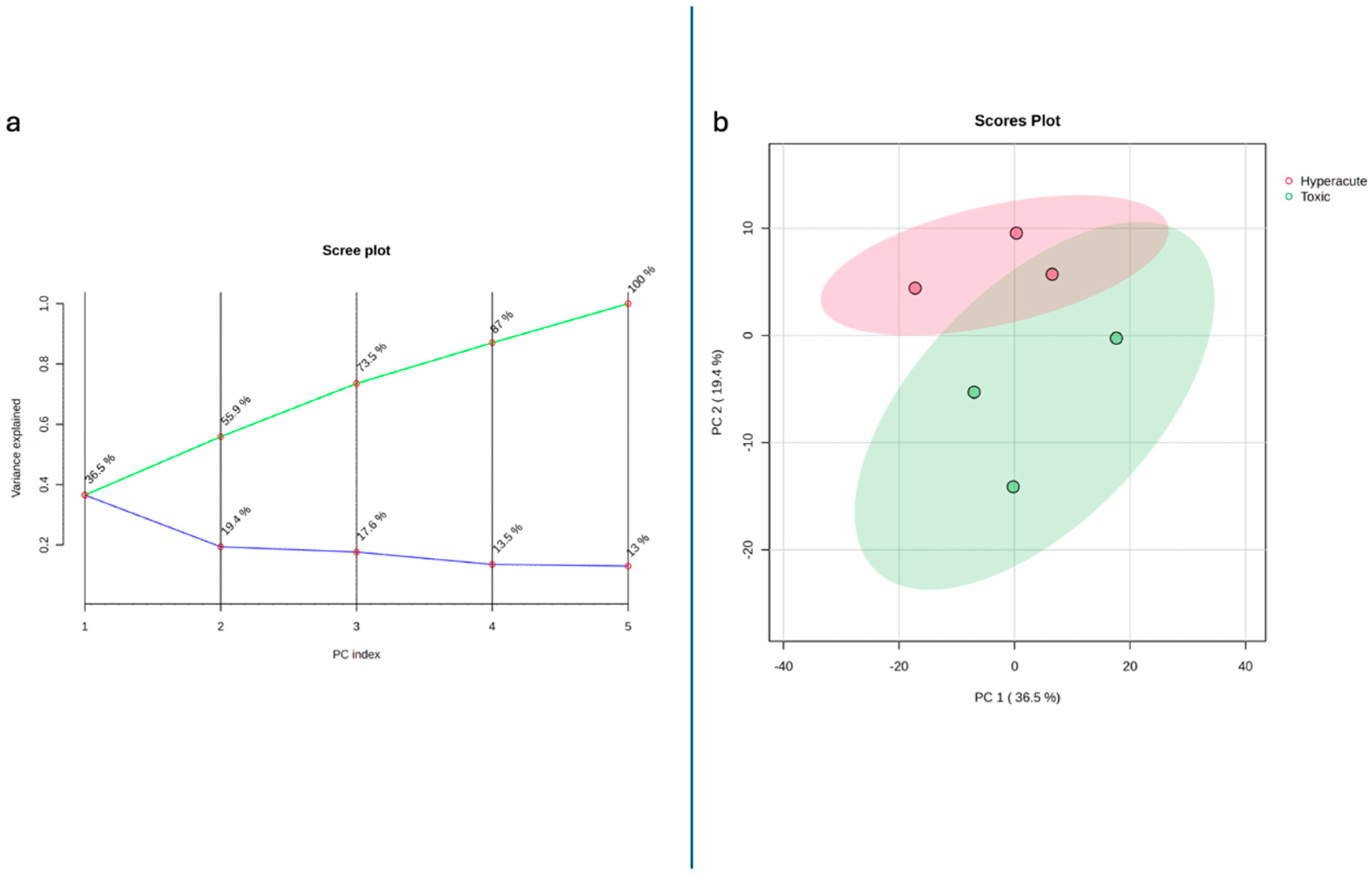



| ID | Sex | Age | Coronary Artery | Histology | Pharmacological Anamnesis | Toxicological Analyses | Cause of Death |
|---|---|---|---|---|---|---|---|
| 1 | F | 50 | Critical stenosis of LCA and AD (85%) | Waviness, contraction band necrosis, fibrosis | None | Negative | Acute ischemic heart disease |
| 2 | M | 50 | Critical stenosis of LCA (80%) and RCA | Fibrosis | None | Negative | Acute ischemic heart disease |
| 3 | F | 36 | Unremarkable | Contraction band necrosis | Methadone | Methadone, benzodiazepines | Drug abuse |
| 4 | M | 32 | Stenosis < 50% | Contraction band necrosis, fibrosis | None | Morphine, cocaine | Drug abuse |
| 5 | M | 35 | Unremarkable | Contraction band necrosis, fibrosis | Levetiracetam, lacosamide | Cocaine, levetiracetam, lacosamide | Drug abuse |
| 6 | M | 50 | Critical stenosis of LCA (90%) | Contraction band necrosis, fibrosis | Levothyroxine | Negative | Acute ischemic heart disease |
| PC1 | PC2 | PC3 | PC4 | PC5 | PC6 | |
|---|---|---|---|---|---|---|
| PC 18:1_22:5 | 0.08268 | −0.001322 | 0.01877 | −0.0032998 | 0.019833 | 0.0047293 |
| PE 18:1_22:6 | 0.082675 | −0.0018474 | 0.018954 | −0.0038093 | 0.019489 | −0.010676 |
| PC O-36:1_A | 0.082265 | −0.018953 | 0.017814 | −0.0092145 | 0.0047633 | 0.0054953 |
| PC O-36:2_C | 0.081399 | 0.016645 | 0.00069367 | −0.03186 | −0.0025386 | 0.012184 |
| PS 18:0_22:5_B | 0.081365 | 0.016019 | 0.00068467 | −0.032586 | −0.0019647 | 0.02206 |
| PC O-32:0_B | 0.081114 | 0.0095399 | 0.0253 | 0.021802 | −0.010351 | 0.016975 |
| DG 18:1/18:1 | 0.08104 | 0.0097585 | 0.025643 | 0.021807 | −0.010522 | 0.01667 |
| PC O-40:4_A | 0.080877 | 0.014356 | 0.001999 | −0.01839 | 0.032414 | 0.0053446 |
| PC 16:0/16:0_A | 0.080873 | 0.014213 | 0.0017864 | −0.018386 | 0.032557 | 0.017219 |
| PC 16:0_17:0 | 0.079403 | 0.031759 | −0.024691 | −0.0061435 | 0.0026323 | −0.0042648 |
| PC O-40:5_D | 0.0794 | 0.031783 | −0.024647 | −0.0064169 | 0.0023449 | 0.0034258 |
| SM 34:2;2O | 0.079356 | 0.031872 | −0.024789 | −0.0065367 | 0.0023463 | 0.00348 |
| PC 18:0_18:1_A | 0.0787 | 0.019454 | 0.0092217 | 0.01544 | 0.042007 | 0.036046 |
| TG 18:1_20:4_22:6 | 0.078681 | 0.019355 | 0.0090687 | 0.01536 | 0.042248 | 0.012975 |
| PE 17:1_18:1 | 0.078677 | 0.019372 | 0.0091107 | 0.015384 | 0.042236 | 0.020715 |
| PI 40:6 | 0.078587 | −0.01689 | 0.033612 | −0.020057 | −0.019065 | 0.020643 |
| LPC 16:0 | 0.078521 | −0.017407 | 0.033926 | −0.019703 | −0.01876 | 0.024681 |
| PI 34:2 | 0.078172 | −0.0078061 | 0.035193 | 0.018927 | 0.027916 | −0.0059157 |
| PC O-32:1_B | 0.078117 | −0.0079559 | 0.035441 | 0.019066 | 0.027765 | 0.013363 |
| DG O-16:0_16:0 | 0.078086 | −0.0081328 | 0.035356 | 0.019247 | 0.027949 | 0.0057207 |
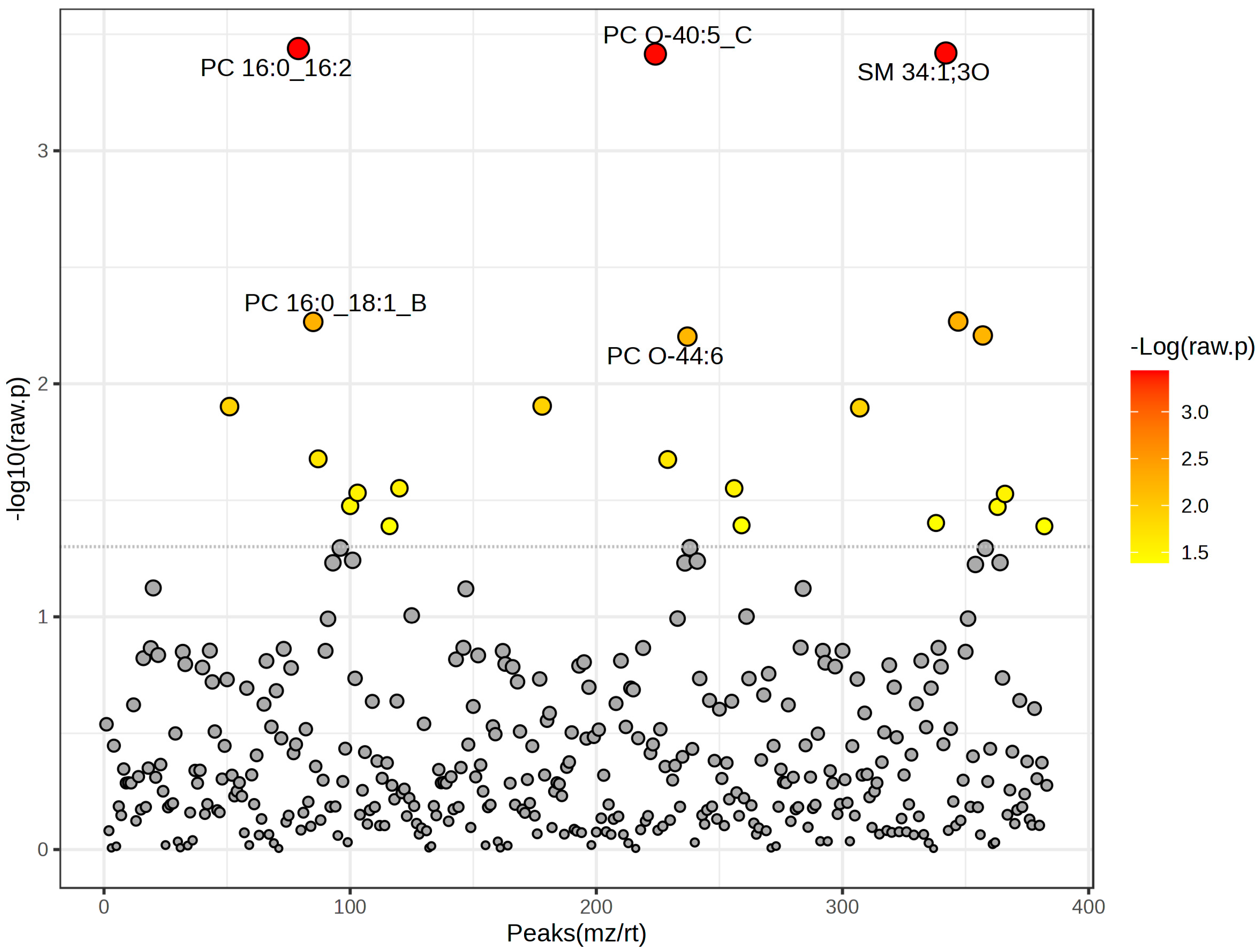
| Lipids | t-Stat | p-Value | −log10(p) | FDR |
|---|---|---|---|---|
| PC 16:0_16:2 | 11.182 | 0.00036411 | 3.4388 | 0.049095 |
| SM 34:1;3O | 11.056 | 0.00038053 | 3.4196 | 0.049095 |
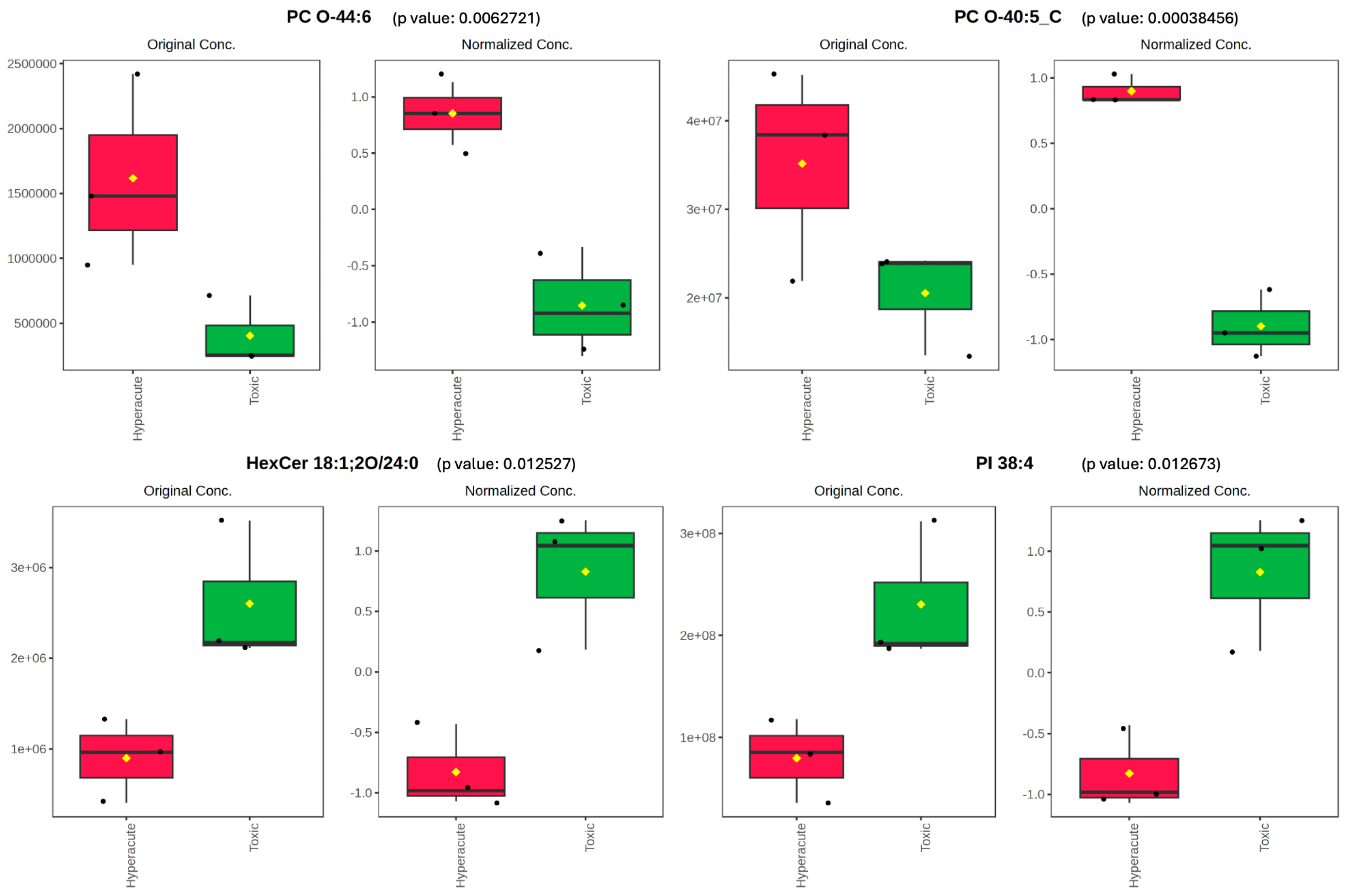
| PC O-40:5_C | 11.026 | 0.00038456 | 3.415 | 0.049095 |
| SM 40:2;2O_A | 5.4796 | 0.0054 | 2.2676 | 0.34317 |
| PC 16:0_18:1_B | 5.4733 | 0.0054225 | 2.2658 | 0.34317 |
| TG 16:0_18:1_20:4 | 5.2722 | 0.0062027 | 2.2074 | 0.34317 |
| PC O-44:6 | 5.2559 | 0.0062721 | 2.2026 | 0.34317 |
| PC O-34:0_B | −4.3202 | 0.012446 | 1.905 | 0.48536 |
| HexCer 18:1;2O/24:0 | −4.3119 | 0.012527 | 1.9021 | 0.48536 |
| PI 38:4 | −4.2973 | 0.012673 | 1.8971 | 0.48536 |
| PC 16:0_20:4_B | 3.6917 | 0.020988 | 1.678 | 0.67426 |
| PC O-40:8_A | 3.6842 | 0.021125 | 1.6752 | 0.67426 |
| PC 18:1_22:4 | −3.3692 | 0.028064 | 1.5518 | 0.71071 |
| PE 18:1_20:4_B | −3.3679 | 0.028098 | 1.5513 | 0.71071 |
| PC 17:1_18:1 | 3.3204 | 0.029368 | 1.5321 | 0.71071 |
| TG 18:0_19:1_22:6 | 3.3087 | 0.02969 | 1.5274 | 0.71071 |
| PC 17:0_18:0 | 3.1829 | 0.033442 | 1.4757 | 0.71163 |
| TG 18:0_18:1_22:3 | 3.174 | 0.033729 | 1.472 | 0.71163 |
| SM 20:1;2O/13:0_A | 3.0096 | 0.039564 | 1.4027 | 0.71163 |
| PE 22:1_17:2 | −2.9863 | 0.040484 | 1.3927 | 0.71163 |
| PC 18:1_18:2_B | −2.9779 | 0.040825 | 1.3891 | 0.71163 |
| TG O-18:1_22:3_22:5 | −2.9766 | 0.040877 | 1.3885 | 0.71163 |
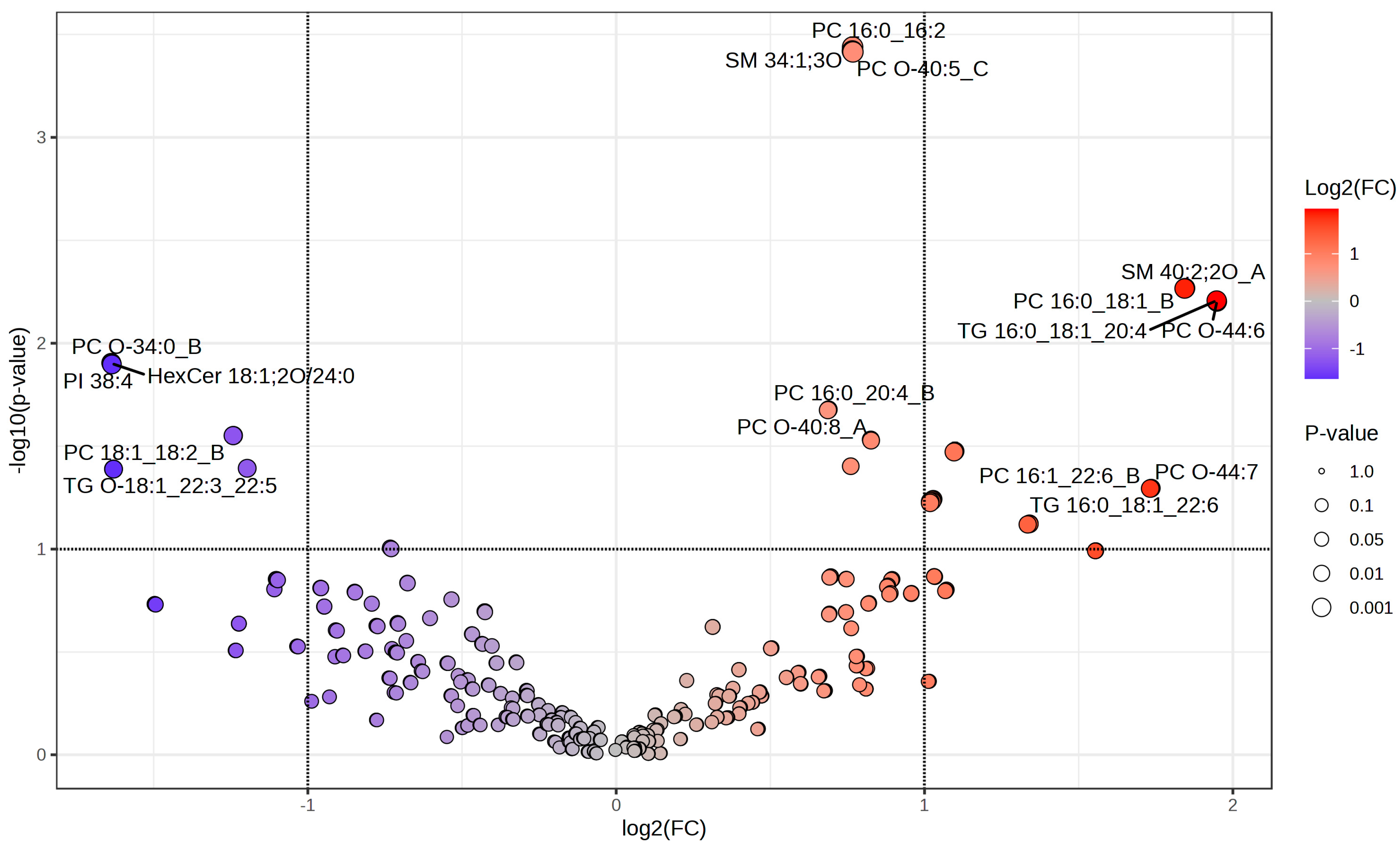
| FC | log2(FC) | Raw p-Val | −log10(p) | |
|---|---|---|---|---|
| SM 40:2;2O_A | 3.5923 | 1.8449 | 0.0054 | 2.2676 |
| PC 16:0_18:1_B | 3.5887 | 1.8434 | 0.0054225 | 2.2658 |
| TG 16:0_18:1_20:4 | 3.8569 | 1.9474 | 0.0062027 | 2.2074 |
| PC O-44:6 | 3.8589 | 1.9482 | 0.0062721 | 2.2026 |
| PC O-34:0_B | 0.32141 | −1.6375 | 0.012446 | 1.905 |
| HexCer 18:1;2O/24:0 | 0.3215 | −1.6371 | 0.012527 | 1.9021 |
| PI 38:4 | 0.32186 | −1.6355 | 0.012673 | 1.8971 |
| PC 18:1_22:4 | 0.42293 | −1.2415 | 0.028064 | 1.5518 |
| PE 18:1_20:4_B | 0.42272 | −1.2422 | 0.028098 | 1.5513 |
| PC 17:0_18:0 | 2.1415 | 1.0986 | 0.033442 | 1.4757 |
| TG 18:0_18:1_22:3 | 2.1372 | 1.0957 | 0.033729 | 1.472 |
| PE 22:1_17:2 | 0.43626 | −1.1967 | 0.040484 | 1.3927 |
| PC 18:1_18:2_B | 323 | −1.6304 | 0.040825 | 1.3891 |
| TG O-18:1_22:3_22:5 | 0.32306 | −1.6301 | 0.040877 | 1.3885 |
| PC O-44:7 | 3.3297 | 1.7354 | 0.050556 | 1.2962 |
| PC 16:1_22:6_B | 3.327 | 1.7342 | 0.050653 | 1.2954 |
| TG 16:0_18:1_22:6 | 3.3216 | 1.7319 | 0.050772 | 1.2944 |
| PC 17:0_18:1 | 2.0392 | 1.028 | 0.057225 | 1.2424 |
| PE 16:0_18:1 | 2.0349 | 1.0249 | 0.057639 | 1.2393 |
| TG 18:0_18:1_22:4 | 2.0342 | 1.0245 | 0.058514 | 1.2327 |
| PC 16:1_18:1 | 2.0255 | 1.0182 | 0.058641 | 1.2318 |
| PC O-42:7 | 2.0265 | 1.019 | 0.058718 | 1.2312 |
| TG 14:0_20:3_21:1 | 2.0251 | 1.018 | 0.059619 | 1.2246 |
| Cer 16:1;2O/16:0 | 2.5321 | 1.3404 | 0.07518 | 1.1239 |
| PE P-18:1_20:1 | 2.5322 | 1.3404 | 0.075568 | 1.1217 |
| PC 38:5 | 2.5219 | 1.3345 | 0.075859 | 1.12 |
| Pathway Name | Pathway Lipids | Converted Lipids (Number) | Converted Lipids (Percentage) | Converted Lipids (List) | p-Value | Benjamini Correction | Bonferroni Correction |
|---|---|---|---|---|---|---|---|
| Glycosylphosphatidylinositol (GPI)-anchor biosynthesis | 3 | 2 | 28.57142857 | C00350, C01194 | 0.000435028 | 0.003480223 | 0.006960447 |
| Inositol phosphate metabolism | 9 | 1 | 14.28571429 | C01194 | 0.112178644 | 0.163168937 | 1 |
| Ether lipid metabolism | 16 | 1 | 14.28571429 | C05212 | 0.191801237 | 0.236063061 | 1 |
| Linoleic acid metabolism | 25 | 1 | 14.28571429 | C00157 | 0.285119368 | 0.304127326 | 1 |
| Glycerophospholipid metabolism | 26 | 3 | 42.85714286 | C00157, C00350, C01194 | 0.003109731 | 0.012438924 | 0.049755697 |
| Alpha-linolenic acid metabolism | 23 | 1 | 14.28571429 | C00157 | 0.265223024 | 0.303112028 | 1 |
| Autophagy-other | 3 | 2 | 28.57142857 | C01194, C00350 | 0.000435028 | 0.003480223 | 0.006960447 |
| Arachidonic acid metabolism | 75 | 1 | 14.28571429 | C00157 | 0.653348996 | 0.653348996 | 1 |
| Autophagy-animal | 4 | 2 | 28.57142857 | C00350, C01194 | 0.000864632 | 0.004611368 | 0.013834105 |
| Ferroptosis | 11 | 2 | 28.57142857 | C21480, C21481 | 0.007585493 | 0.020227982 | 0.121367892 |
| Phosphatidylinositol signaling system | 11 | 1 | 14.28571429 | C01194 | 0.135585354 | 0180780472 | 1 |
| Retrograde endocannabinoid signaling | 8 | 2 | 28.57142857 | C00157, C00350 | 0.003935112 | 0.012592358 | 0.062961791 |
| Pathogenic Escherichia coli infection | 1 | 1 | 14.28571429 | C00350 | 0.013035382 | 0.029795158 | 0.208566108 |
| Tuberculosis | 5 | 1 | 14.28571429 | C01194 | 0.06373131 | 0.101970097 | 1 |
| Kaposi’s sarcoma-associated herpesvirus infection | 3 | 1 | 14.28571429 | C00350 | 0.038669754 | 0.077339507 | 0.618716058 |
| Choline metabolism in cancer | 5 | 1 | 14.28571429 | C00157 | 0.06373131 | 0.101970097 | 1 |
| Pathway Name | Pathway Lipids | Converted Lipids (Number) | Converted Lipids (Percentage) | Converted Lipids (List) | p-Value | Benjamini Correction | Bonferroni Correction |
|---|---|---|---|---|---|---|---|
| Glycerophospholipid metabolism | 26 | 1 | 33.33333333 | C00157 | 0.138577917 | 0.153975463 | 1 |
| Ether lipid metabolism | 16 | 1 | 33.33333333 | C05212 | 0.086905835 | 0.153975463 | 0.869058353 |
| Linoleic acid metabolism | 25 | 1 | 33.33333333 | C00157 | 0.133500773 | 0.153975463 | 1 |
| Sphingolipid metabolism | 21 | 1 | 33.33333333 | C00550 | 0.112992704 | 0.153975463 | 1 |
| Arachidonic acid metabolism | 75 | 1 | 33.33333333 | C00157 | 0.363779183 | 0.363779183 | 1 |
| Alpha-linolenic acid metabolism | 23 | 1 | 33.33333333 | C00157 | 0.123286715 | 0.153975463 | 1 |
| Sphingolipid signaling pathway | 9 | 1 | 33.33333333 | C00550 | 0.049532165 | 0.123830412 | 0.495321648 |
| Necroptosis | 4 | 1 | 33.33333333 | C00550 | 0.022221452 | 0.123830412 | 0.222214516 |
| Retrograde endocannabinoid signaling | 8 | 1 | 33.33333333 | C00157 | 0.044111246 | 0.123830412 | 0.441112456 |
| Choline metabolism in cancer | 5 | 1 | 33.33333333 | C00157 | 0.027724896 | 0.123830412 | 0.277248956 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Radaelli, D.; Concato, M.; Bruscagin, T.; Sinagra, G.; Stornaiuolo, M.; D’Errico, S. Lipidomic Profiling of Hyperacute Ischemic Heart Disease and Toxic Deaths: A Forensic Investigation into Metabolic Biomarkers. Int. J. Mol. Sci. 2025, 26, 9031. https://doi.org/10.3390/ijms26189031
Radaelli D, Concato M, Bruscagin T, Sinagra G, Stornaiuolo M, D’Errico S. Lipidomic Profiling of Hyperacute Ischemic Heart Disease and Toxic Deaths: A Forensic Investigation into Metabolic Biomarkers. International Journal of Molecular Sciences. 2025; 26(18):9031. https://doi.org/10.3390/ijms26189031
Chicago/Turabian StyleRadaelli, Davide, Monica Concato, Tommaso Bruscagin, Gianfranco Sinagra, Mariano Stornaiuolo, and Stefano D’Errico. 2025. "Lipidomic Profiling of Hyperacute Ischemic Heart Disease and Toxic Deaths: A Forensic Investigation into Metabolic Biomarkers" International Journal of Molecular Sciences 26, no. 18: 9031. https://doi.org/10.3390/ijms26189031
APA StyleRadaelli, D., Concato, M., Bruscagin, T., Sinagra, G., Stornaiuolo, M., & D’Errico, S. (2025). Lipidomic Profiling of Hyperacute Ischemic Heart Disease and Toxic Deaths: A Forensic Investigation into Metabolic Biomarkers. International Journal of Molecular Sciences, 26(18), 9031. https://doi.org/10.3390/ijms26189031







