Pc-AIF1 Is Expressed in Hemocyte-Rich and Neural Tissues and Links Immune Response and Regeneration in the Snail Model Pomacea canaliculata
Abstract
1. Introduction
2. Results
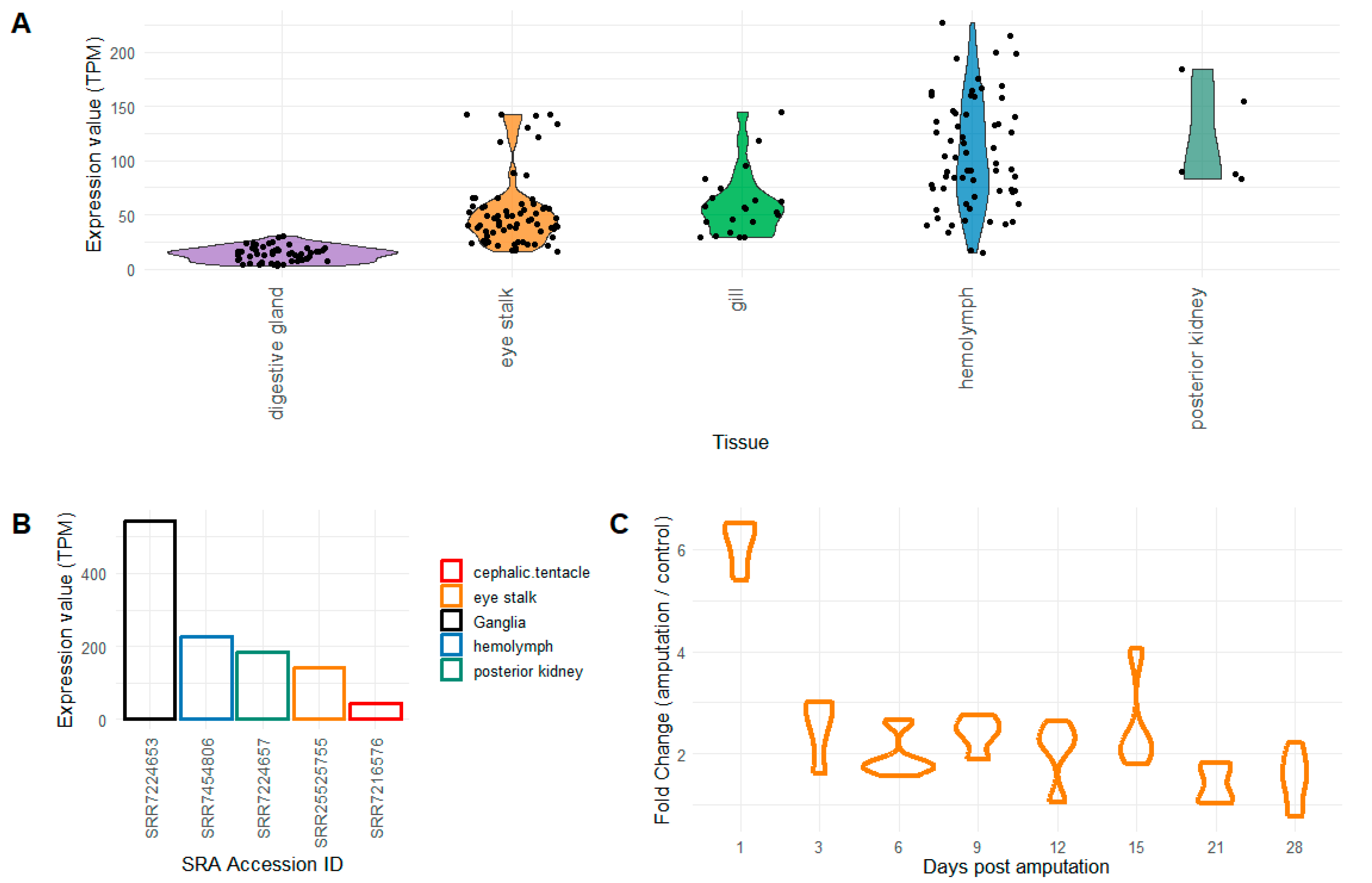
2.1. Evaluation of Pc-AIF1 Expression Levels and Validation
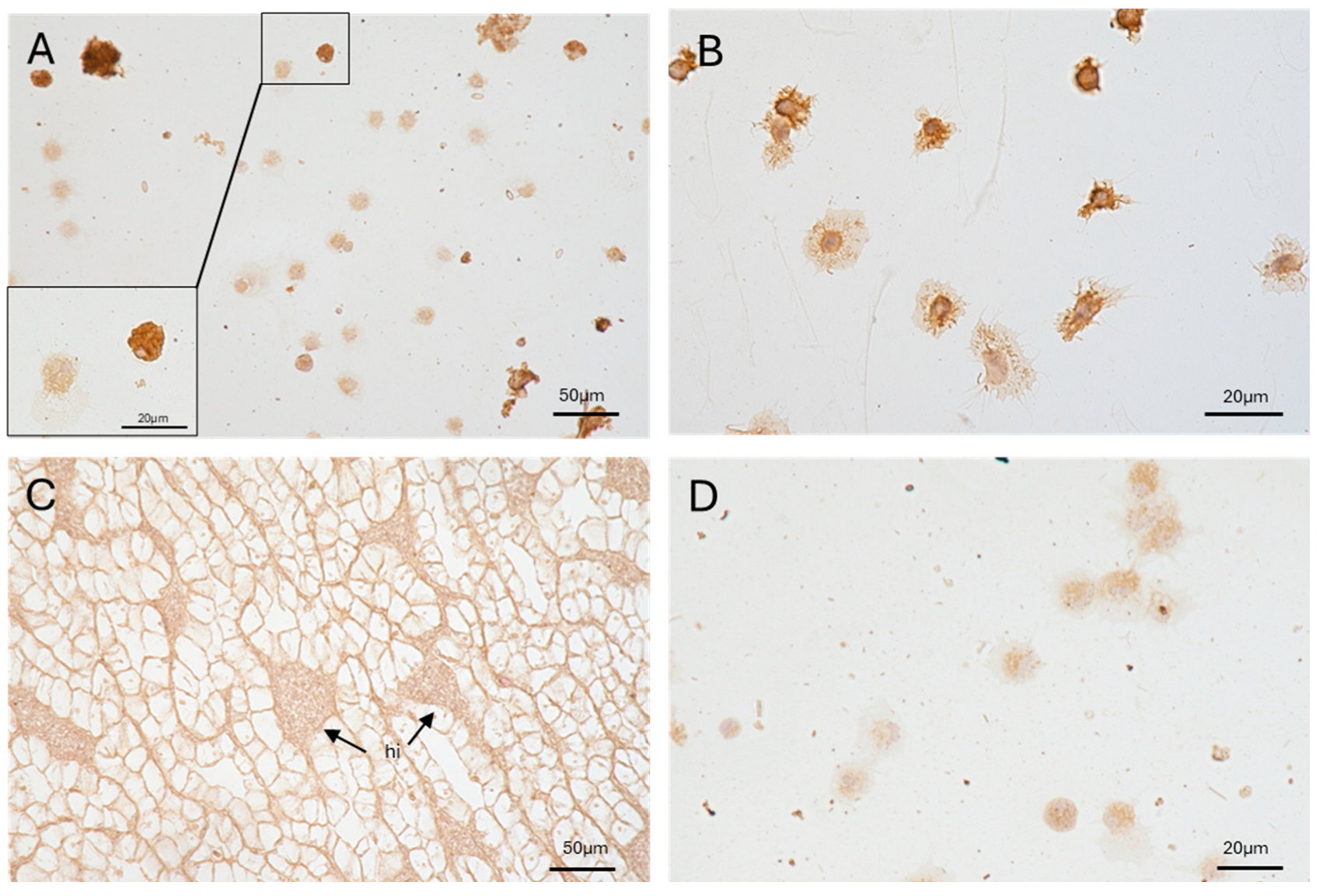
2.2. Pc-AIF1 Is Expressed by Circulating and Resident Hemocytes
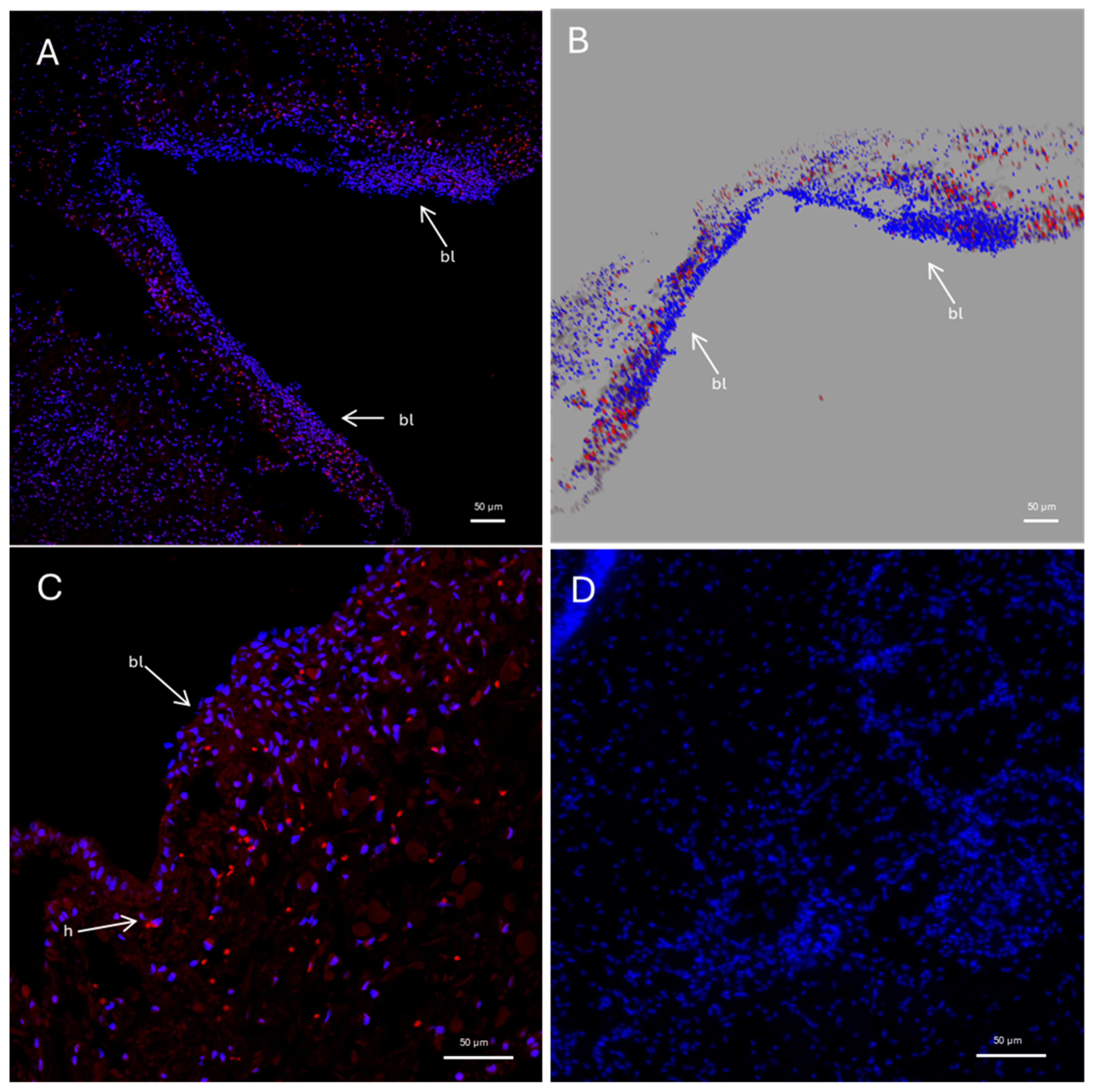
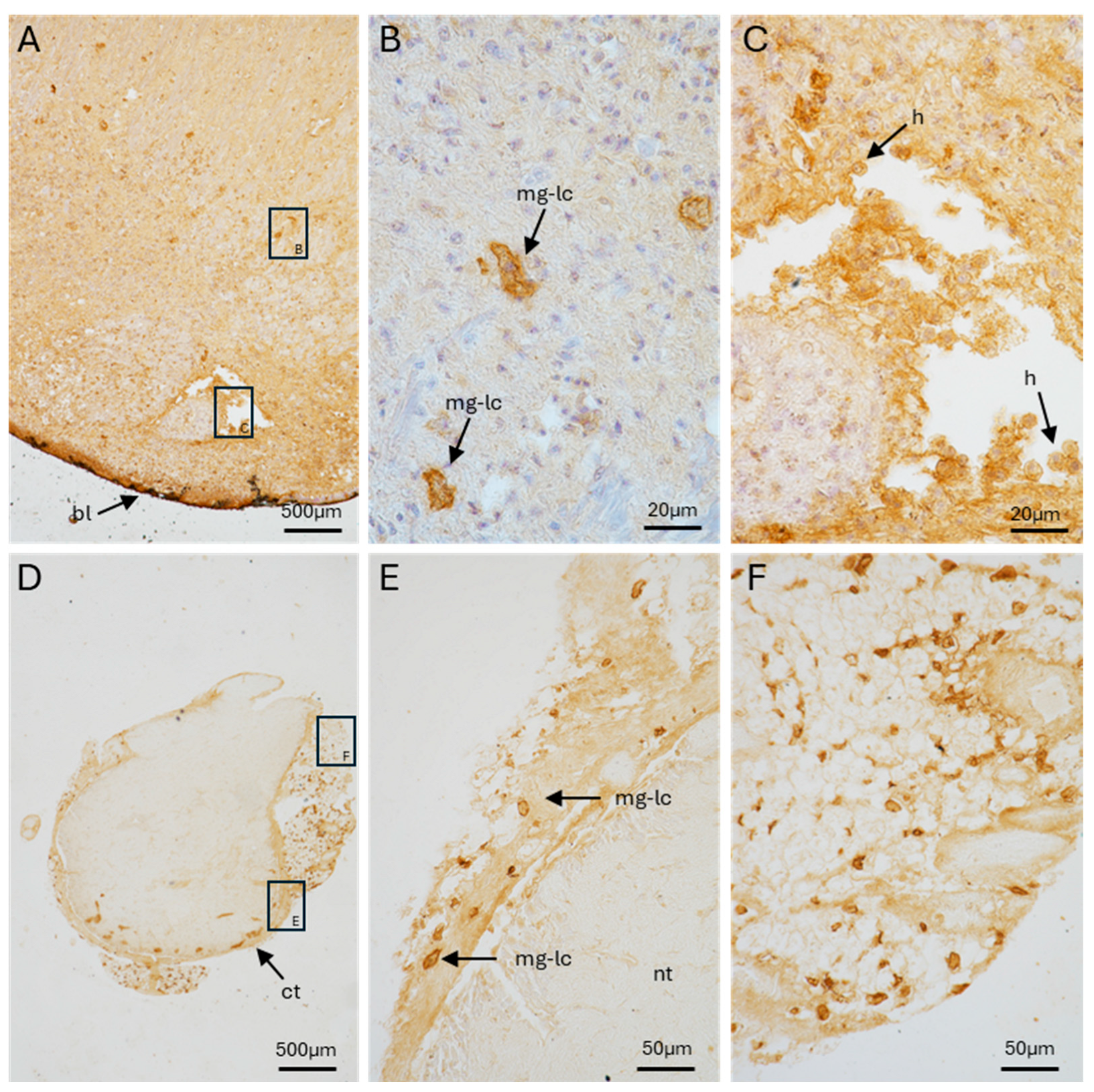
2.3. Pc-AIF1 Is Expressed in Cells Surrounding the Control Cerebral Ganglia and by Cells Infiltrating Cephalic Tentacle Blastema
2.4. Presence of Microglia-like Cells in P. canaliculata Ganglia
2.5. Pc-AIF1 Expression Is Up-Regulated in Cerebral Ganglia During Tentacle Regeneration
3. Discussion
4. Materials and Methods
4.1. Samples Retrieval and Whole Transcriptome Expression Analysis
4.2. Animals
4.3. Treatment, Animal Dissection, and Organ Collection
4.4. RT-PCR, Gel Extraction, and Sequencing
4.5. Slide Preparation and Staining for Histological Analysis
4.6. Riboprobe Synthesis
4.7. FISH Protocol
4.8. Immunohystochemistry
4.9. Imaging Acquisition
4.10. RT-qPCR and Expression Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Utans, U.; Arceci, R.J.; Yamashita, Y.; Russell, M.E. Cloning and Characterization of Allograft Inflammatory Factor-1: A Novel Macrophage Factor Identified in Rat Cardiac Allografts with Chronic Rejection. J. Clin. Investig. 1995, 95, 2954–2962. [Google Scholar] [CrossRef]
- Sasaki, Y.; Ohsawa, K.; Kanazawa, H.; Kohsaka, S.; Imai, Y. Iba1 Is an Actin-Cross-Linking Protein in Macrophages/Microglia. Biochem. Biophys. Res. Commun. 2001, 286, 292–297. [Google Scholar] [CrossRef]
- Ohsawa, K.; Imai, Y.; Kanazawa, H.; Sasaki, Y.; Kohsaka, S. Involvement of Iba1 in Membrane Ruffling and Phagocytosis of Macrophages/Microglia. J. Cell Sci. 2000, 113, 3073–3084. [Google Scholar] [CrossRef]
- Ohsawa, K.; Imai, Y.; Sasaki, Y.; Kohsaka, S. Microglia/Macrophage-specific Protein Iba1 Binds to Fimbrin and Enhances Its Actin-bundling Activity. J. Neurochem. 2004, 88, 844–856. [Google Scholar] [CrossRef]
- Iris, F.J.M.; Bougueleret, L.; Prieur, S.; Caterina, D.; Primas, G.; Perrot, V.; Jurka, J.; Rodriguez-Tome, P.; Claverie, J.M.; Dausset, J.; et al. Dense Alu Clustering and a Potential New Member of the NFκB Family within a 90 Kilobase HLA Class III Segment. Nat. Genet. 1993, 3, 137–145. [Google Scholar] [CrossRef]
- Utans, U.; Quist, W.C.; McManus, B.M.; Wilson, J.E.; Arceci, R.J.; Wallace, A.F.; Russell, M.E. Allograft inflammatory factory-1. A cytokine-responsive macrophage molecule expressed in transplanted human hearts. Transplantation 1996, 61, 1387–1392. [Google Scholar] [CrossRef]
- Deininger, M.H.; Meyermann, R.; Schluesener, H.J. The Allograft Inflammatory Factor-1 Family of Proteins. FEBS Lett. 2002, 514, 115–121. [Google Scholar] [CrossRef] [PubMed]
- Yamada, M.; Ohsawa, K.; Imai, Y.; Kohsaka, S.; Kamitori, S. X-Ray Structures of the Microglia/Macrophage-Specific Protein Iba1 from Human and Mouse Demonstrate Novel Molecular Conformation Change Induced by Calcium Binding. J. Mol. Biol. 2006, 364, 449–457. [Google Scholar] [CrossRef] [PubMed]
- Fukasawa, M.; Nishio, K.; Oikawa, D.; Itou, T.; Iinuma, T.; Asano, M. Allograft Inflammatory Factor-1 Released from the Cerebral Microglia Affect Several Organs in the Body. J. Mol. Histol. 2023, 54, 147–156. [Google Scholar] [CrossRef] [PubMed]
- Leon-Oliva, D.D.; Garcia-Montero, C.; Fraile-Martinez, O.; Boaru, D.L.; García-Puente, L.; Rios-Parra, A.; Garrido-Gil, M.J.; Casanova-Martín, C.; García-Honduvilla, N.; Bujan, J.; et al. AIF1: Function and Connection with Inflammatory Diseases. Biology 2023, 12, 694. [Google Scholar] [CrossRef]
- Liu, G.; Ma, H.; Jiang, L.; Zhao, Y. Allograft Inflammatory Factor-1 and Its Immune Regulation. Autoimmunity 2007, 40, 95–102. [Google Scholar] [CrossRef]
- Autieri, M.V.; Kelemen, S.E.; Wendt, K.W. AIF-1 Is an Actin-Polymerizing and Rac1-Activating Protein That Promotes Vascular Smooth Muscle Cell Migration. Circ. Res. 2003, 92, 1107–1114. [Google Scholar] [CrossRef]
- Yang, Z.F.; Ho, D.W.; Lau, C.K.; Lam, C.T.; Lum, C.T.; Poon, R.T.P.; Fan, S.T. Allograft Inflammatory Factor-1 (AIF-1) Is Crucial for the Survival and pro-Inflammatory Activity of Macrophages. Int. Immunol. 2005, 17, 1391–1397. [Google Scholar] [CrossRef]
- Zhao, Y.-Y.; Yan, D.-J.; Chen, Z.-W. Role of AIF-1 in the Regulation of Inflammatory Activation and Diverse Disease Processes. Cell. Immunol. 2013, 284, 75–83. [Google Scholar] [CrossRef]
- Zhou, X.; He, Z.; Henegar, J.; Allen, B.; Bigler, S. Expression of Allograft Inflammatory Factor-1 (AIF-1) in Acute Cellular Rejection of Cardiac Allografts. Cardiovasc. Pathol. 2011, 20, e177–e184. [Google Scholar] [CrossRef] [PubMed]
- Chang, X.; Hao, J.; Wang, X.; Liu, J.; Ni, J.; Hao, L. The Role of AIF-1 in the Aldosterone-Induced Vascular Calcification Related to Chronic Kidney Disease: Evidence from Mice Model and Cell Co-Culture Model. Front. Endocrinol. 2022, 13, 917356. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Wang, D.; Li, N.; Sheng, J.; Xie, M.; Zhou, Z.; Cheng, G.; Fan, Y. The Novel Tumor Microenvironment-Related Prognostic Gene AIF1 May Influence Immune Infiltrates and Is Correlated with TIGIT in Esophageal Cancer. Ann. Surg. Oncol. 2022, 29, 2930–2940. [Google Scholar] [CrossRef] [PubMed]
- Broglio, L.; Erne, B.; Tolnay, M.; Schaeren-Wiemers, N.; Fuhr, P.; Steck, A.J.; Renaud, S. Allograft Inflammatory Factor-1: A Pathogenetic Factor for Vasculitic Neuropathy. Muscle Nerve 2008, 38, 1272–1279. [Google Scholar] [CrossRef]
- Sikora, M.; Kopeć, B.; Piotrowska, K.; Pawlik, A. Role of Allograft Inflammatory Factor-1 in Pathogenesis of Diseases. Immunol. Lett. 2020, 218, 1–4. [Google Scholar] [CrossRef]
- Prinz, M.; Priller, J. Microglia and Brain Macrophages in the Molecular Age: From Origin to Neuropsychiatric Disease. Nat. Rev. Neurosci. 2014, 15, 300–312. [Google Scholar] [CrossRef]
- Vizioli, J.; Verri, T.; Pagliara, P. Allograft Inflammatory Factor-1 in Metazoans: Focus on Invertebrates. Biology 2020, 9, 355. [Google Scholar] [CrossRef] [PubMed]
- Schorn, T.; Drago, F.; Tettamanti, G.; Valvassori, R.; de Eguileor, M.; Vizioli, J.; Grimaldi, A. Homolog of Allograft Inflammatory Factor-1 Induces Macrophage Migration during Innate Immune Response in Leech. Cell Tissue Res. 2015, 359, 853–864. [Google Scholar] [CrossRef] [PubMed]
- Baranzini, N.; Monti, L.; Vanotti, M.; Orlandi, V.T.; Bolognese, F.; Scaldaferri, D.; Girardello, R.; Tettamanti, G.; de Eguileor, M.; Vizioli, J.; et al. AIF-1 and RNASET2 Play Complementary Roles in the Innate Immune Response of Medicinal Leech. J. Innate Immun. 2019, 11, 150–167. [Google Scholar] [CrossRef] [PubMed]
- Drago, F.; Sautière, P.; Marrec-Croq, F.L.; Accorsi, A.; Camp, C.V.; Salzet, M.; Lefebvre, C.; Vizioli, J. Microglia of Medicinal Leech (Hirudo medicinalis) Express a Specific Activation Marker Homologous to Vertebrate Ionized Calcium-binding Adapter Molecule 1 (Iba1/Alias Aif-1). Dev. Neurobiol. 2014, 74, 987–1001. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, J.; Yu, F.; He, X.; Yu, Z. Allograft Inflammatory Factor-1 Stimulates Hemocyte Immune Activation by Enhancing Phagocytosis and Expression of Inflammatory Cytokines in Crassostrea gigas. Fish Shellfish Immunol. 2013, 34, 1071–1077. [Google Scholar] [CrossRef]
- Zoysa, M.D.; Nikapitiya, C.; Kim, Y.; Oh, C.; Kang, D.-H.; Whang, I.; Kim, S.-J.; Lee, J.-S.; Choi, C.Y.; Lee, J. Allograft Inflammatory Factor-1 in Disk Abalone (Haliotis discus discus): Molecular Cloning, Transcriptional Regulation against Immune Challenge and Tissue Injury. Fish Shellfish Immunol. 2010, 29, 319–326. [Google Scholar] [CrossRef]
- Li, J.; Chen, J.; Zhang, Y.; Yu, Z. Expression of Allograft Inflammatory Factor-1 (AIF-1) in Response to Bacterial Challenge and Tissue Injury in the Pearl Oyster, Pinctada martensii. Fish Shellfish Immunol. 2013, 34, 365–371. [Google Scholar] [CrossRef]
- Coelho, F.S.; Rodpai, R.; Miller, A.; Karinshak, S.E.; Mann, V.H.; dos Santos Carvalho, O.; Caldeira, R.L.; de Moraes Mourão, M.; Brindley, P.J.; Ittiprasert, W. Diminished Adherence of Biomphalaria glabrata Embryonic Cell Line to Sporocysts of Schistosoma mansoni Following Programmed Knockout of the Allograft Inflammatory Factor. Parasites Vectors 2020, 13, 511. [Google Scholar] [CrossRef]
- Zhang, L.; Zhao, J.; Li, C.; Su, X.; Chen, A.; Li, T.; Qin, S. Cloning and Characterization of Allograft Inflammatory Factor-1 (AIF-1) from Manila Clam Venerupis philippinarum. Fish Shellfish Immunol. 2011, 30, 148–153. [Google Scholar] [CrossRef]
- Bergamini, G.; Sacchi, S.; Ferri, A.; Franchi, N.; Montanari, M.; Ahmad, M.; Losi, C.; Nasi, M.; Cocchi, M.; Malagoli, D. Clodronate Liposome-Mediated Phagocytic Hemocyte Depletion Affects the Regeneration of the Cephalic Tentacle of the Invasive Snail, Pomacea canaliculata. Biology 2023, 12, 992. [Google Scholar] [CrossRef]
- Accorsi, A.; Pardo, B.; Ross, E.; Corbin, T.J.; McClain, M.; Weaver, K.; Delventhal, K.; Gattamraju, A.; Morrison, J.A.; McKinney, M.C.; et al. A Genetically Tractable Non-Vertebrate System to Study Complete Camera-Type Eye Regeneration. Nat. Commun. 2025, 16, 6698. [Google Scholar] [CrossRef] [PubMed]
- Tascedda, F.; Malagoli, D.; Accorsi, A.; Rigillo, G.; Blom, J.M.C.; Ottaviani, E. Molluscs as Models for Translational Medicine. Med. Sci. Monit Basic Res. 2015, 21, 96–99. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Zhang, Y.; Ren, Y.; Wang, H.; Li, S.; Jiang, F.; Yin, L.; Qiao, X.; Zhang, G.; Qian, W.; et al. The Genome of the Golden Apple Snail Pomacea canaliculata Provides Insight into Stress Tolerance and Invasive Adaptation. GigaScience 2018, 7, giy101. [Google Scholar] [CrossRef]
- Montanari, A.; Bergamini, G.; Ferrari, A.; Ferri, A.; Nasi, M.; Simonini, R.; Malagoli, D. The Immune Response of the Invasive Golden Apple Snail to a Nematode-Based Molluscicide Involves Different Organs. Biology 2020, 9, 371. [Google Scholar] [CrossRef]
- Malagoli, D.; Franchi, N.; Sacchi, S. The Eco-Immunological Relevance of the Anti-Oxidant Response in Invasive Molluscs. Antioxidants 2023, 12, 1266. [Google Scholar] [CrossRef]
- Song, L.; Wang, X.; Yang, Z.; Lv, Z.; Wu, Z. Angiostrongylus cantonensis in the Vector Snails Pomacea canaliculata and Achatina fulica in China: A Meta-Analysis. Parasitol. Res. 2016, 115, 913–923. [Google Scholar] [CrossRef] [PubMed]
- Boraldi, F.; Lofaro, F.D.; Accorsi, A.; Ross, E.; Malagoli, D. Toward the Molecular Deciphering of Pomacea canaliculata Immunity: First Proteomic Analysis of Circulating Hemocytes. Proteomics 2019, 19, e1800314. [Google Scholar] [CrossRef]
- Yang, L.; Cheng, T.; Zhao, F. Comparative Profiling of Hepatopancreas Transcriptomes in Satiated and Starving Pomacea canaliculata. BMC Genet. 2017, 18, 18. [Google Scholar] [CrossRef][Green Version]
- Xiao, Q.; Lin, Y.; Li, H.; Chen, Y.; Wei, W.; Li, P.; Chen, L. Transcriptome Sequencing Reveals the Differentially Expressed lncRNAs and mRNAs in Response to Cold Acclimation and Cold Stress in Pomacea canaliculata. BMC Genom. 2022, 23, 382. [Google Scholar] [CrossRef]
- Sun, J.; Mu, H.; Ip, J.C.H.; Li, R.; Xu, T.; Accorsi, A.; Alvarado, A.S.; Ross, E.; Lan, Y.; Sun, Y.; et al. Signatures of Divergence, Invasiveness, and Terrestrialization Revealed by Four Apple Snail Genomes. Mol. Biol. Evol. 2019, 36, 1507–1520. [Google Scholar] [CrossRef]
- Malagoli, D. Going beyond a Static Picture: The Apple Snail Pomacea canaliculata Can Tell Us the Life History of Molluscan Hemocytes. Inv. Surviv. J. 2018, 1, 61–65. [Google Scholar] [CrossRef]
- Sacchi, S.; Malagoli, D.; Franchi, N. The Invertebrate Immunocyte: A Complex and Versatile Model for Immunological, Developmental, and Environmental Research. Cells 2024, 13, 2106. [Google Scholar] [CrossRef]
- Rodriguez, C.; Prieto, G.I.; Vega, I.A.; Castro-Vazquez, A. Assessment of the Kidney and Lung as Immune Barriers and Hematopoietic Sites in the Invasive Apple Snail Pomacea canaliculata. PeerJ 2018, 6, e5789. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, C.; Simon, V.; Conget, P.; Vega, I.A. Both Quiescent and Proliferating Cells Circulate in the Blood of the Invasive Apple Snail Pomacea canaliculata. Fish Shellfish Immunol. 2020, 107, 95–103. [Google Scholar] [CrossRef] [PubMed]
- Bergamini, G.; Ahmad, M.; Cocchi, M.; Malagoli, D. A New Protocol of Computer-Assisted Image Analysis Highlights the Presence of Hemocytes in the Regenerating Cephalic Tentacles of Adult Pomacea canaliculata. Int. J. Mol. Sci. 2021, 22, 5023. [Google Scholar] [CrossRef] [PubMed]
- Stankov, A.; Belakaposka-Srpanova, V.; Bitoljanu, N.; Cakar, L.; Cakar, Z.; Rosoklija, G. Visualisation of Microglia with the Use of Immunohistochemical Double Staining Method for CD-68 and Iba-1 of Cerebral Tissue Samples in Cases of Brain Contusions. Prilozi 2015, 36, 141–145. [Google Scholar] [CrossRef]
- Lier, J.; Streit, W.J.; Bechmann, I. Beyond Activation: Characterizing Microglial Functional Phenotypes. Cells 2021, 10, 2236. [Google Scholar] [CrossRef]
- Hess, D.C.; Abe, T.; Hill, W.D.; Studdard, A.M.; Carothers, J.; Masuya, M.; Fleming, P.A.; Drake, C.J.; Ogawa, M. Hematopoietic Origin of Microglial and Perivascular Cells in Brain. Exp. Neurol. 2004, 186, 134–144. [Google Scholar] [CrossRef]
- Malagoli, D.; Paolo, I.D.; Ottaviani, E. Presence of and Stress-Related Changes in Urocortin-like Molecules in Neurons and Immune Cells from the Mussel Mytilus galloprovincialis. Peptides 2007, 28, 1545–1552. [Google Scholar] [CrossRef]
- Accorsi, A.; Box, A.C.; Peuß, R.; Wood, C.; Alvarado, A.S.; Rohner, N. Image3C, a Multimodal Image-Based and Label-Independent Integrative Method for Single-Cell Analysis. eLife 2021, 10, e65372. [Google Scholar] [CrossRef]
- Cueto, J.A.; Rodriguez, C.; Vega, I.A.; Castro-Vazquez, A. Immune Defenses of the Invasive Apple Snail Pomacea canaliculata (Caenogastropoda, Ampullariidae): Phagocytic Hemocytes in the Circulation and the Kidney. PLoS ONE 2015, 10, e0123964. [Google Scholar] [CrossRef]
- Chiaramonte, M.; Arizza, V.; Rosa, S.L.; Queiroz, V.; Mauro, M.; Vazzana, M.; Inguglia, L. Allograft Inflammatory Factor AIF-1: Early Immune Response in the Mediterranean Sea Urchin Paracentrotus lividus. Zoology 2020, 142, 125815. [Google Scholar] [CrossRef] [PubMed]
- Parisi, M.G.; Baranzini, N.; Dara, M.; Corte, C.L.; Vizioli, J.; Cammarata, M. AIF-1 and RNASET2 Are Involved in the Inflammatory Response in the Mediterranean Mussel Mytilus galloprovincialis Following Vibrio Infection. Fish Shellfish Immunol. 2022, 127, 109–118. [Google Scholar] [CrossRef]
- Zeng, C.W.; Tsai, H.J. The Promising Role of a Zebrafish Model Employed in Neural Regeneration Following a Spinal Cord Injury. Int. J. Mol. Sci. 2023, 24, 13938. [Google Scholar] [CrossRef] [PubMed]
- García, M.; Vecino, E. Role of Müller Glia in Neuroprotection and Regeneration in the Retina. Histol. Histopathol. 2003, 18, 1205–1218. [Google Scholar] [CrossRef]
- Gallo, V.; Deneen, B. Glial Development: The Crossroads of Regeneration and Repair in the CNS. Neuron 2014, 83, 283–308. [Google Scholar] [CrossRef] [PubMed]
- Toy, D.; Namgung, U. Role of Glial Cells in Axonal Regeneration. Exp. Neurobiol. 2013, 22, 68–76. [Google Scholar] [CrossRef]
- Suzumura, A.; Takeuchi, H.; Zhang, G.; Kuno, R.; Mizuno, T. Roles of Glia-Derived Cytokines on Neuronal Degeneration and Regeneration. Ann. N. Y. Acad. Sci. 2006, 1088, 219–229. [Google Scholar] [CrossRef]
- Vizioli, J.; Drago, F.; Lefebvre, C. Chapter 5—Neuroprotection and Immunity in the Medicinal Leech Hirudo medicinalis: What About Microglia? In Lessons in Immunity; Elsevier: Amsterdam, The Netherlands, 2016; pp. 67–78. [Google Scholar] [CrossRef]
- Louveau, A.; Smirnov, I.; Keyes, T.J.; Eccles, J.D.; Rouhani, S.J.; Peske, J.D.; Derecki, N.C.; Castle, D.; Mandell, J.W.; Lee, K.S.; et al. Corrigendum: Structural and Functional Features of Central Nervous System Lymphatic Vessels. Nature 2016, 533, 278. [Google Scholar] [CrossRef]
- Marrec-Croq, F.L.; Drago, F.; Vizioli, J.; Sautière, P.-E.; Lefebvre, C. The Leech Nervous System: A Valuable Model to Study the Microglia Involvement in Regenerative Processes. Clin. Dev. Immunol. 2013, 2013, 274019. [Google Scholar] [CrossRef]
- Malagoli, D.; Ottaviani, E. Life Is a Huge Compromise: Is the Complexity of the Vertebrate Immune-Neuroendocrine System an Advantage or the Price to Pay? Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2010, 155, 134–138. [Google Scholar] [CrossRef] [PubMed]
- Tasiemski, A.; Salzet, M. Neuro-Immune Lessons from an Annelid: The Medicinal Leech. Dev. Comp. Immunol. 2017, 66, 33–42. [Google Scholar] [CrossRef]
- Malagoli, D.; Ottaviani, E. Cross-Talk among Immune and Neuroendocrine Systems in Molluscs and Other Invertebrate Models. Horm. Behav. 2017, 88, 41–44. [Google Scholar] [CrossRef]
- Chaves da Silva, P.G.; Hsu, K.; Benton, J.L.; Beltz, B.S.; Allodi, S. A Balancing Act: The Immune System Supports Neurodegeneration and Neurogenesis. Cell. Mol. Neurobiol. 2020, 40, 967–989. [Google Scholar] [CrossRef] [PubMed]
- Benton, J.L.; Kery, R.; Li, J.; Noonin, C.; Söderhäll, I.; Beltz, B.S. Cells from the Immune System Generate Adult-Born Neurons in Crayfish. Dev. Cell 2014, 30, 322–333. [Google Scholar] [CrossRef]
- Mezey, É.; Brownstein, M.J. Do Circulating Cells Transdifferentiate and Replenish Stem Cell Pools in the Brain and Periphery? BioEssays 2015, 37, 398–402. [Google Scholar] [CrossRef]
- Makhijani, K.; Brückner, K. Of Blood Cells and the Nervous System: Hematopoiesis in the Drosophila Larva. Fly 2012, 6, 254–260. [Google Scholar] [CrossRef]
- Rishal, I.; Fainzilber, M. Axon–Soma Communication in Neuronal Injury. Nat. Rev. Neurosci. 2014, 15, 32–42. [Google Scholar] [CrossRef]
- Siebert, J.R.; Kennedy, K.; Osterhout, D.J. Neurons Are Not All the Same: Diversity in Neuronal Populations and Their Intrinsic Responses to Spinal Cord Injury. ASN Neuro 2025, 17, 2440299. [Google Scholar] [CrossRef]
- Chiu, C.-C.; Liao, Y.-E.; Yang, L.-Y.; Wang, J.-Y.; Tweedie, D.; Karnati, H.K.; Greig, N.H.; Wang, J.-Y. Neuroinflammation in Animal Models of Traumatic Brain Injury. J. Neurosci. Methods 2016, 272, 38–49. [Google Scholar] [CrossRef] [PubMed]
- Bortoletto, E.; Frizzo, R.; Venier, P.; Rosani, U. Srahunter: A User-Friendly Tool to Speed up and Simplify Data Downloading from NCBI SRA. bioRxiv 2024, preprint. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An Ultra-Fast All-in-One FASTQ Preprocessor. bioRxiv 2018, preprint. [Google Scholar] [CrossRef]
- Ferri, A.; Costa, P.M.; Simonini, R. Secretory Cells in Halla parthenopeia (Oenonidae): Potential Implications for the Feeding and Defence Strategies of a Carnivorous Burrowing Polychaete. J. Morphol. 2024, 285, e21781. [Google Scholar] [CrossRef]
- King, R.S.; Newmark, P.A. In Situ Hybridization Protocol for Enhanced Detection of Gene Expression in the Planarian schmidtea mediterranea. BMC Dev. Biol. 2013, 13, 8. [Google Scholar] [CrossRef]
- Ottaviani, E.; Cossarizza, A. Immunocytochemical Evidence of Vertebrate Bioactive Peptide-like Molecules in the Immuno Cell Types of the Freshwater Snail Plianorbarius corneus (L.) (Gastropoda, Pulmonata). FEBS Lett. 1990, 267, 250–252. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Hammer, Ø.; Harper, D.A.T.; Ryan, P.D. PAST: Paleontological statistics software package for education and data analysis. Palaeontol. Electron. 2001, 4, 1–9. Available online: http://palaeo-electronica.org/2001_1/past/issue1_01.htm (accessed on 4 May 2025).



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ferri, A.; Sacchi, S.; Franchi, N.; Rosani, U.; Malagoli, D. Pc-AIF1 Is Expressed in Hemocyte-Rich and Neural Tissues and Links Immune Response and Regeneration in the Snail Model Pomacea canaliculata. Int. J. Mol. Sci. 2025, 26, 9022. https://doi.org/10.3390/ijms26189022
Ferri A, Sacchi S, Franchi N, Rosani U, Malagoli D. Pc-AIF1 Is Expressed in Hemocyte-Rich and Neural Tissues and Links Immune Response and Regeneration in the Snail Model Pomacea canaliculata. International Journal of Molecular Sciences. 2025; 26(18):9022. https://doi.org/10.3390/ijms26189022
Chicago/Turabian StyleFerri, Anita, Sandro Sacchi, Nicola Franchi, Umberto Rosani, and Davide Malagoli. 2025. "Pc-AIF1 Is Expressed in Hemocyte-Rich and Neural Tissues and Links Immune Response and Regeneration in the Snail Model Pomacea canaliculata" International Journal of Molecular Sciences 26, no. 18: 9022. https://doi.org/10.3390/ijms26189022
APA StyleFerri, A., Sacchi, S., Franchi, N., Rosani, U., & Malagoli, D. (2025). Pc-AIF1 Is Expressed in Hemocyte-Rich and Neural Tissues and Links Immune Response and Regeneration in the Snail Model Pomacea canaliculata. International Journal of Molecular Sciences, 26(18), 9022. https://doi.org/10.3390/ijms26189022





