Transcriptomic Analysis Reveals the Regulatory Mechanism of Cold Tolerance in Saussurea involucrata: The Gene Expression and Function Characterization of Dehydrins
Abstract
1. Introduction
2. Results
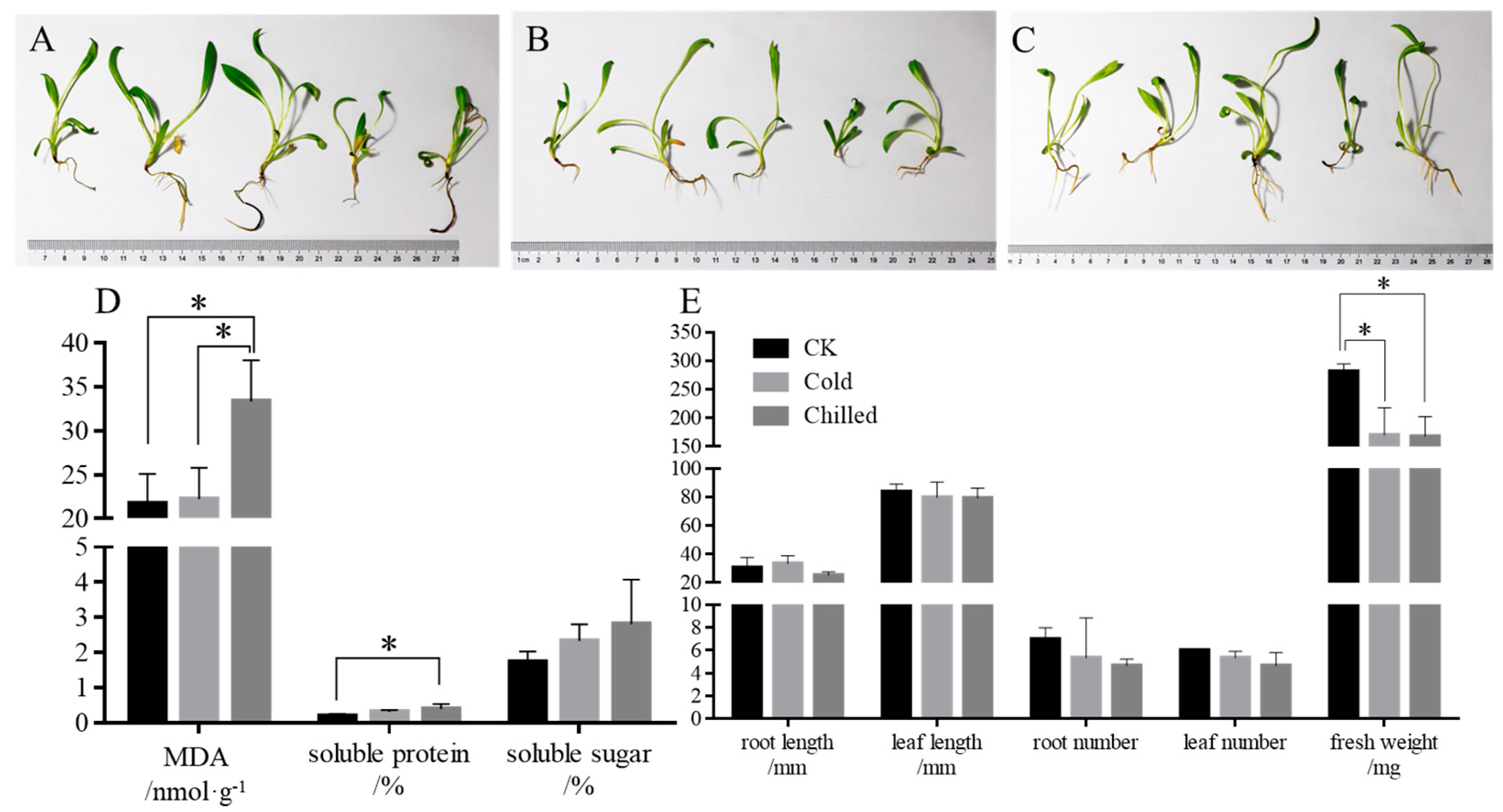
2.1. Morphological Profiles of S. involucrata at Low Temperature
2.2. Physiological Profiles of S. involucrata at Low Temperature
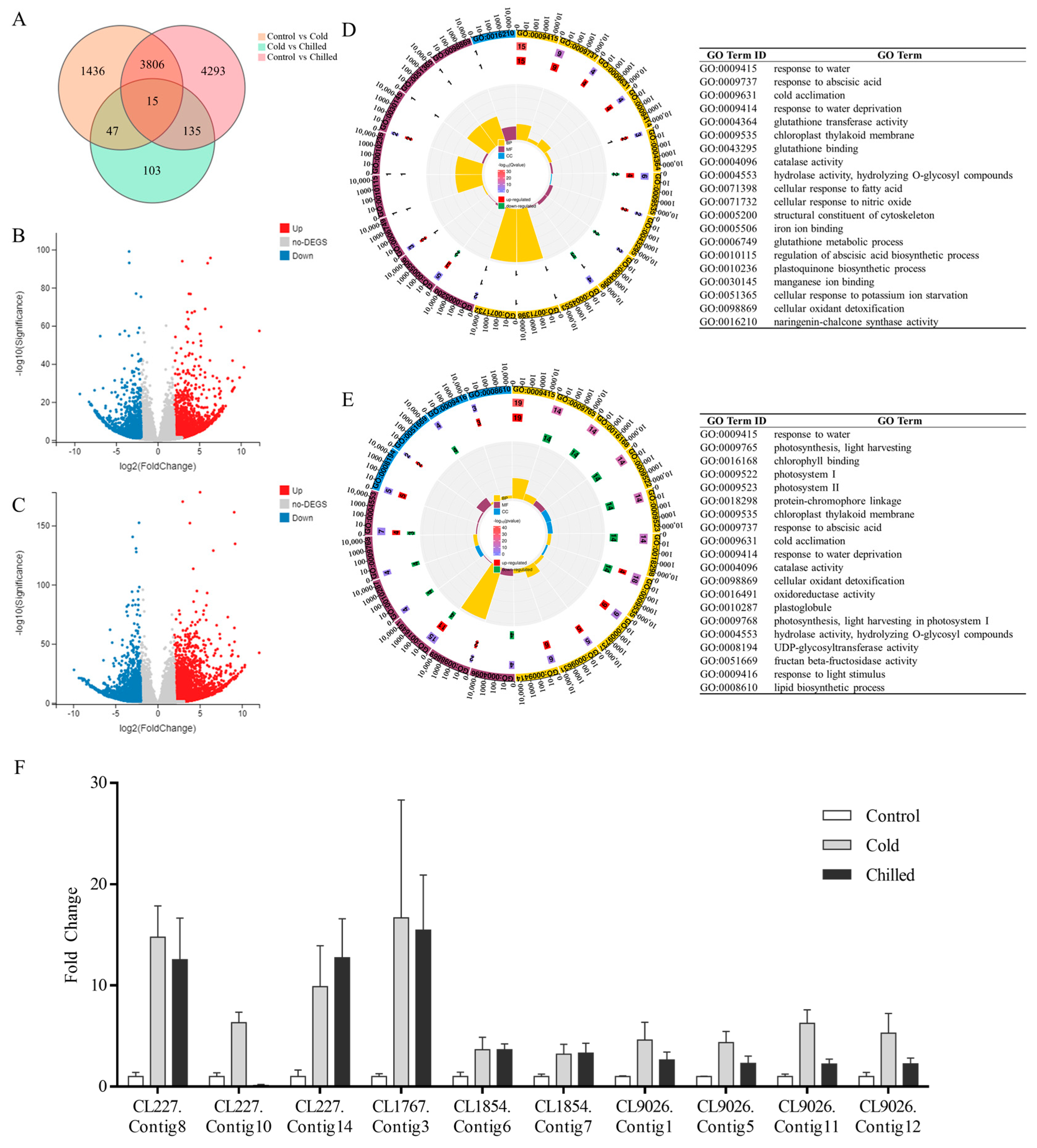
2.3. RNA-Seq Profiles of S. involucrata at Low Temperature
2.4. Annotation of S. involucrata Transcripts
2.5. Expression Analysis of DEGs Related to Cold Stress
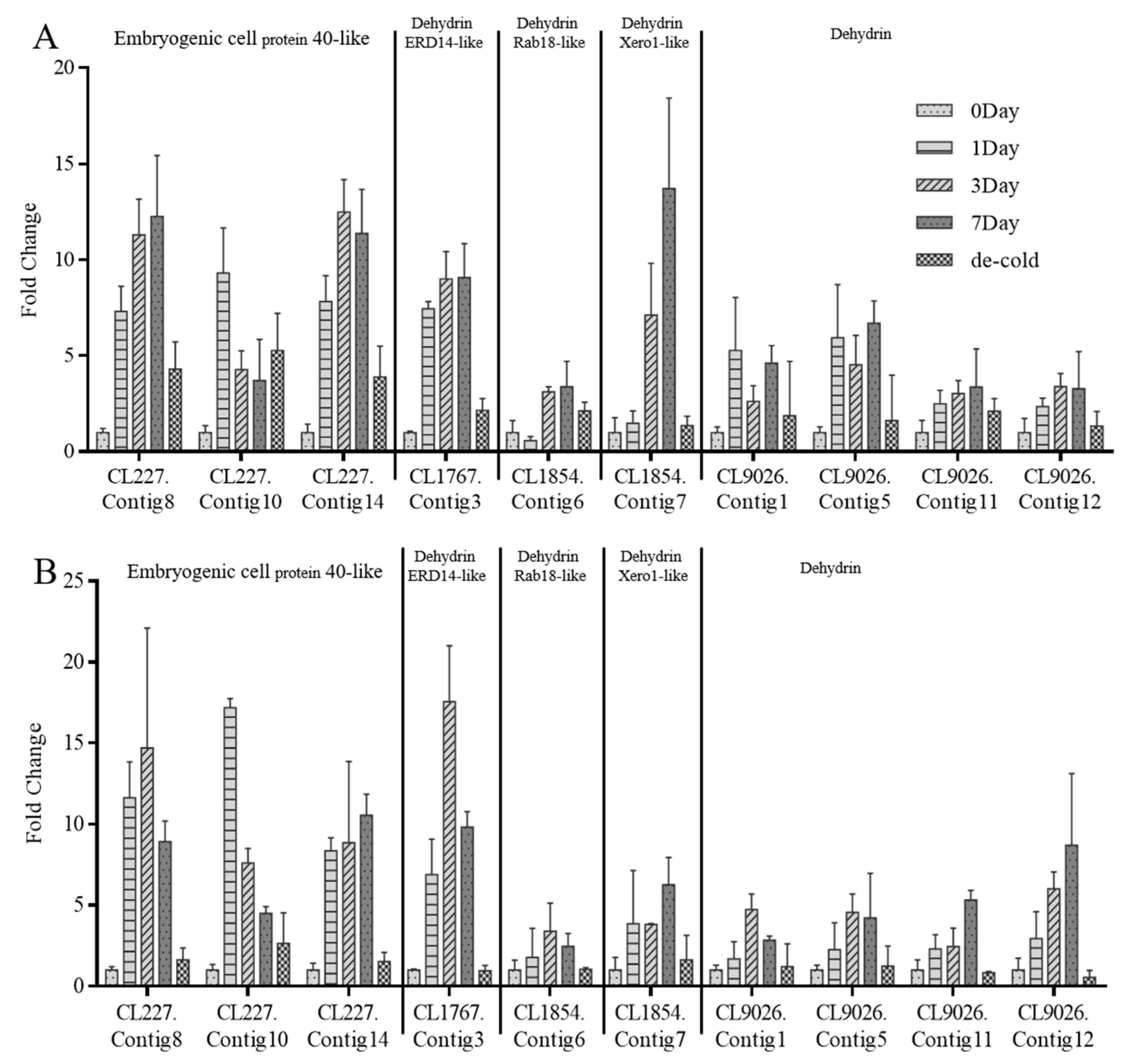
2.6. qPCR Analysis of Putative DHNs
2.7. DHN Gene Expression in Different Periods of Treatment at Multiple Low Temperatures
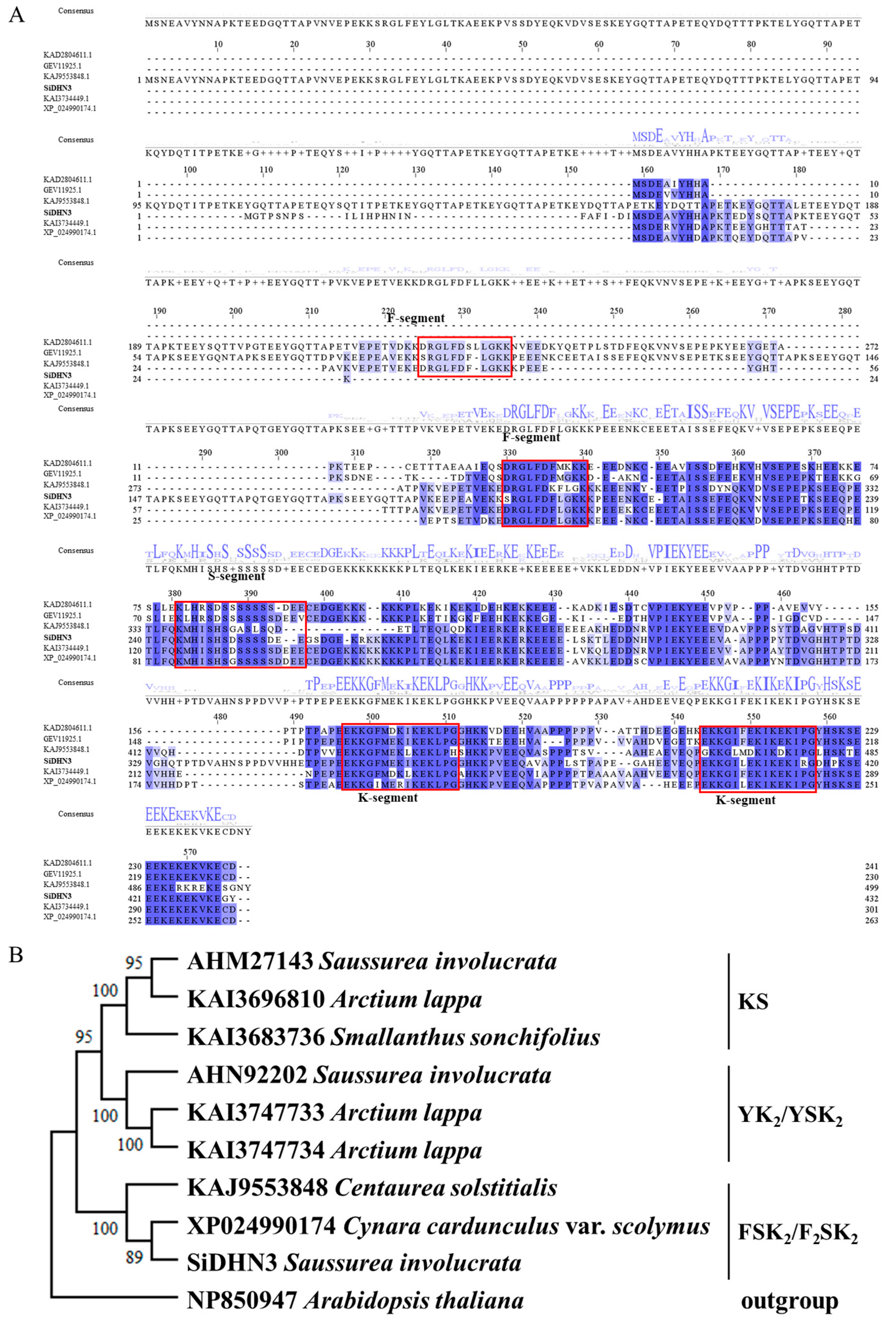
2.8. Sequencing Analysis of SiDHN3
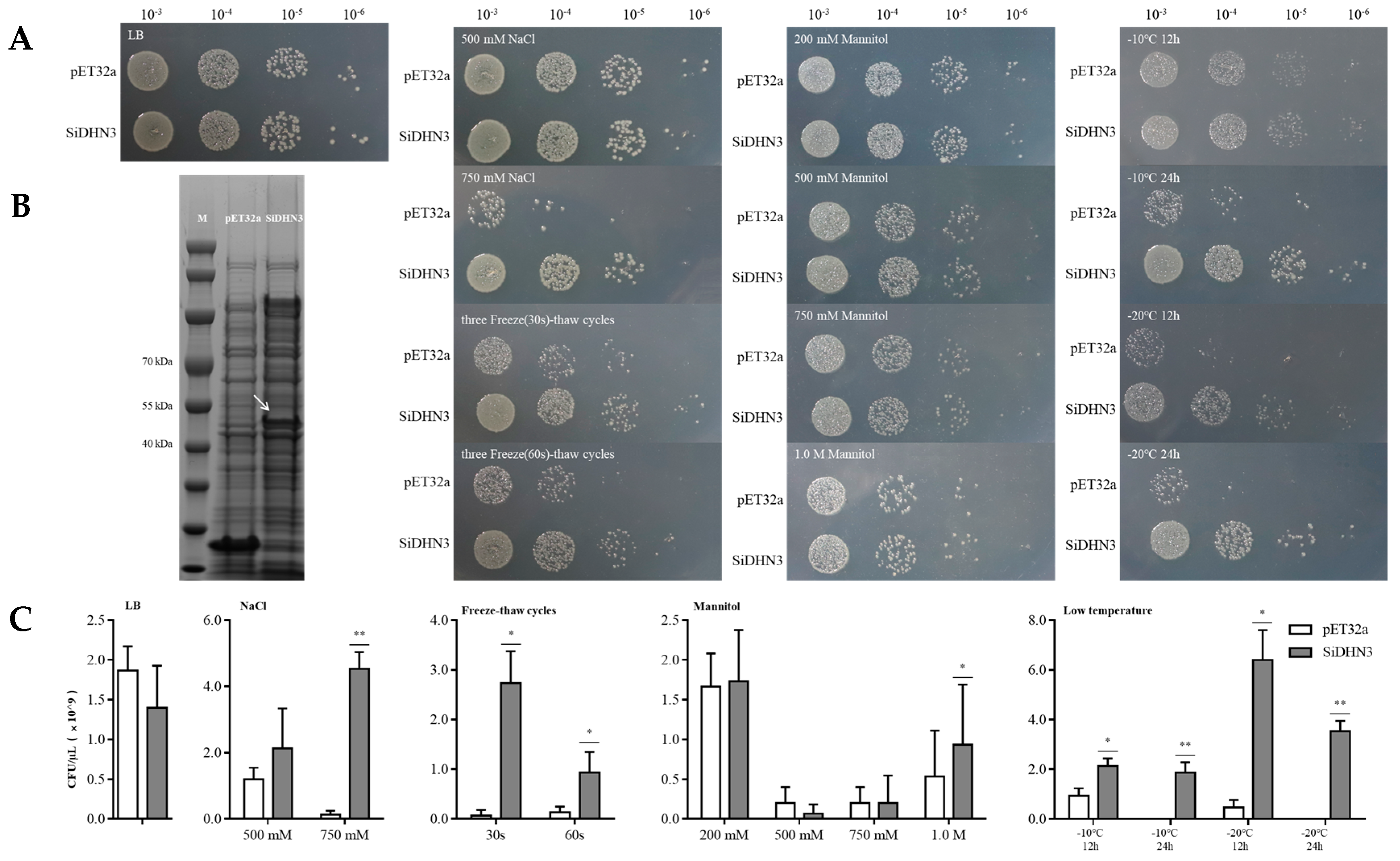
2.9. Expression and Protective Effect of SiDHN3 Proteins on E. coli
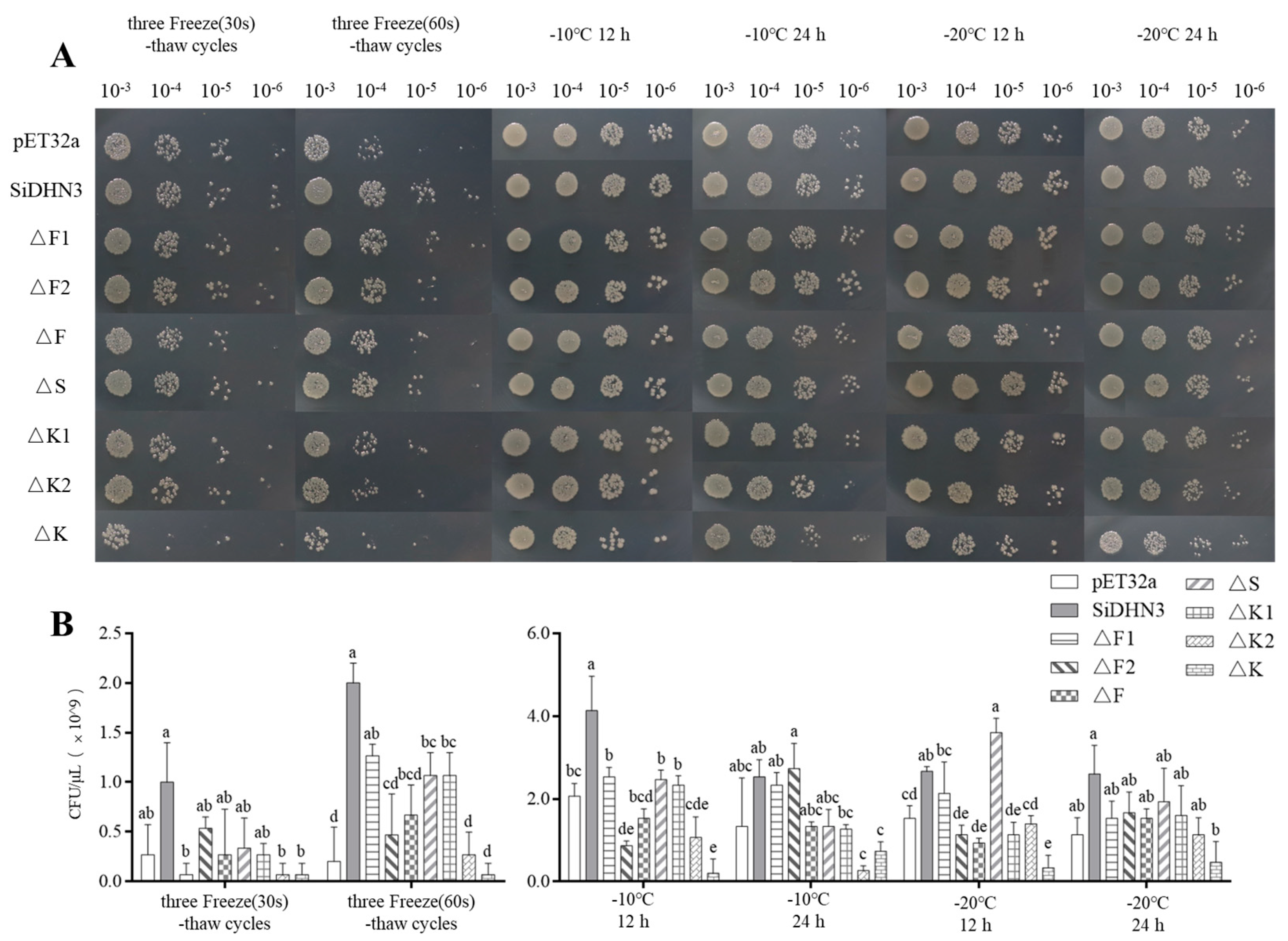
2.10. Functional Validation of SiDHN3 Protective Domains
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Determining the Physiological Characteristics of S. involucrata
4.3. Transcriptomic Analysis
4.4. Expression of DHNs from Different Groups
4.5. Expression of DHNs in Different Periods of Treatment at Multiple Low Temperatures
4.6. Cloning of Full-Length Coding Sequence of SiDHN3
4.7. Sequence Analysis of SiDHN3
4.8. Construction and Expression of SiDHN3 and Its Truncated Derivatives
4.9. Abiotic Stress Tolerance Assays
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Jia, J.M.; Wu, C.F.; Liu, W.; Yu, H.; Hao, Y.; Zheng, J.H.; Ji, Y.R. Antiinflammatory and analgesic activities of the tissue culture of Saussurea involucrata (pharmacology). Biol. Pharm. Bull. 2005, 28, 1612–1614. [Google Scholar] [CrossRef]
- Xu, M.H.; Guo, Q.Y.; Wang, S.J.; Wang, N.; Wei, L.R.; Wang, J.B. Anti-rheumatoid arthritic effects of Saussurea involucrata on type II collagen-induced arthritis in rats. Food Funct. 2016, 2, 763–770. [Google Scholar] [CrossRef] [PubMed]
- Liou, C.J.; Wu, S.J.; Chen, C.L.; Yeh, K.W.; Chen, C.Y.; Huang, W.C. Acacetin from traditionally used Saussurea involucrata Kar. et Kir. suppressed adipogenesis in 3T3-L1 adipocytes and attenuated lipid accumulation in obese mice. Front. Pharmacol. 2017, 8, 589. [Google Scholar] [CrossRef]
- Gong, G.W.; Xie, F.; Zheng, Y.Z.; Hu, W.H.; Qi, B.H.; He, H.; Dong, T.T.; Tsim, K.W. The effect of methanol extract from Saussurea involucrata in the lipopolysaccharide-stimulated inflammation in cultured RAW 264.7 cells. J. Ethnopharmacol. 2020, 251, 112532. [Google Scholar] [CrossRef]
- Zhang, Q.; He, L.Y.; Jiang, Q.Q.; Zhu, H.Q.; Kong, D.H.; Zhang, H.; Cheng, Z.Q.; Deng, H.T.; Zheng, Y.X.; Ying, X. Systems Pharmacology-Based Dissection of Anti-Cancer Mechanism of Traditional Chinese Herb Saussurea involucrata. Front. Pharmacol. 2021, 12, 678203. [Google Scholar] [CrossRef]
- Yin, L.K. Rare Endangered Endemic Higher Plants in Xinjiang of China; Xinjiang Science and Technology Press: Urumqi, China, 2006; pp. 126–127. [Google Scholar]
- Kidokoro, S.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Transcriptional regulatory network of plant cold-stress responses. Trends Plant Sci. 2022, 27, 922–935. [Google Scholar] [CrossRef]
- Lu, C.F.; Jian, L.C.; Kuang, T.Y. Freezing hardiness in alpine plants. Chin. Bull. Bot. 1998, 15, 17–22. [Google Scholar] [CrossRef]
- Peng, D.L.; Zhang, Z.Q.; Niu, Y.; Yang, Y.; Song, B.; Sun, H.; Li, Z.M. Advances in the studies of reproductive strategies of alpine plants. Biodivers. Sci. 2012, 20, 286–299. [Google Scholar] [CrossRef]
- Li, J.; Xia, W.W.; Zang, H.X.; Dai, B.; Zhang, Y.; Feng, Y.J.; Wang, A.W.; Lin, Z.P.; Liu, H.L.; Zhu, J.B. Expression analysis of aquaporin genes in Saussurea involucrata rosette leaves and functional analysis of upregulated SiPIP1;5A under low-temperature stress. Environ. Exp. Bot. 2020, 171, 103958. [Google Scholar] [CrossRef]
- Xia, W.W.; Liu, X.Y.; Xin, H.L.; Wu, X.Y.; Liu, R.N.; Li, J.; Zhu, J.B. Saussurea involucrata PIP2;7 improves photosynthesis and drought resistance by decreasing the stomatal density and increasing intracellular osmotic pressure. Environ. Exp. Bot. 2021, 191, 104605. [Google Scholar] [CrossRef]
- Guo, X.; Zhang, L.; Zhu, J.B.; Liu, H.L.; Wang, A.Y. Cloning and characterization of SiDHN, a novel dehydrin gene from Saussurea involucrata Kar. et Kir. that enhances cold and drought tolerance in tobacco. Plant Sci. 2017, 256, 160–169. [Google Scholar] [CrossRef]
- Guo, X.Y.; Zhang, L.; Wang, X.Z.; Zhang, M.H.; Xi, Y.X.; Wang, A.Y.; Zhu, J.B. Overexpression of Saussurea involucrata dehydrin gene SiDHN promotes cold and drought tolerance in transgenic tomato plants. PLoS ONE 2019, 14, e0225090. [Google Scholar] [CrossRef]
- Gusain, S.; Joshi, S.; Joshi, R. Sensing, signalling, and regulatory mechanism of cold-stress tolerance in plants. Plant Physiol. Biochem. 2023, 197, 107646. [Google Scholar] [CrossRef]
- Hanin, M.; Brini, F.; Ebel, C.; Toda, Y.; Takeda, S.; Masmoudi, K. Plant dehydrins and stress tolerance versatile proteins for complex mechanisms. Plant Signal. Behav. 2011, 6, 1503–1509. [Google Scholar] [CrossRef]
- Yang, Y.Z.; He, M.Y.; Zhu, Z.G.; Li, S.X.; Xu, Y.; Zhang, C.H.; Singer, S.D.; Wang, Y.J. Identification of the dehydrin gene family from grapevine species and analysis of their responsiveness to various forms of abiotic and biotic stress. BMC Plant Biol. 2012, 12, 140. [Google Scholar] [CrossRef]
- Sun, Z.P.; Li, S.Y.; Chen, W.Y.; Zhang, J.Q.; Zhang, L.X.; Sun, W.; Wang, Z.L. Plant dehydrins: Expression, regulatory networks, and protective roles in plants challenged by abiotic stress. Int. J. Mol. Sci. 2021, 22, 12619. [Google Scholar] [CrossRef]
- Rorat, T.; Grygorowicz, W.J.; Irzykowski, W.; Rey, P. Expression of KS-type dehydrins is primarily regulated by factors related to organ type and leaf developmental stage during vegetative growth. Planta 2004, 218, 878–885. [Google Scholar] [CrossRef] [PubMed]
- Zhu, B.; Choi, D.W.; Fenton, R.; Close, T.J. Expression of the barley dehydrin multigene family and the development of freezing tolerance. Mol. Gen. Genet. 2000, 264, 145–153. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Ouyang, B.; Zhang, J.H.; Wang, T.T.; Li, H.X.; Zhang, Y.Y.; Yu, C.Y.; Ye, Z.B. Differential modulation of photosynthesis, signaling, and transcriptional regulation between tolerant and sensitive tomato genotypes under cold stress. PLoS ONE 2012, 11, e50785. [Google Scholar] [CrossRef] [PubMed]
- Hara, M.; Terashima, S.; Kuboi, T. Characterization and cryoprotective activity of cold-responsive dehydrin from Citrus unshiu. J. Plant Physiol. 2001, 158, 1333–1339. [Google Scholar] [CrossRef]
- Hara, M.; Terashima, S.; Fukaya, T.; Kuboi, T. Enhancement of cold tolerance and inhibition of lipid peroxidation by citrus dehydrin in transgenic tobacco. Planta 2003, 217, 290–298. [Google Scholar] [CrossRef]
- Hara, M.; Monna, S.; Murata, T.; Nakano, T.; Amano, S.; Nachbar, M.; Wätzig, H. The Arabidopsis KS-type dehydrin recovers lactate dehydrogenase activity inhibited by copper with the contribution of His residues. Plant Sci. 2016, 245, 135–142. [Google Scholar] [CrossRef]
- Zhang, H.; Zheng, J.X.; Su, H.X.; Xia, K.F.; Jian, S.G.; Zhang, M. Molecular cloning and functional characterization of the Dehydrin (IpDHN) gene from Ipomoea pes-caprae. Front. Plant Sci. 2018, 9, 1454. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.X.; Xiang, X.; Geng, M.T.; You, Q.; Huang, X. Effect of HbDHN1 and HbDHN2 Genes on Abiotic Stress Responses in Arabidopsis. Front. Plant Sci. 2017, 8, 470. [Google Scholar] [CrossRef]
- Wang, X.Z.; Xin, X.; Zhang, F.Y.Z.; Yu, Q.Y.; Chen, X.; Pang, Q.Y.; Zhang, A.Q. Advances in biological function and regulation mechanism of plant dehydrins. Plant Physiol. J. 2022, 58, 1617–1628. [Google Scholar] [CrossRef]
- Zhang, H.; Zhao, Y.; Zhu, J.K. Thriving under stress: How plants balance growth and the stress response. Dev. Cell 2020, 55, 529–543. [Google Scholar] [CrossRef]
- Li, L.; Wang, Y.F.; Guo, W.X.; Ma, W.M.; Jiang, C.L. Response of resource allocation Saussurea leontodontoides during its fruiting stage to the elevation. Chin. J. Plant Ecol. 2020, 44, 1164–1171. [Google Scholar] [CrossRef]
- Rorat, T. Plant dehydrins-tissue location, structure and function. Cell. Mol. Biol. Lett. 2006, 11, 536–556. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, P.; Chakrabarty, D. Dehydrin in the past four decades: From chaperones to transcription co-regulators in regulating abiotic stress response. Curr. Res. Biotechnol. 2021, 3, 249–259. [Google Scholar] [CrossRef]
- Jing, H.; Li, C.; Ma, F.; Ma, J.H.; Khan, A.; Wang, X.; Zhao, L.Y.; Gong, Z.H.; Chen, R.G. Genome-Wide identification, expression diversication of dehydrin gene family and characterization of CaDHN3 in pepper (Capsicum annuum L.). PLoS ONE 2016, 11, e0161073. [Google Scholar] [CrossRef]
- Kosová, K.; Klíma, M.; Prášil, I.T.; Vítámvás, P. COR/LEA proteins as indicators of frost tolerance in Triticeae: A comparison of controlled versus field conditions. Plants 2021, 10, 789. [Google Scholar] [CrossRef]
- Vítámvás, P.; Kosová, K.; Musilová, J.; Holková, L.; Mařík, P.; Smutná, P.; Klíma, M.; Prášil, I.T. Relationship between dehydrin accumulation and winter survival in winter wheat and barley grown in the field. Front. Plant Sci. 2019, 10, 7. [Google Scholar] [CrossRef]
- Ochoa-Alfaro, A.E.; Rodríguez-Kessler, M.; Pérez-Morales, M.B.; Delgado-Sánchez, B.; Cuevas-Velazquez, C.L.; Gómez-Anduro, G.; Jiménez-Bremont, J.F. Functional characterization of an acidic SK3 dehydrin isolated from an Opuntia streptacantha cDNA library. Planta 2011, 235, 565–578. [Google Scholar] [CrossRef]
- Shin, S.Y.; Kim, H.Y.; Kim, I.S.; Kim, J.J.; Kim, Y.S.; Yoon, H.S. The dehydrin gene of the Arctic plant Cerastium arcticum, CaDHN, increases tolerance to multiple stresses in Arabidopsis thaliana. Plant Biotechnol. Rep. 2020, 14, 387–395. [Google Scholar] [CrossRef]
- Qiu, H.L.; Zhang, L.H.; Liu, C.; He, L.; Wang, A.Y.; Liu, H.L.; Zhu, J.B. Cloning and characterization of a novel dehydrin gene, SiDhn2, from Saussurea involucrata Kar. et Kir. Plant Mol. Biol. 2013, 84, 707–718. [Google Scholar] [CrossRef] [PubMed]
- Wei, H.; Yang, Y.; Himmel, M.E.; Tucker, M.P.; Ding, S.Y.; Yang, S.H.; Arora, R. Identification and characterization of five cold stress-related rhododendron dehydrin genes: Spotlight on a FSK-type dehydrin with multiple f-segments. Front. Bioeng. Biotechnol. 2019, 7, 30. [Google Scholar] [CrossRef]
- Qin, Y.X.; Qin, F.Y. Dehydrins from wheat x Thinopyrum ponticum amphiploid increase salinity and drought tolerance under their own inducible promoters without growth retardation. Plant Physiol. Biochem. 2016, 99, 142–149. [Google Scholar] [CrossRef]
- Luo, D.; Hou, X.M.; Zhang, Y.M.; Meng, Y.C.; Zhang, H.F.; Liu, S.Y.; Wang, X.K.; Chen, R.G. CaDHN5, a dehydrin gene from pepper, plays an important role in salt and osmotic stress responses. Int. J. Mol. Sci. 2019, 20, 1989. [Google Scholar] [CrossRef]
- Aduse Poku, S.; Nkachukwu Chukwurah, P.; Aung, H.H.; Nakamura, I. Over-Expression of a Melon Y3SK2-Type LEA Gene Confers Drought and Salt Tolerance in Transgenic Tobacco Plants. Plants 2020, 9, 1749. [Google Scholar] [CrossRef]
- Meng, Y.C.; Zhang, H.F.; Pan, X.X.; Chen, N.; Hu, H.F.; Haq, S.; Khan, A.; Chen, R.G. CaDHN3, a pepper (Capsicum annuum L.) dehydrin gene enhances the tolerance against salt and drought stresses by reducing ROS accumulation. Int. J. Mol. Sci. 2021, 22, 3205. [Google Scholar] [CrossRef] [PubMed]
- Halder, T.; Upadhyaya, G.; Ray, S. YSK2 type dehydrin (SbDhn1) from Sorghum bicolor showed improved protection under high temperature and osmotic stress condition. Front. Plant Sci. 2017, 8, 918. [Google Scholar] [CrossRef] [PubMed]
- Close, T.J. Dehydrins: Emergence of a biochemical role of a family of plant dehydration proteins. Physiol. Plant. 1996, 97, 795–803. [Google Scholar] [CrossRef]
- Close, T.J. Dehydrins: A commonalty in the response of plants to dehydration and low temperature. Physiol. Plant. 1997, 100, 291–296. [Google Scholar] [CrossRef]
- Alsheikh, M.K.; Heyen, B.J.; Randall, S.K. Ion binding properties of the dehydrin ERD14 are dependent upon phosphorylation. J. Biol. Chem. 2003, 278, 40882–40889. [Google Scholar] [CrossRef]
- Drira, M.; Saibi, W.; Brini, F.; Gargouri, A.; Masmoudi, K.; Hanin, M. The K-segments of the wheat dehydrin DHN-5 are essential for the protection of lactate dehydrogenase and β-glucosidase activities in vitro. Mol. Biotechnol. 2013, 54, 643–650. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Song, Q.P.; Li, D.X.; Yang, X.H.; Li, D.Q. Multifunctional roles of plant dehydrins in response to environmental stresses. Front. Plant Sci. 2017, 8, 1018. [Google Scholar] [CrossRef] [PubMed]
- Malik, A.A.; Veltri, M.; Boddington, K.F.; Singh, K.K.; Graether, S.P. Genome analysis of conserved dehydrin motifs in vascular plants. Front. Plant Sci. 2017, 8, 709. [Google Scholar] [CrossRef]
- Petersen, J.; Eriksson, S.K.; Harryson, P.; Pierog, S.; Colby, T.; Bartels, D.; Rohrig, H. The lysine-rich motif of intrinsically disordered stress protein CDeT11-24 from Craterostigma plantagineum is responsible for phosphatidic acid binding and protection of enzymes from damaging effects caused by desiccation. J. Exp. Bot. 2012, 63, 4919–4929. [Google Scholar] [CrossRef]
- Koag, M.C.; Fenton, R.D.; Wilkens, S.; Close, T.J. The binding of maize DHN1 to lipid vesicles. Gain of structure and lipid specificity. Plant Physiol. 2003, 131, 309–316. [Google Scholar] [CrossRef]
- Strimbeck, G.R. Hiding in plain sight: The F-segment and other conserved features of seed plant SKn dehydrins. Planta 2017, 245, 1061–1066. [Google Scholar] [CrossRef]
- Ohkubo, T.; Kameyama, A.; Kamiya, K.; Kondo, M.; Hara, M. F-segments of Arabidopsis dehydrins show cryoprotective activities for lactate dehydrogenase depending on the hydrophobic residues. Phytochemistry 2020, 173, 112300. [Google Scholar] [CrossRef]
- Mohanty, D.; Sharma, G.S. Function in disorder: A review on the roles of the disordered dehydrin proteins in conferring stress tolerance. Int. J. Biol. Macromol. 2025, 311, 143672. [Google Scholar] [CrossRef]
- Ahari, D.; Sahil, K.; Kaushal, S.; Sharma, A.; Rangan, L.; Swaminathan, R. Structural transitions of dehydrin in response to temperature, the presence of trifluoroethanol and sodium dodecyl sulfate, and its protective role in heat and cold stress. Biochemistry 2025, 64, 3045–3062. [Google Scholar] [CrossRef] [PubMed]
- Szlachtowska, Z.; Rurek, M. Plant dehydrins and dehydrin-like proteins: Characterization and participation in abiotic stress response. Front. Plant Sci. 2023, 14, 1213188. [Google Scholar] [CrossRef] [PubMed]
- Sheng, J.; Wang, C.; Liu, T.; Yang, T.C.; Zhang, D. The cytoprotective function of NnRab18 dehydrin, a late embryogenesis abundant protein specifically accumulated in seeds of Nelumbo nucifera. Plant Physiol. Biochem. 2025, 110261. [Google Scholar] [CrossRef]
- Murray, M.R.; Graether, S.P. Physiological, structural, and functional insights into the cryoprotection of membranes by the dehydrins. Front. Plant Sci. 2022, 13, 886525. [Google Scholar] [CrossRef]
- Melgar, A.E.; Rizzo, A.J.; Moyano, L.; Cenizo, R.; Palacios, M.B.; Zelada, A.M. Genome-wide identification and salt stress-expression analysis of the dehydrin gene family in Chenopodium quinoa. Curr. Plant Biol. 2024, 38, 100340. [Google Scholar] [CrossRef]
- Li, J.; Liu, H.L.; Xia, W.W.; Mu, J.Q.; Feng, Y.J.; Liu, R.N.; Yan, P.Y.; Wang, A.Y.; Lin, Z.P.; Guo, Y.; et al. De novo transcriptome sequencing and the hypothetical cold response mode of Saussurea involucrata in extreme cold environments. Int. J. Mol. Sci. 2017, 18, 1155. [Google Scholar] [CrossRef]
- Chen, Y.X.; Chen, Y.S.; Shi, C.M.; Huang, Z.B.; Zhang, Y.; Li, S.K.; Li, Y.; Ye, J.; Yu, C.; Li, Z.; et al. SOAPnuke: A MapReduce acceleration-supported software for integrated quality control and preprocessing of high-throughput sequencing data. Gigascience 2018, 7, gix120. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levine, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.D.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Saizberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef]
- Shan, T.T.; Zhou, L.S.; Li, B.; Chen, X.M.; Guo, S.X.; Wang, A.R.; Tian, L.X.; Liu, J.T. The plant growth-promoting fungus MF23 (Mycena sp.) increases production of Dendrobium officinale (Orchidaceae) by affecting nitrogen uptake and NH4+ assimilation. Front. Plant Sci. 2021, 12, 693561. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.Z.; Li, F.L. Effects of cold-hardening on freezing tolerance and antioxidant enzyme activities in plantlets of Saussurea laniceps Hand.-Mazz. J. Plant Physiol. Mol. Biol. 2005, 31, 437–440. [Google Scholar] [CrossRef]
- Waterhouse, A.M.; Procter, J.B.; Martin, D.M.A.; Clamp, M.; Barton, G.J. Jalview Version 2—A multiple sequence alignment editor and analysis workbench. Bioinformatics 2009, 25, 1189–1191. [Google Scholar] [CrossRef] [PubMed]
| No. | Gene id | Gene | Reference Species | log2 (Cold/Control) | log2 (Chilled/Control) |
|---|---|---|---|---|---|
| 1 | CL227.Contig10 | embryogenic cell protein 40-like | C. cardunculus var. scolymus | 5.45 | 5.09 |
| 2 | CL227.Contig4 | 5.76 | 5.11 | ||
| 3 | CL227.Contig8 | 4.22 | 3.95 | ||
| 4 | CL227.Contig14 | 3.88 | 3.13 | ||
| 5 | CL227.Contig6 | 2.98 | 1.93 | ||
| 6 | CL1854.Contig4 | dehydrin Xero 1-like | 3.63 | 4.02 | |
| 7 | CL1854.Contig7 | 3.58 | 4.00 | ||
| 8 | CL1854.Contig6 | dehydrin Rab18-like | 2.91 | 2.95 | |
| 9 | CL1767.Contig3 | dehydrin ERD14-like | 5.40 | 6.00 | |
| 10 | CL1767.Contig1 | 3.62 | 4.23 | ||
| 11 | CL1767.Contig4 | 2.73 | 3.57 | ||
| 12 | CL9026.Contig6 | dehydrin | S. involucrata | 6.01 | 5.78 |
| 13 | CL9026.Contig4 | 3.09 | 3.44 | ||
| 14 | CL9026.Contig10 | 1.70 | 3.19 | ||
| 15 | CL9026.Contig5 | 1.54 | 3.11 | ||
| 16 | CL9026.Contig11 | 3.07 | 3.10 | ||
| 17 | CL9026.Contig1 | 2.14 | 2.27 | ||
| 18 | CL9026.Contig8 | 4.88 | 6.11 | ||
| 19 | CL9026.Contig12 | 2.34 | 2.72 | ||
| 20 | CL9026.Contig7 | 2.11 | 1.40 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, T.; Zhou, L.; Zhu, J.; Guo, S.; Liu, C.; Wang, A.; Zeng, X.; Chen, X. Transcriptomic Analysis Reveals the Regulatory Mechanism of Cold Tolerance in Saussurea involucrata: The Gene Expression and Function Characterization of Dehydrins. Int. J. Mol. Sci. 2025, 26, 9030. https://doi.org/10.3390/ijms26189030
Chen T, Zhou L, Zhu J, Guo S, Liu C, Wang A, Zeng X, Chen X. Transcriptomic Analysis Reveals the Regulatory Mechanism of Cold Tolerance in Saussurea involucrata: The Gene Expression and Function Characterization of Dehydrins. International Journal of Molecular Sciences. 2025; 26(18):9030. https://doi.org/10.3390/ijms26189030
Chicago/Turabian StyleChen, Tongyao, Lisi Zhou, Jun Zhu, Shunxing Guo, Chengcheng Liu, Airong Wang, Xu Zeng, and Xiaomei Chen. 2025. "Transcriptomic Analysis Reveals the Regulatory Mechanism of Cold Tolerance in Saussurea involucrata: The Gene Expression and Function Characterization of Dehydrins" International Journal of Molecular Sciences 26, no. 18: 9030. https://doi.org/10.3390/ijms26189030
APA StyleChen, T., Zhou, L., Zhu, J., Guo, S., Liu, C., Wang, A., Zeng, X., & Chen, X. (2025). Transcriptomic Analysis Reveals the Regulatory Mechanism of Cold Tolerance in Saussurea involucrata: The Gene Expression and Function Characterization of Dehydrins. International Journal of Molecular Sciences, 26(18), 9030. https://doi.org/10.3390/ijms26189030





