Transcriptomic Identification of Immune-Related Hubs as Candidate Predictor Biomarkers of Therapeutic Response in Psoriasis
Abstract
1. Introduction
2. Results
2.1. Patient Demographics
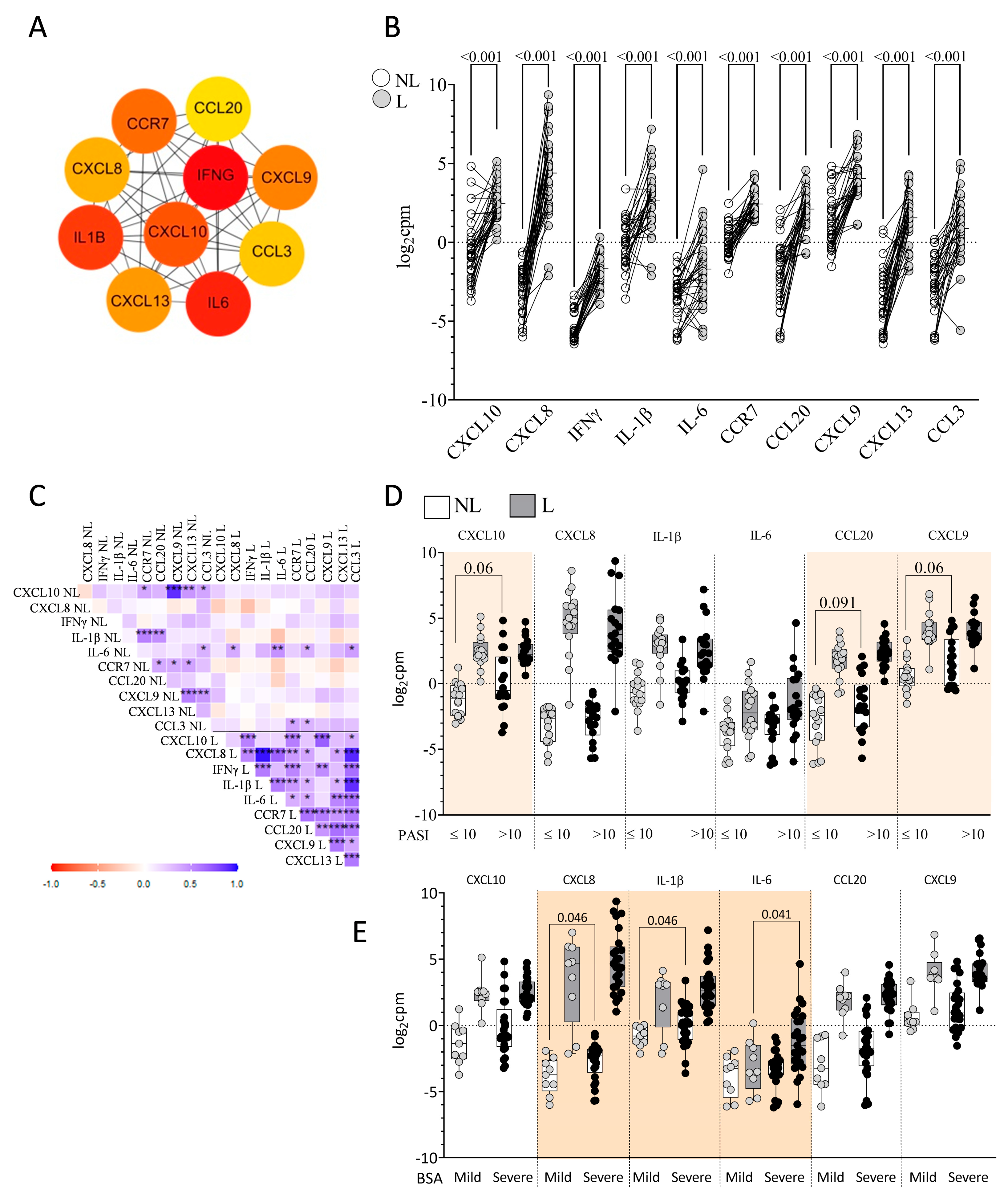
2.2. Identification of DEGs in L vs. NL Skin
2.3. Identification of Skin Immune Hub Genes
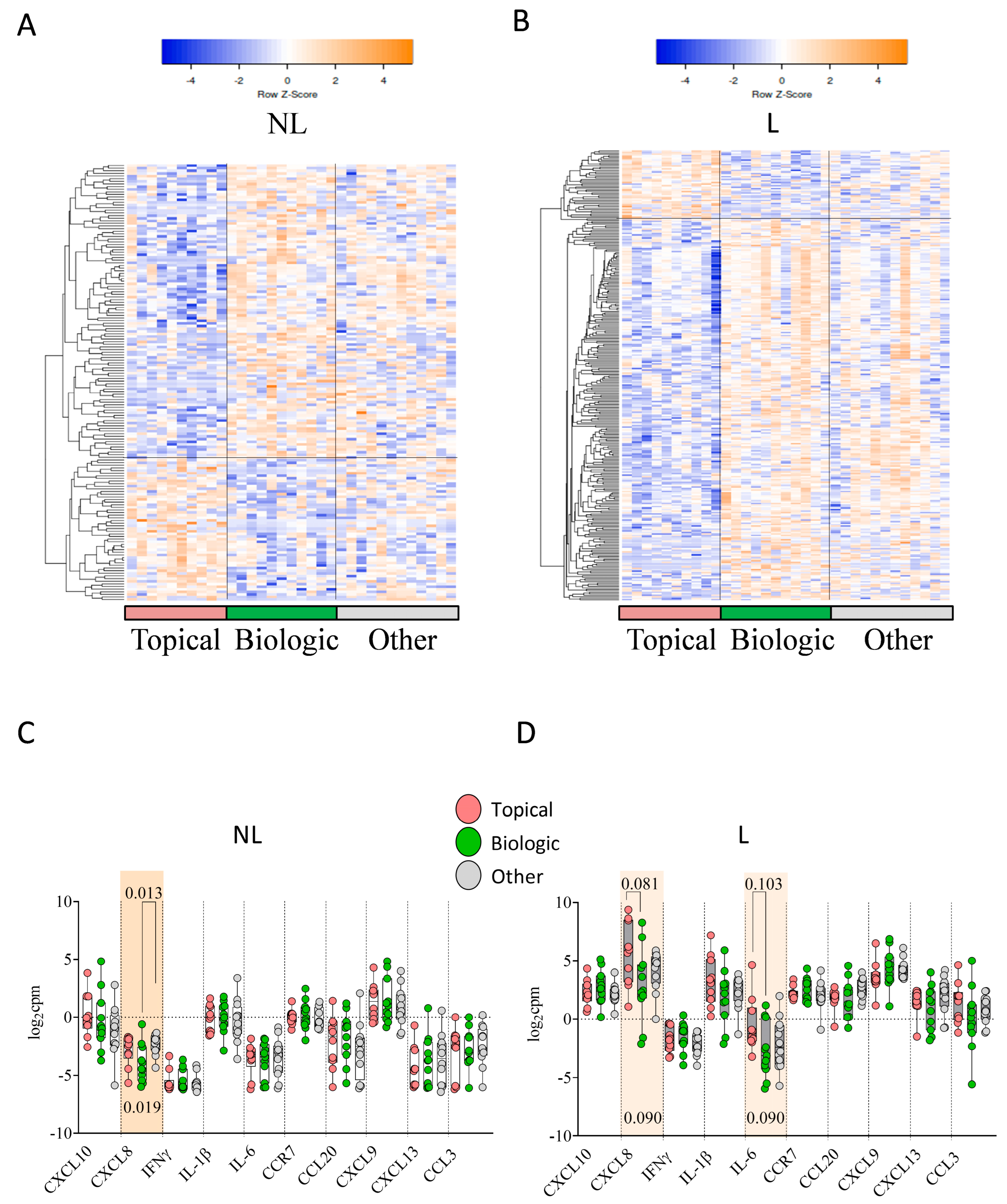
2.4. Previous Treatment Conditioned Differential Skin DEGs and Skin Immune Hubs Expression
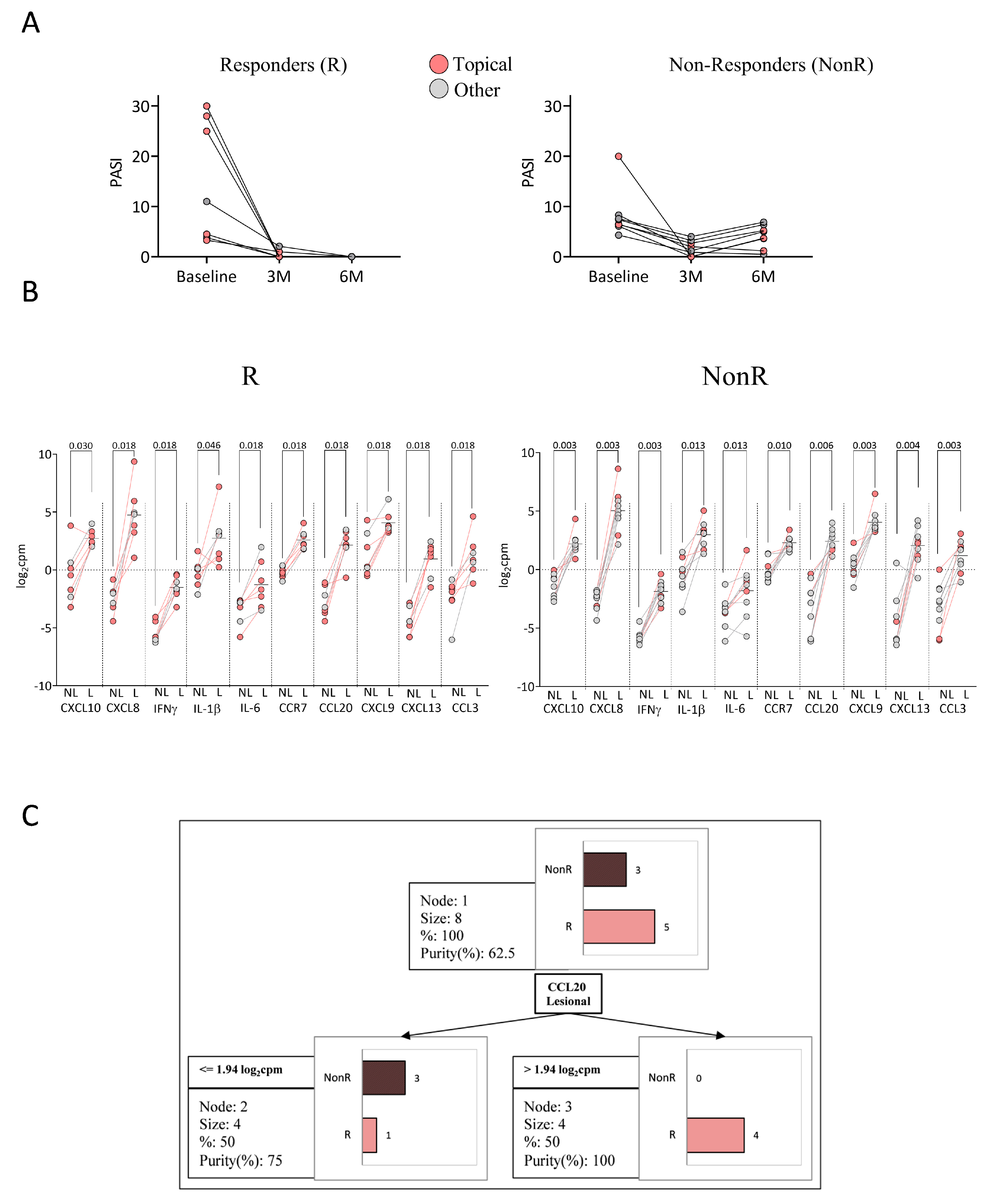
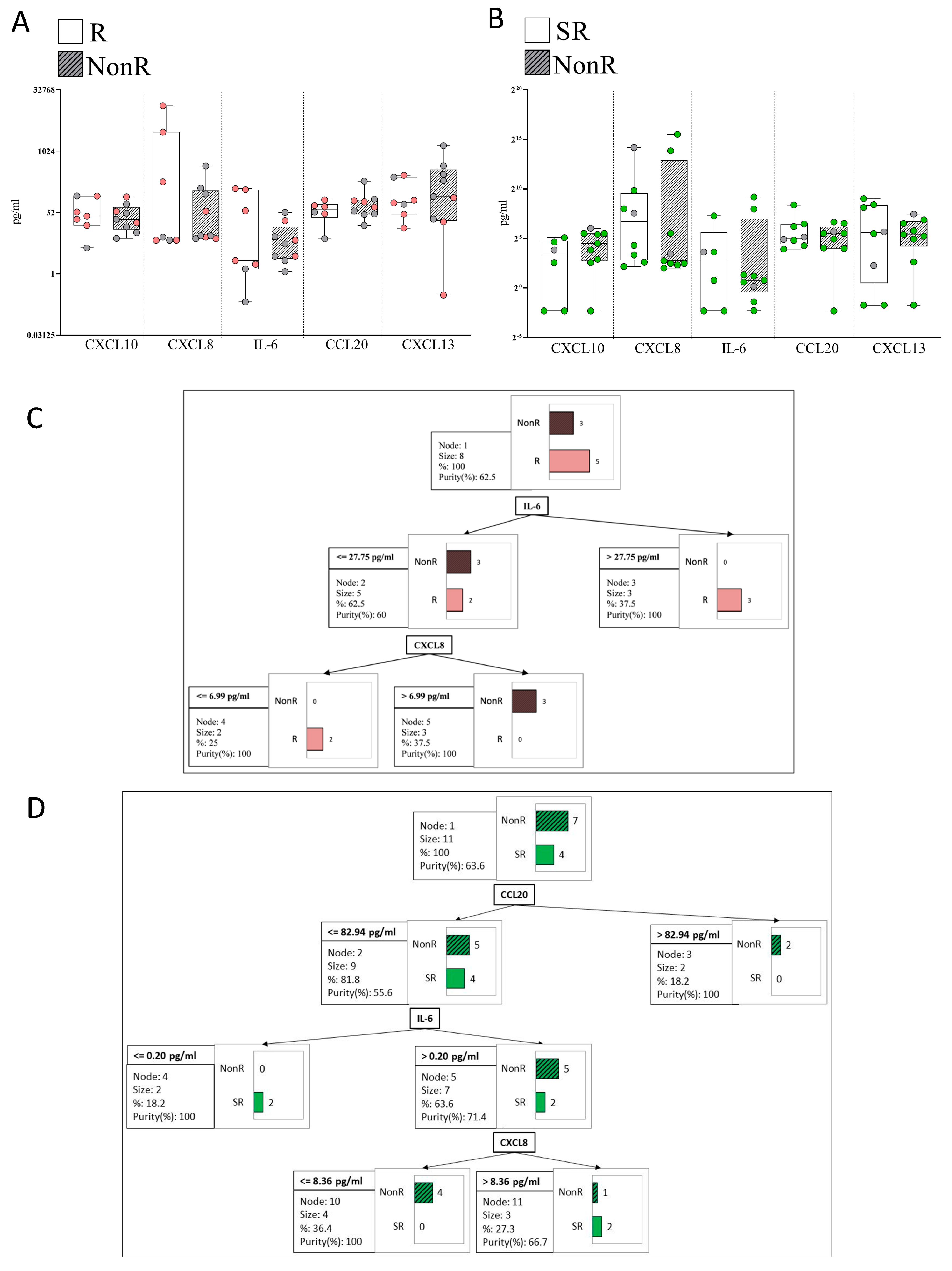
2.5. Response to Anti-TNFα
2.6. Response to Anti-IL-23
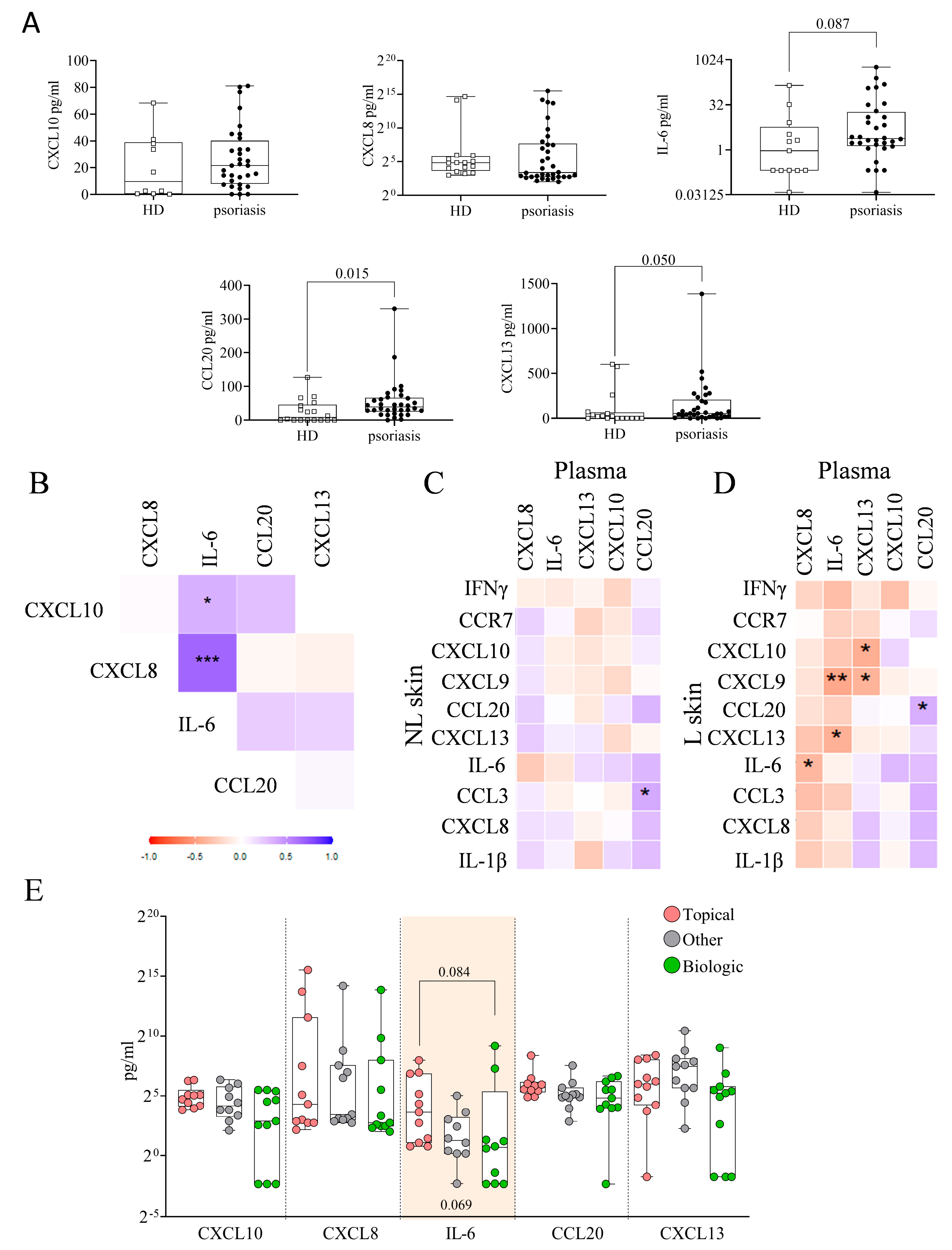
2.7. Plasma Immune Hubs and Their Relationships with Clinical Psoriasis Characteristics
2.8. Plasma Immune Hub Expression Conditions Anti-TNFα and Anti-IL-23 Response
3. Discussion
4. Materials and Methods
4.1. Patient Cohort and Samples
4.2. RNA Sequencing
4.3. Functional Enrichment Analysis
4.4. Protein–Protein Interaction (PPI) Network Construction and Immune-Related Hub Genes
4.5. ELISAs
4.6. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kalayciyan, A.; Aydemir, E.H.; Kotogyan, A. Experimental Koebner Phenomenon in Patients with Psoriasis. Dermatology 2007, 215, 114–117. [Google Scholar] [CrossRef]
- Dorschner, R.A.; Pestonjamasp, V.K.; Tamakuwala, S.; Ohtake, T.; Rudisill, J.; Nizet, V.; Agerberth, B.; Gudmundsson, G.H.; Gallo, R.L. Cutaneous Injury Induces the Release of Cathelicidin AntiMicrobial Peptides Active Against Group A Streptococcus. J. Investig. Dermatol. 2001, 117, 91–97. [Google Scholar] [CrossRef]
- Winterfield, L.S.; Menter, A.; Gordon, K.; Gottlieb, A. Psoriasis treatment: Current and emerging directed therapies. Ann. Rheum. Dis. 2005, 64 (Suppl. 2), ii87–ii90; discussion ii91–ii92. [Google Scholar] [CrossRef]
- Chow, M.; Lai, K.; Ahn, R.; Gupta, R.; Arron, S. Effect of adalimumab on gene expression profiles of psoriatic. J. Drugs Dermatol. 2016, 15, 988–994. [Google Scholar]
- Teng, M.W.; Bowman, E.P.; McElwee, J.J.; Smyth, M.J.; Casanova, J.L.; Cooper, A.M.; Cua, D.J. IL-12 and IL-23 cytokines: From discovery to targeted therapies for immune-mediated inflammatory diseases. Nat. Med. 2015, 21, 719–729. [Google Scholar] [CrossRef] [PubMed]
- Lande, R.; Gregorio, J.; Facchinetti, V.; Chatterjee, B.; Wang, Y.H.; Homey, B.; Cao, W.; Wang, Y.H.; Su, B.; Nestle, F.O.; et al. Plasmacytoid dendritic cells sense self-DNA coupled with antimicrobial peptide. Nature 2007, 449, 564–569. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Wang, Y.; Li, Y.; Zhong, X.; Gong, Y.; Ding, Y.; Yu, N.; Shi, Y. Based on Gene Expression Analysis: Low-Density Neutrophil Expression Is a Characteristic of the Fast Responders Treated With Guselkumab for Psoriasis. Front. Immunol. 2022, 13, 865875. [Google Scholar] [CrossRef] [PubMed]
- Tsoi, L.C.; Patrick, M.T.; Shuai, S.; Sarkar, M.K.; Chi, S.; Ruffino, B.; Billi, A.C.; Xing, X.; Uppala, R.; Zang, C.; et al. Cytokine responses in nonlesional psoriatic skin as clinical predictor to anti-TNF agents. J. Allergy. Clin. Immunol. 2022, 149, 640–649.e5. [Google Scholar] [CrossRef]
- Armesto, S.; Coto-Segura, P.; Mayorga, J.; Illaro, A.; Santos-Juanes, J. Efficacy of adalimumab in the treatment of moderate-to-severe psoriasis: A retrospective study of 100 patients in daily practice. J. Dermatol. Treat 2015, 26, 49–53. [Google Scholar] [CrossRef]
- Ruiz-Villaverde, R.; Vasquez-Chinchay, F.; Rodriguez-Fernandez-Freire, L.; C. Armario-Hita, J.; Perez-Gil, A.; Galan-Gutierrez, M. Super-Responders in Moderate-Severe Psoriasis under Guselkumab Treatment: Myths, Realities and Future Perspectives. Life 2022, 12, 1412. [Google Scholar] [CrossRef]
- Albanesi, C.; Madonna, S.; Gisondi, P.; Girolomoni, G. The Interplay Between Keratinocytes and Immune Cells in the Pathogenesis of Psoriasis. Front. Immunol. 2018, 9, 1549. [Google Scholar] [CrossRef] [PubMed]
- Su, W.; Wei, Y.; Huang, B.; Ji, J. Identification of Hub Genes and Immune Infiltration in Psoriasis by Bioinformatics Method. Front. Genet. 2021, 12, 606065. [Google Scholar] [CrossRef]
- Li, Y.; Li, L.; Tian, Y.; Luo, J.; Huang, J.; Zhang, L.; Zhang, J.; Li, X.; Hu, L. Identification of novel immune subtypes and potential hub genes of patients with psoriasis. J. Transl. Med. 2023, 21, 182. [Google Scholar] [CrossRef]
- Zhang, Y.J.; Sun, Y.Z.; Gao, X.H.; Qi, R.Q. Integrated bioinformatic analysis of differentially expressed genes and signaling pathways in plaque psoriasis. Mol. Med. Rep. 2019, 20, 225–235. [Google Scholar] [CrossRef]
- Deng, W.; Yan, Y.; Shi, C.; Sui, D. Single-cell and bulk RNAseq unveils the immune infiltration landscape and targeted therapeutic biomarkers of psoriasis. Front. Genet. 2024, 15, 1365273. [Google Scholar] [CrossRef]
- Choudhary, S.; Anand, R.; Pradhan, D.; Bastia, B.; Kumar, S.N.; Singh, H.; Puri, P.; Thomas, G.; Jain, A.K. Transcriptomic landscaping of core genes and pathways of mild and severe psoriasis vulgaris. Int. J. Mol. Med. 2021, 47, 219–231. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Chang, H.W.; Huang, Z.M.; Nakamura, M.; Sekhon, S.; Ahn, R.; Munoz-Sandoval, P.; Bhattarai, S.; Beck, K.M.; Sanchez, I.M.; et al. Single-cell RNA sequencing of psoriatic skin identifies pathogenic Tc17 cell subsets and reveals distinctions between CD8(+) T cells in autoimmunity and cancer. J. Allergy Clin. Immunol. 2021, 147, 2370–2380. [Google Scholar] [CrossRef]
- Chen, S.C.; de Groot, M.; Kinsley, D.; Laverty, M.; McClanahan, T.; Arreaza, M.; Gustafson, E.L.; Teunissen, M.B.; de Rie, M.A.; Fine, J.S.; et al. Expression of chemokine receptor CXCR3 by lymphocytes and plasmacytoid dendritic cells in human psoriatic lesions. Arch. Dermatol. Res. 2010, 302, 113–123. [Google Scholar] [CrossRef]
- Krueger, G.G.; Chambers, D.A.; Shelby, J. Involved and Uninvolved Skin from Psoriatic Subjects. J. Clin. Investig. 1981, 68, 1548–1557. [Google Scholar] [CrossRef] [PubMed]
- Gudjonsson, J.E.; Ding, J.; Li, X.; Nair, R.P.; Tejasvi, T.; Qin, Z.S.; Ghosh, D.; Aphale, A.; Gumucio, D.L.; Voorhees, J.J.; et al. Global gene expression analysis reveals evidence for decreased lipid biosynthesis and increased innate immunity in uninvolved psoriatic skin. J. Investig. Dermatol. 2009, 129, 2795–2804. [Google Scholar] [CrossRef]
- Kim, B.E.; Howell, M.D.; Guttman-Yassky, E.; Gilleaudeau, P.M.; Cardinale, I.R.; Boguniewicz, M.; Krueger, J.G.; Leung, D.Y. TNF-alpha downregulates filaggrin and loricrin through c-Jun N-terminal kinase: Role for TNF-alpha antagonists to improve skin barrier. J. Investig. Dermatol. 2011, 131, 1272–1279. [Google Scholar] [CrossRef] [PubMed]
- Szel, E.; Bozo, R.; Hunyadi-Gulyas, E.; Manczinger, M.; Szabo, K.; Kemeny, L.; Bata-Csorgo, Z.; Groma, G. Comprehensive Proteomic Analysis Reveals Intermediate Stage of Non-Lesional Psoriatic Skin and Points out the Importance of Proteins Outside this Trend. Sci. Rep. 2019, 9, 11382. [Google Scholar] [CrossRef]
- Visvanathan, S.; Baum, P.; Vinisko, R.; Schmid, R.; Flack, M.; Lalovic, B.; Kleiner, O.; Fuentes-Duculan, J.; Garcet, S.; Davis, J.W.; et al. Psoriatic skin molecular and histopathologic profiles after treatment with risankizumab versus ustekinumab. J. Allergy Clin. Immunol. 2019, 143, 2158–2169. [Google Scholar] [CrossRef]
- Liu, L.; Liu, W.; Zheng, Y.; Chen, J.; Zhou, J.; Dai, H.; Cai, S.; Liu, J.; Zheng, M.; Ren, Y. Anti-TNF-α treatment-related pathways and biomarkers revealed by transcriptome analysis in Chinese psoriasis patients. BMC Syst. Biol. 2019, 13 (Suppl. 2), 29. [Google Scholar] [CrossRef]
- Onderdijk, A.J.; Balak, D.M.; Baerveldt, E.M.; Florencia, E.F.; Kant, M.; Laman, J.D.; van IJcken, W.F.; Racz, E.; de Ridder, D.; Thio, H.B.; et al. Regulated genes in psoriatic skin during treatment with fumaric acid esters. Br. J. Dermatol. 2014, 171, 732–741. [Google Scholar] [CrossRef] [PubMed]
- Dowlatshahi, E.A.; van der Voort, E.A.; Arends, L.R.; Nijsten, T. Markers of systemic inflammation in psoriasis: A systematic review and meta-analysis. Br. J. Dermatol. 2013, 169, 266–282. [Google Scholar] [CrossRef]
- Furue, K.; Ito, T.; Tsuji, G.; Nakahara, T.; Furue, M. The CCL20 and CCR6 axis in psoriasis. Scand. J. Immunol 2020, 91, e12846. [Google Scholar] [CrossRef]
- Ainali, C.; Valeyev, N.; Perera, G.; Williams, A.; Gudjonsson, J.E.; Ouzounis, C.A.; Nestle, F.O.; Tsoka, S. Transcriptome classification reveals molecular subtypes in psoriasis. BMC Genom. 2012, 13, 472. [Google Scholar] [CrossRef] [PubMed]
- Harper, E.G.; Guo, C.; Rizzo, H.; Lillis, J.V.; Kurtz, S.E.; Skorcheva, I.; Purdy, D.; Fitch, E.; Iordanov, M.; Blauvelt, A. Th17 cytokines stimulate CCL20 expression in keratinocytes in vitro and in vivo: Implications for psoriasis pathogenesis. J. Investig. Dermatol. 2009, 129, 2175–2183. [Google Scholar] [CrossRef] [PubMed]
- Ferrari, S.M.; Ruffilli, I.; Colaci, M.; Antonelli, A.; Ferri, C.; Fallahi, P. CXCL10 in psoriasis. Adv. Med. Sci. 2015, 60, 349–354. [Google Scholar] [CrossRef]
- Gagliani, N.; Amezcua Vesely, M.C.; Iseppon, A.; Brockmann, L.; Xu, H.; Palm, N.W.; de Zoete, M.R.; Licona-Limon, P.; Paiva, R.S.; Ching, T.; et al. Th17 cells transdifferentiate into regulatory T cells during resolution of inflammation. Nature 2015, 523, 221–225. [Google Scholar] [CrossRef]
- Jager, A.; Kuchroo, V.K. Effector and regulatory T-cell subsets in autoimmunity and tissue inflammation. Scand. J. Immunol. 2010, 72, 173–184. [Google Scholar] [CrossRef]
- Sofen, H.; Smith, S.; Matheson, R.T.; Leonardi, C.L.; Calderon, C.; Brodmerkel, C.; Li, K.; Campbell, K.; Marciniak, S.J., Jr.; Wasfi, Y.; et al. Guselkumab (an IL-23-specific mAb) demonstrates clinical and molecular response in patients with moderate-to-severe psoriasis. J. Allergy Clin. Immunol. 2014, 133, 1032–1040. [Google Scholar] [CrossRef]
- Babicki, S.; Arndt, D.; Marcu, A.; Liang, Y.; Grant, J.R.; Maciejewski, A.; Wishart, D.S. Heatmapper: Web-enabled heat mapping for all. Nucleic. Acids. Res. 2016, 44, W147–W153. [Google Scholar] [CrossRef] [PubMed]
- Ge, S.X.; Jung, D.; Yao, R. ShinyGO: A graphical gene-set enrichment tool for animals and plants. Bioinformatics 2020, 36, 2628–2629. [Google Scholar] [CrossRef] [PubMed]
- Bindea, G.; Mlecnik, B.; Hackl, H.; Charoentong, P.; Tosolini, M.; Kirilovsky, A.; Fridman, W.H.; Pages, F.; Trajanoski, Z.; Galon, J. ClueGO: A Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 2009, 25, 1091–1093. [Google Scholar] [CrossRef]
- Antonatos, C.; Koskeridis, F.; Ralliou, C.M.; Evangelou, E.; Grafanaki, K.; Georgiou, S.; Tsilidis, K.K.; Tzoulaki, I.; Vasilopoulos, Y. A polygenic risk score model for psoriasis based on the protein interactions of psoriasis susceptibility loci. Front. Genet. 2024, 15, 1451679. [Google Scholar] [CrossRef] [PubMed]
- Chin, C.H.; Chen, S.H.; Wu, H.H.; Ho, C.H.; Ko, M.T.; Lin, C.Y. cytoHubba: Identifying hub objects and subnetworks from complex interactome. BMC Syst. Biol. 2014, 8 (Suppl. 4), S11. [Google Scholar] [CrossRef]
- Wickham, H.; Navarro, D.; Pedersen, T.L. Ggplot2: Elegant Graphics for Data Analysis (3e); Springer: Cham, Switzerland, 2016. [Google Scholar]


| Patients | Anti-IL-23 (n = 18) | Anti-TNFα (n = 16) | p-Value | |
|---|---|---|---|---|
| Gender Male/Female | 23/11 | 13/5 | 10/6 | n.s |
| Age at baseline | 51.00 (36.50–56.50) | 53.00 (37.00–65.00) | 49.00 (35.50–53.50) | n.s |
| Age at diagnosis | 32.00 (17.75–52.50) | 39.00 (28.50–64.50) | 17.50 (11.25–34.25) | 0.02 |
| PASI Baseline | 10.80 (6.95–14.95) | 11.70 (9.80–14.83) | 6.95 (4.90–10.75) | n.s |
| BSA Baseline | 14.20 (8.85–27.15) | 16.25 (9.75–25.75) | 11.55 (7.52–23.20) | n.s |
| PGA Baseline | 0.01 | |||
| 2 | 3 | 0 | 3 | |
| 3 | 12 | 3 | 9 | |
| 4 | 10 | 9 | 1 | |
| 5 | 9 | 6 | 3 | |
| Previous treatments | n.s | |||
| Topical | 11 | 3 | 8 | |
| Apremilast | 3 | 1 | 2 | |
| Anti-TNFα | 4 | 4 | 0 | |
| Anti-IL-17 | 2 | 2 | 0 | |
| Anti-p40 | 5 | 5 | 0 | |
| Anti-p19 | 1 | 1 | 0 | |
| Methotrexate | 6 | 2 | 4 | |
| Cyclosporine | 2 | 0 | 2 | |
| Fumarate | 1 | 0 | 1 |
| Topical (n = 10) | Biologic (n = 11) | Other (n = 12) | p-Value | |
|---|---|---|---|---|
| Gender M/F | 7/3 | 6/5 | 8/4 | n.s |
| Age at baseline | 48.50 (37.00–54.25) | 60.00 (46.00–75.00) | 48.00 (35.25–53.00) | n.s |
| Age at diagnosis | 24.00 (15.00–47.00) | 51.00 (29.00–64.00) | 25.00 (12.25–35.75) | 0.056 |
| PASI Baseline | 12.85 (5.92–25.25) | 10.80 (8.40–12.60) | 7.95 (6.20–12.50) | n.s |
| BSA Baseline | 14.70 (10.00–57.50) | 13.00 (8.50–17.00) | 15.80 (7.52–30.25) | n.s |
| PGA Baseline | n.s | |||
| 2 | 1 | 0 | 2 | |
| 3 | 3 | 3 | 6 | |
| 4 | 2 | 6 | 1 | |
| 5 | 4 | 2 | 3 |
| Anti-TNFα | p-Value | ||
|---|---|---|---|
| R | NonR | ||
| Gender M/F | 2/5 | 7/2 | n.s |
| Age at baseline | 52.50 (35.50–69.00) | 48.00 (31.75–51.75) | n.s |
| Age at diagnosis | 22.50 (12.75–48.25) | 17.50 (9.75–41.00) | n.s |
| PASI Baseline | 11.00 (3.80–28.00) | 6.50 (6.25–7.95) | n.s |
| BSA Baseline | 24.60 (6.30–80.00) | 10.50 (7.95–14.70) | n.s |
| PGA Baseline | 0.08 | ||
| 2 | 2 | 1 | |
| 3 | 1 | 7 | |
| 4 | 1 | 0 | |
| 5 | 3 | 1 | |
| Previous treatments | n.s | ||
| Topical | 5 | 3 | |
| Apremilast | 0 | 2 | |
| Anti-TNFα | 0 | 0 | |
| Anti-IL-17 | 0 | 0 | |
| Anti-p40 | 0 | 0 | |
| Methotrexate | 1 | 2 | |
| Cyclosporine | 0 | 2 | |
| Fumarate | 1 | 0 | |
| Anti-IL-23 | p-Value | ||
|---|---|---|---|
| SR | NonR | ||
| Gender M/F | 7/1 | 5/4 | n.s |
| Age at baseline | 52.00 (37.25–73.25) | 62.00 (41.50–73.00) | n.s |
| Age at diagnosis | 31.50 (19.00–58.75) | 51.00 (32.00–61.00) | n.s |
| PASI Baseline | 11.80 (8.55–16.53) | 11.70 (10.00–14.95) | n.s |
| BSA Baseline | 19.75 (9.25–31.75) | 16.00 (9.25–20.50) | n.s |
| PGA Baseline | n.s | ||
| 2 | 0 | 0 | |
| 3 | 2 | 1 | |
| 4 | 2 | 6 | |
| 5 | 4 | 2 | |
| Previous treatments | n.s | ||
| Topical | 2 | 1 | |
| Apremilast | 1 | 0 | |
| Anti-TNFα | 2 | 2 | |
| Anti-IL-17 | 0 | 2 | |
| Anti-p40 | 2 | 3 | |
| Methotrexate | 1 | 1 | |
| Cyclosporine | 0 | 0 | |
| Fumarate | 0 | 0 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cantó, E.; del Prado, M.E.; Vilarrasa, E.; López-Ferrer, A.; García Latasa de Araníbar, F.J.; Ortiz, M.A.; Gut, M.; Mulet, M.; Esteve-Codina, A.; Osuna-Gómez, R.; et al. Transcriptomic Identification of Immune-Related Hubs as Candidate Predictor Biomarkers of Therapeutic Response in Psoriasis. Int. J. Mol. Sci. 2025, 26, 8118. https://doi.org/10.3390/ijms26178118
Cantó E, del Prado ME, Vilarrasa E, López-Ferrer A, García Latasa de Araníbar FJ, Ortiz MA, Gut M, Mulet M, Esteve-Codina A, Osuna-Gómez R, et al. Transcriptomic Identification of Immune-Related Hubs as Candidate Predictor Biomarkers of Therapeutic Response in Psoriasis. International Journal of Molecular Sciences. 2025; 26(17):8118. https://doi.org/10.3390/ijms26178118
Chicago/Turabian StyleCantó, Elisabet, María Elena del Prado, Eva Vilarrasa, Anna López-Ferrer, Francisco Javier García Latasa de Araníbar, Maria Angels Ortiz, Marta Gut, Maria Mulet, Anna Esteve-Codina, Ruben Osuna-Gómez, and et al. 2025. "Transcriptomic Identification of Immune-Related Hubs as Candidate Predictor Biomarkers of Therapeutic Response in Psoriasis" International Journal of Molecular Sciences 26, no. 17: 8118. https://doi.org/10.3390/ijms26178118
APA StyleCantó, E., del Prado, M. E., Vilarrasa, E., López-Ferrer, A., García Latasa de Araníbar, F. J., Ortiz, M. A., Gut, M., Mulet, M., Esteve-Codina, A., Osuna-Gómez, R., Guinart-Cuadra, A., Puig, L., & Vidal, S. (2025). Transcriptomic Identification of Immune-Related Hubs as Candidate Predictor Biomarkers of Therapeutic Response in Psoriasis. International Journal of Molecular Sciences, 26(17), 8118. https://doi.org/10.3390/ijms26178118







