Mutations and Recombination at G4 DNA-Forming Sequences Exacerbated by CPT-Resistant Mutant Topoisomerase 1 Is Dependent on SUMOylation
Abstract
1. Introduction
2. Results
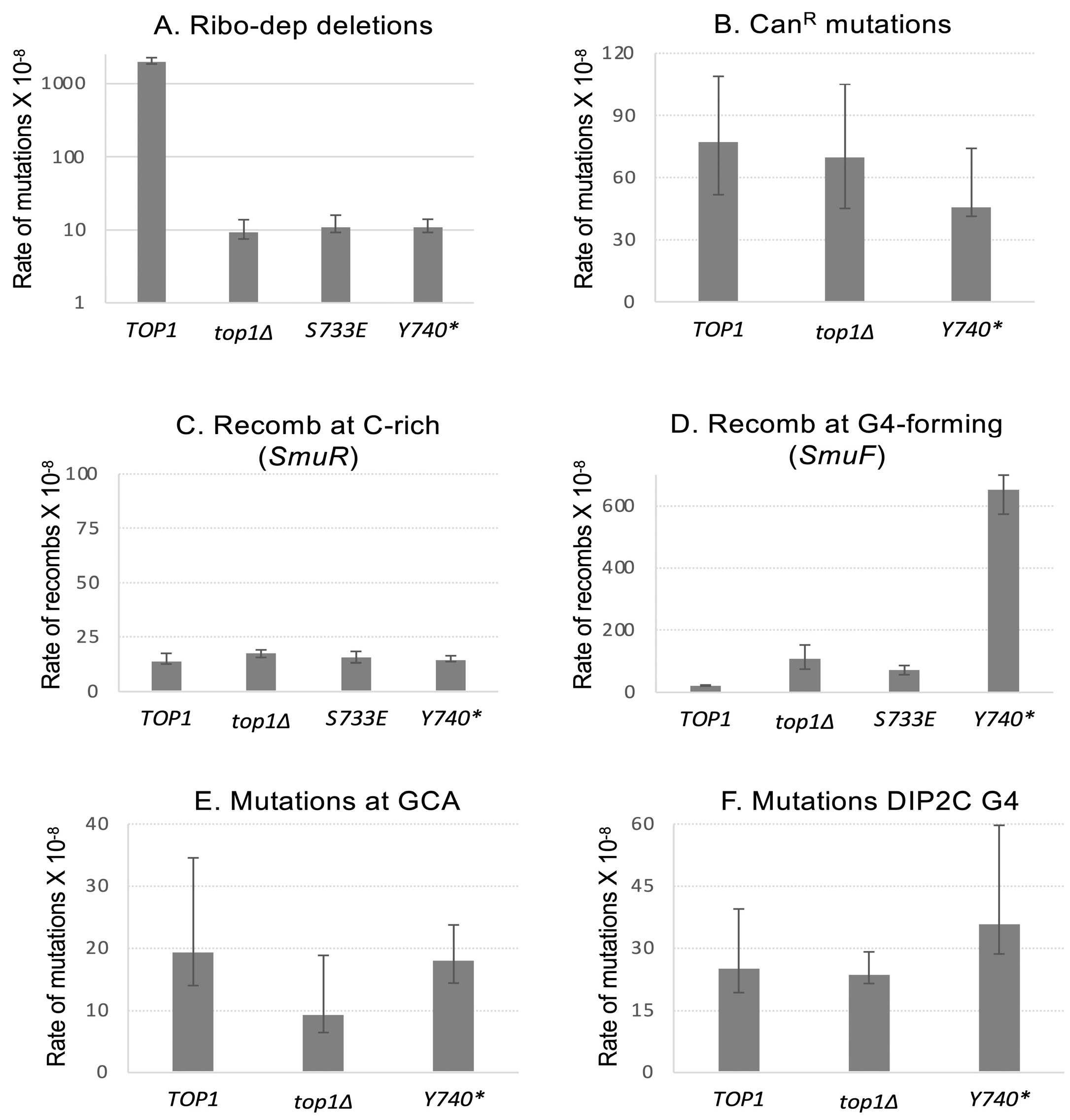
2.1. The Expression of Top1Y740* Mutant Affects Genome Instability Specifically at G4-Forming Sequences
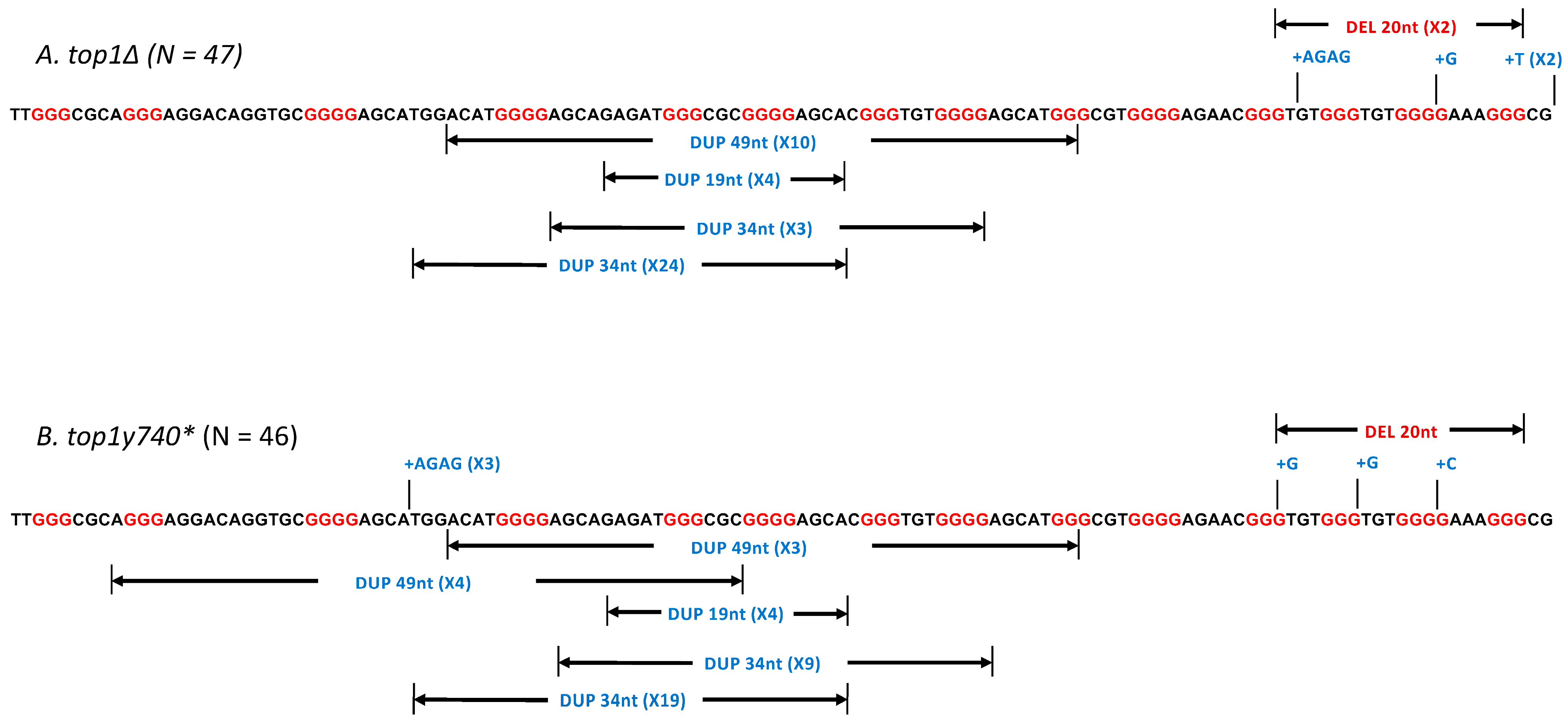
2.2. The Expression of Top1Y740* Mutant Elevates Duplication Events Proximal to G4-Forming Sequences
| Genotype | Total Rate of Mutation (×10−8) | Total Mutants Sequenced | # of >10 nt Duplications | Rate of >10 nt Duplications (×10−8) |
|---|---|---|---|---|
| TOP1 | 25.0 | 45 * | 38 * | 21.1 |
| top1∆ | 23.6 | 47 | 41 | 20.6 |
| top1Y740* | 35.8 | 46 | 39 | 30.4 |
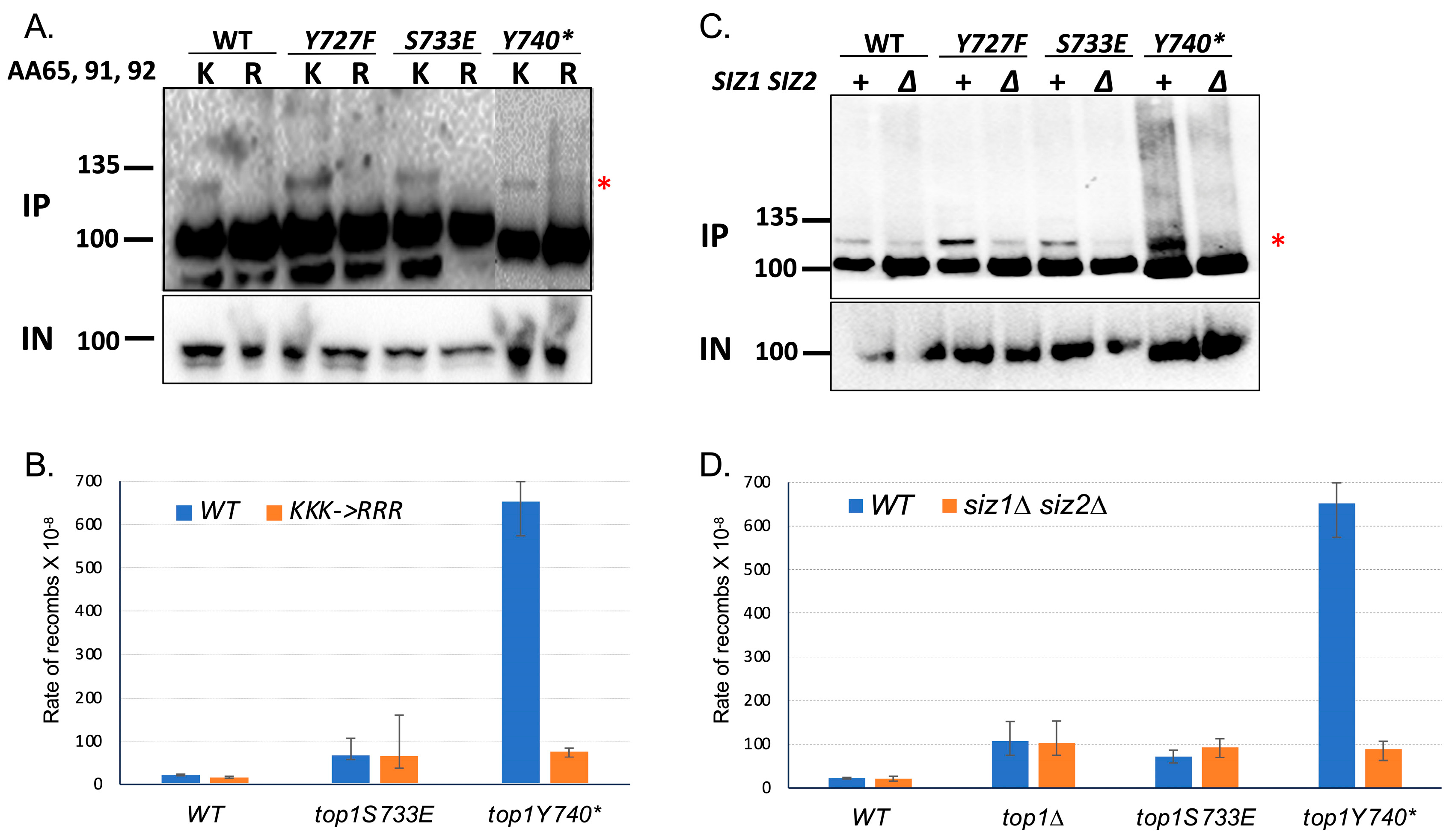
2.3. Recombination at G4 Is Significantly Reduced When the SUMOylation of Top1Y740* Is Abolished
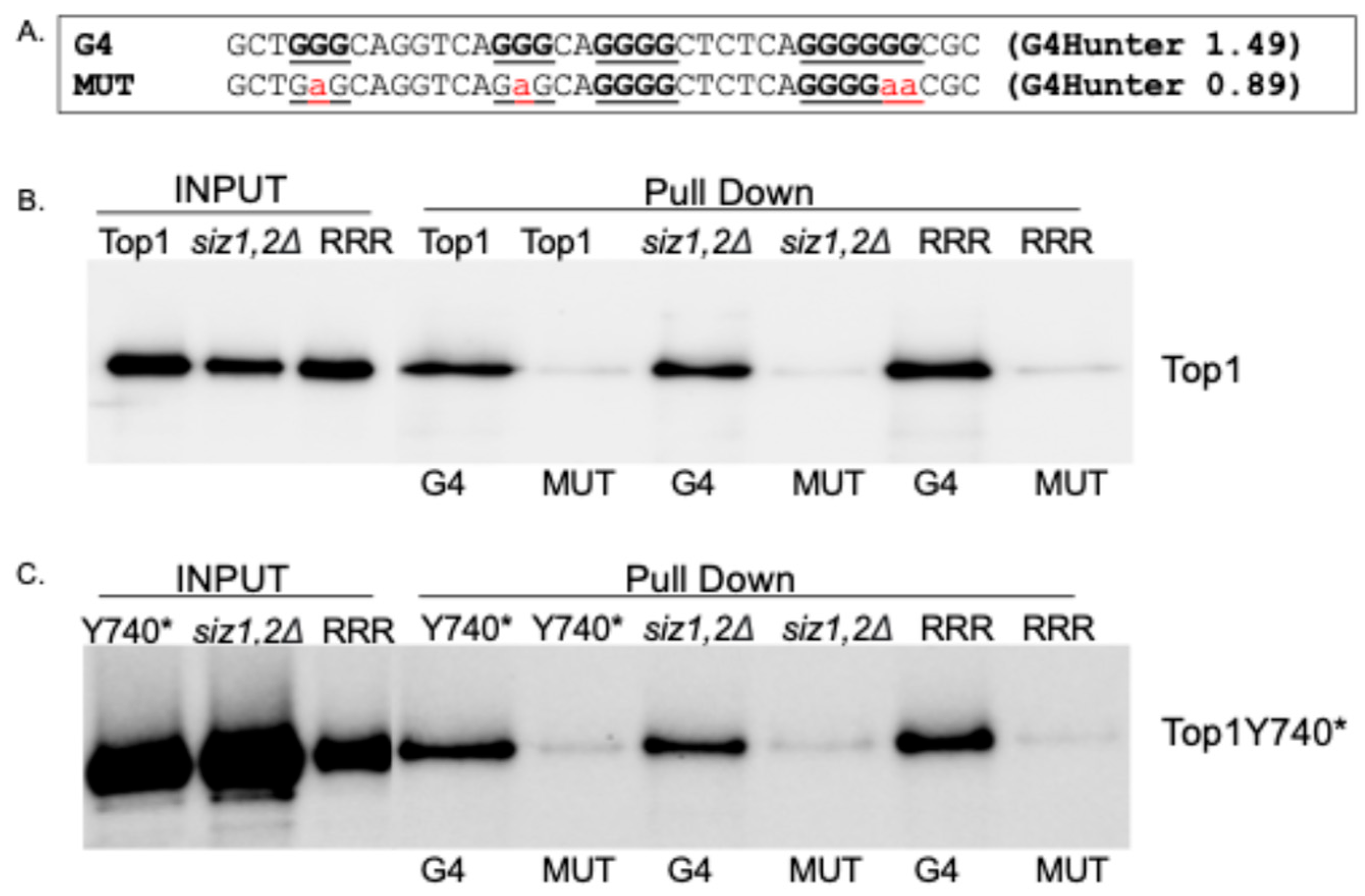
2.4. Binding to G4 DNA Is Not Affected When the SUMOylation Sites of Top1Y740* Are Mutated
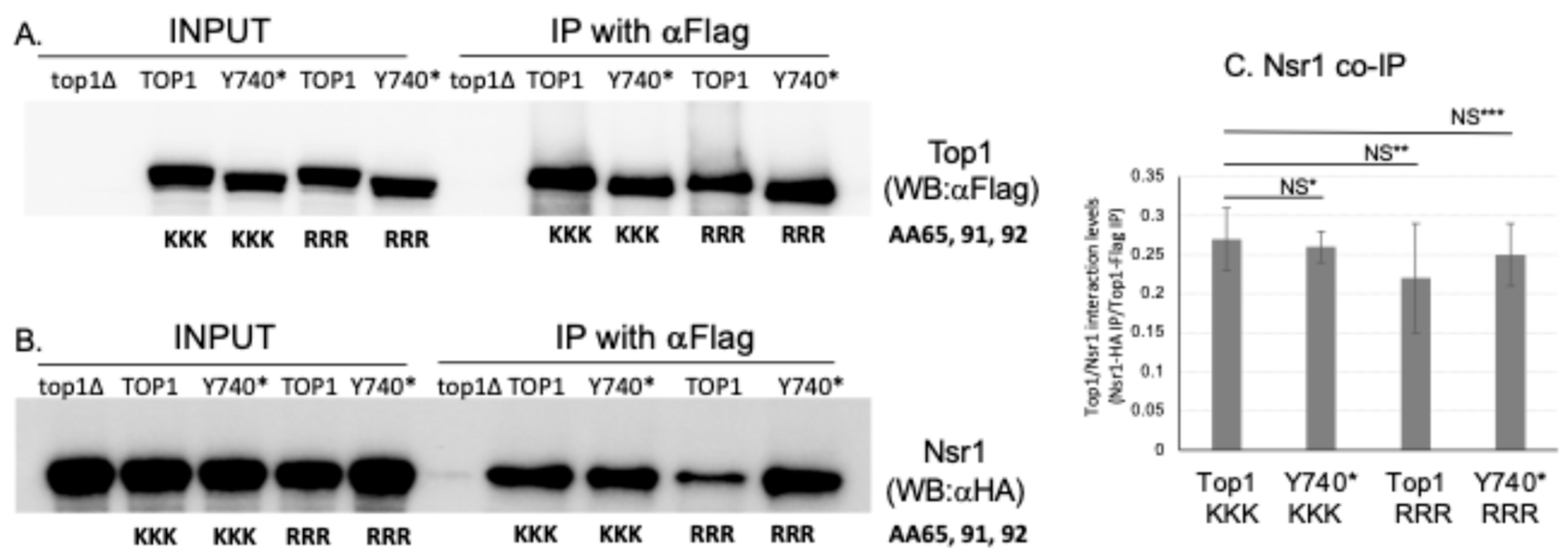
2.5. Interaction with Yeast Nucleolin Nsr1 Is Not Affected When the SUMOylation Sites of Top1Y740* Are Mutated
2.6. Mutations at the C-Terminal Residues of Human Top1 Correlate with High Incidence of Mutations at G4-Containing Loci Genome-Wide
3. Discussion
4. Materials and Methods
4.1. Yeast Strain and Plasmid Construction
4.2. Determination of Recombination and Mutation Rates
4.3. Mutation Spectra
4.4. Western Blotting
4.5. The Oligo Pull-Down Assay
4.6. Top1-SUMO Pull-Down
4.7. Co-IP
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| CPT | Camptothecin |
| G4 | G quadruplex |
References
- Pommier, Y.; Leo, E.; Zhang, H.; Marchand, C. DNA topoisomerases and their poisoning by anticancer and antibacterial drugs. Chem. Biol. 2010, 17, 421–433. [Google Scholar] [CrossRef]
- Kim, N.; Jinks-Robertson, S. Guanine repeat-containing sequences confer transcription-dependent instability in an orientation-specific manner in yeast. DNA Repair 2011, 10, 953–960. [Google Scholar] [CrossRef]
- Pavlova, A.V.; Kubareva, E.A.; Monakhova, M.V.; Zvereva, M.I.; Dolinnaya, N.G. Impact of G-Quadruplexes on the Regulation of Genome Integrity, DNA Damage and Repair. Biomolecules 2021, 11, 1284. [Google Scholar] [CrossRef] [PubMed]
- Rider, S.D., Jr.; Gadgil, R.Y.; Hitch, D.C.; Damewood, F.J.T.; Zavada, N.; Shanahan, M.; Alhawach, V.; Shrestha, R.; Shin-Ya, K.; Leffak, M. Stable G-quadruplex DNA structures promote replication-dependent genome instability. J. Biol. Chem. 2022, 298, 101947. [Google Scholar] [CrossRef] [PubMed]
- Sato, K.; Knipscheer, P. G-quadruplex resolution: From molecular mechanisms to physiological relevance. DNA Repair 2023, 130, 103552. [Google Scholar] [CrossRef] [PubMed]
- Yadav, P.; Harcy, V.; Argueso, J.L.; Dominska, M.; Jinks-Robertson, S.; Kim, N. Topoisomerase I plays a critical role in suppressing genome instability at a highly transcribed G-quadruplex-forming sequence. PLoS Genet. 2014, 10, e1004839. [Google Scholar] [CrossRef][Green Version]
- Husain, A.; Begum, N.A.; Taniguchi, T.; Taniguchi, H.; Kobayashi, M.; Honjo, T. Chromatin remodeller SMARCA4 recruits topoisomerase 1 and suppresses transcription-associated genomic instability. Nat. Commun. 2016, 7, 10549. [Google Scholar] [CrossRef]
- Arimondo, P.B.; Riou, J.; Mergny, J.; Tazi, J.; Sun, J.; Garestier, T.; Hélène, C. Interaction of human topoisomerase I with G-quartet structures. Nucleic Acids Res. 2000, 28, 4832–4838. [Google Scholar] [CrossRef]
- Marchand, C.; Pourquier, P.; Laco, G.S.; Jing, N.; Pommier, Y. Interaction of human nuclear topoisomerase I with guanosine quartet-forming and guanosine-rich single-stranded DNA and RNA oligonucleotides. J. Biol. Chem. 2002, 277, 8906–8911. [Google Scholar] [CrossRef]
- Berroyer, A.; Kim, N. The Functional Consequences of Eukaryotic Topoisomerase 1 Interaction with G-Quadruplex DNA. Genes 2020, 11, 193. [Google Scholar] [CrossRef]
- Berroyer, A.; Bacolla, A.; Tainer, J.A.; Kim, N. Cleavage-defective Topoisomerase I mutants sharply increase G-quadruplex-associated genomic instability. Microb. Cell 2022, 9, 52–68. [Google Scholar] [CrossRef]
- Keller, J.G.; Hymoller, K.M.; Thorsager, M.E.; Hansen, N.Y.; Erlandsen, J.U.; Tesauro, C.; Simonsen, A.K.W.; Andersen, A.B.; Vandso Petersen, K.; Holm, L.L.; et al. Topoisomerase 1 inhibits MYC promoter activity by inducing G-quadruplex formation. Nucleic Acids Res. 2022, 50, 6332–6342. [Google Scholar] [CrossRef]
- Liang, H.T.; Yan, J.Y.; Yao, H.J.; Zhang, X.N.; Xing, Z.M.; Liu, L.; Chen, Y.Q.; Li, G.R.; Huang, J.; He, Y.D.; et al. G-quadruplexes on chromosomal DNA negatively regulates topoisomerase 1 activity. Nucleic Acids Res. 2024, 52, 2142–2156. [Google Scholar] [CrossRef]
- Chen, S.H.; Chan, N.L.; Hsieh, T.S. New mechanistic and functional insights into DNA topoisomerases. Annu. Rev. Biochem. 2013, 82, 139–170. [Google Scholar] [CrossRef]
- Woo, M.H.; Vance, J.R.; Bjornsti, M.A. Studying DNA topoisomerase I-targeted drugs in the yeast. Saccharomyces cerevisiae. Methods Mol. Biol. 2001, 95, 303–313. [Google Scholar] [CrossRef] [PubMed]
- Wright, C.M.; van der Merwe, M.; DeBrot, A.H.; Bjornsti, M.A. DNA topoisomerase I domain interactions impact enzyme activity and sensitivity to camptothecin. J. Biol. Chem. 2015, 290, 12068–12078. [Google Scholar] [CrossRef]
- Bjornsti, M.A.; Benedetti, P.; Viglianti, G.A.; Wang, J.C. Expression of human DNA topoisomerase I in yeast cells lacking yeast DNA topoisomerase I: Restoration of sensitivity of the cells to the antitumor drug camptothecin. Cancer Res. 1989, 49, 6318–6323. [Google Scholar] [PubMed]
- Tsurutani, J.; Nitta, T.; Hirashima, T.; Komiya, T.; Uejima, H.; Tada, H.; Syunichi, N.; Tohda, A.; Fukuoka, M.; Nakagawa, K. Point mutations in the topoisomerase I gene in patients with non-small cell lung cancer treated with irinotecan. Lung Cancer 2002, 35, 299–304. [Google Scholar] [CrossRef] [PubMed]
- Losasso, C.; Cretaio, E.; Fiorani, P.; D’Annessa, I.; Chillemi, G.; Benedetti, P. A single mutation in the 729 residue modulates human DNA topoisomerase IB DNA binding and drug resistance. Nucleic Acids Res. 2008, 36, 5635–5644. [Google Scholar] [CrossRef]
- Kim, N.; Huang, S.N.; Williams, J.S.; Li, Y.C.; Clark, A.B.; Cho, J.E.; Kunkel, T.A.; Pommier, Y.; Jinks-Robertson, S. Mutagenic processing of ribonucleotides in DNA by yeast topoisomerase I. Science 2011, 332, 1561–1564. [Google Scholar] [CrossRef]
- Huang, S.Y.; Ghosh, S.; Pommier, Y. Topoisomerase I alone is sufficient to produce short DNA deletions and can also reverse nicks at ribonucleotide sites. J. Biol. Chem. 2015, 290, 14068–14076. [Google Scholar] [CrossRef]
- Lang, G.I.; Murray, A.W. Estimating the per-base-pair mutation rate in the yeast Saccharomyces cerevisiae. Genetics 2008, 178, 67–82. [Google Scholar] [CrossRef]
- Yadav, P.; Owiti, N.; Kim, N. The role of topoisomerase I in suppressing genome instability associated with a highly transcribed guanine-rich sequence is not restricted to preventing RNA:DNA hybrid accumulation. Nucleic Acids Res. 2016, 44, 718–729. [Google Scholar] [CrossRef]
- Williams, J.D.; Houserova, D.; Johnson, B.R.; Dyniewski, B.; Berroyer, A.; French, H.; Barchie, A.A.; Bilbrey, D.D.; Demeis, J.D.; Ghee, K.R.; et al. Characterization of long G4-rich enhancer-associated genomic regions engaging in a novel loop:loop ‘G4 Kissing’ interaction. Nucleic Acids Res. 2020, 48, 5907–5925. [Google Scholar] [CrossRef] [PubMed]
- Rallabhandi, P.; Hashimoto, K.; Mo, Y.Y.; Beck, W.T.; Moitra, P.K.; D’Arpa, P. Sumoylation of topoisomerase I is involved in its partitioning between nucleoli and nucleoplasm and its clearing from nucleoli in response to camptothecin. J. Biol. Chem. 2002, 277, 40020–40026. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.L.; Silver, H.R.; Xiong, L.; Belichenko, I.; Adegite, C.; Johnson, E.S. Topoisomerase I-dependent viability loss in saccharomyces cerevisiae mutants defective in both SUMO conjugation and DNA repair. Genetics 2007, 177, 17–30. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, Y.; Kahyo, T.; Toh, E.A.; Yasuda, H.; Kikuchi, Y. Yeast Ull1/Siz1 is a novel SUMO1/Smt3 ligase for septin components and functions as an adaptor between conjugating enzyme and substrates. J. Biol. Chem. 2001, 276, 48973–48977. [Google Scholar] [CrossRef]
- Serbyn, N.; Bagdiul, I.; Noireterre, A.; Michel, A.H.; Suhandynata, R.T.; Zhou, H.; Kornmann, B.; Stutz, F. SUMO orchestrates multiple alternative DNA-protein crosslink repair pathways. Cell Rep. 2021, 37, 110034. [Google Scholar] [CrossRef]
- Lopez, C.R.; Singh, S.; Hambarde, S.; Griffin, W.C.; Gao, J.; Chib, S.; Yu, Y.; Ira, G.; Raney, K.D.; Kim, N. Yeast Sub1 and human PC4 are G-quadruplex binding proteins that suppress genome instability at co-transcriptionally formed G4 DNA. Nucleic Acids Res. 2017, 45, 5850–5862. [Google Scholar] [CrossRef]
- Bedrat, A.; Lacroix, L.; Mergny, J.L. Re-evaluation of G-quadruplex propensity with G4Hunter. Nucleic Acids Res. 2016, 44, 1746–1759. [Google Scholar] [CrossRef]
- Hanakahi, L.A.; Sun, H.; Maizels, N. High affinity interactions of nucleolin with G-G-paired rDNA. J. Biol. Chem. 1999, 274, 15908–15912. [Google Scholar] [CrossRef]
- Tosoni, E.; Frasson, I.; Scalabrin, M.; Perrone, R.; Butovskaya, E.; Nadai, M.; Palu, G.; Fabris, D.; Richter, S.N. Nucleolin stabilizes G-quadruplex structures folded by the LTR promoter and silences HIV-1 viral transcription. Nucleic Acids Res. 2015, 43, 8884–8897. [Google Scholar] [CrossRef]
- Santos, T.; Salgado, G.F.; Cabrita, E.J.; Cruz, C. Nucleolin: A binding partner of G-quadruplex structures. Trends Cell Biol. 2022, 32, 561–564. [Google Scholar] [CrossRef]
- Singh, S.; Berroyer, A.; Kim, M.; Kim, N. Yeast Nucleolin Nsr1 Impedes Replication and Elevates Genome Instability at an Actively Transcribed Guanine-Rich G4 DNA-Forming Sequence. Genetics 2020, 216, 1023–1037. [Google Scholar] [CrossRef] [PubMed]
- Azevedo, C.; Livermore, T.; Saiardi, A. Protein polyphosphorylation of lysine residues by inorganic polyphosphate. Mol. Cell 2015, 58, 71–82. [Google Scholar] [CrossRef] [PubMed]
- Merrouche, Y.; Mugneret, F.; Cahn, J.Y. Secondary acute promyelocytic leukemia following irinotecan and oxaliplatin for advanced colon cancer. Ann. Oncol. 2006, 17, 1025–1026. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Chen, W.; Sun, X.; Wang, Z.; Zou, X.; Wei, H.; Wang, Z.; Chen, W. Metastatic colorectal cancer and severe hypocalcemia following irinotecan administration in a patient with X-linked agammaglobulinemia: A case report. BMC Med. Genet. 2019, 20, 157. [Google Scholar] [CrossRef]
- Redinbo, M.R.; Stewart, L.; Kuhn, P.; Champoux, J.J.; Hol, W.G. Crystal structures of human topoisomerase I in covalent and noncovalent complexes with DNA. Science 1998, 279, 1504–1513. [Google Scholar] [CrossRef]
- Delgado, J.L.; Hsieh, C.M.; Chan, N.L.; Hiasa, H. Topoisomerases as anticancer targets. Biochem. J. 2018, 475, 373–398. [Google Scholar] [CrossRef]
- Balakirev, M.Y.; Mullally, J.E.; Favier, A.; Assard, N.; Sulpice, E.; Lindsey, D.F.; Rulina, A.V.; Gidrol, X.; Wilkinson, K.D. Wss1 metalloprotease partners with Cdc48/Doa1 in processing genotoxic SUMO conjugates. Elife 2015, 4, e06763. [Google Scholar] [CrossRef]
- Mullen, J.R.; Chen, C.F.; Brill, S.J. Wss1 is a SUMO-dependent isopeptidase that interacts genetically with the Slx5-Slx8 SUMO-targeted ubiquitin ligase. Mol. Cell Biol. 2010, 30, 3737–3748. [Google Scholar] [CrossRef]
- Serero, A.; Jubin, C.; Loeillet, S.; Legoix-Ne, P.; Nicolas, A.G. Mutational landscape of yeast mutator strains. Proc. Natl. Acad. Sci. USA 2014, 111, 1897–1902. [Google Scholar] [CrossRef]
- Sikorski, R.S.; Hieter, P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 1989, 122, 19–27. [Google Scholar] [CrossRef]
- Storici, F.; Resnick, M.A. Delitto perfetto targeted mutagenesis in yeast with oligonucleotides. Genet. Eng. 2003, 25, 189–207. [Google Scholar]
- Spell, R.M.; Jinks-Robertson, S. Determination of mitotic recombination rates by fluctuation analysis in Saccharomyces cerevisiae. Methods Mol. Biol. 2004, 262, 3–12. [Google Scholar] [CrossRef]
- Moore, D.S.; Notz, W.I.; Flinger, M.A. The Basic Practice of Statistics; Freeman and Company: Dallas, TX, USA, 2013. [Google Scholar]
- Kushnirov, V.V. Rapid and reliable protein extraction from yeast. Yeast 2000, 16, 857–860. [Google Scholar] [CrossRef]
| Sample Label | Mutation at Top1 | Tissue | # of Mut Total | # of Mut at G4 | % Mut at G4 |
|---|---|---|---|---|---|
| T90 | T747P | Prostate | 49 | 1 | 2.041 |
| APGI-DA-3191 | T747N | Small intestine | 85 | 1 | 1.176 |
| TCGA-AD-6901-01 | R749Q | Large intestine | 319 | 0 | 0 |
| TCGA-ER-A19E-06 | W736C | Skin | 711 | 8 | 1.125 |
| TCGA-D3-A2JP-06 | W754L | Skin | 4857 | 6 | 0.123 |
| TCGA-BS-A0UV-01 | A715V | Endometrium | 9959 | 16 | 0.161 |
| SNU-C2B | I714T | Large intestine | 3413 | 53 | 1.553 |
| TCGA-D8-A27H-01 | Q713H | Breast | 184 | 0 | 0 |
| sysucc-783T | A759T | Large intestine | 1455 | 63 | 4.330 |
| 1117 | R727S | Urinary tract | 182 | 2 | 1.099 |
| CHG-13-27889T | A753S | Liver | 3948 | 109 | 2.761 |
| PD36792a | I743T | Skin | 1978 | 20 | 1.011 |
| TCGA-G2-A2EJ-01 | W736C | Urinary tract | 907 | 14 | 1.544 |
| 163 | W732C | Stomach | 244 | 5 | 2.049 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Singh, S.; Zhu, X.; Kim, N. Mutations and Recombination at G4 DNA-Forming Sequences Exacerbated by CPT-Resistant Mutant Topoisomerase 1 Is Dependent on SUMOylation. Int. J. Mol. Sci. 2025, 26, 9017. https://doi.org/10.3390/ijms26189017
Singh S, Zhu X, Kim N. Mutations and Recombination at G4 DNA-Forming Sequences Exacerbated by CPT-Resistant Mutant Topoisomerase 1 Is Dependent on SUMOylation. International Journal of Molecular Sciences. 2025; 26(18):9017. https://doi.org/10.3390/ijms26189017
Chicago/Turabian StyleSingh, Shivani, Xinji Zhu, and Nayun Kim. 2025. "Mutations and Recombination at G4 DNA-Forming Sequences Exacerbated by CPT-Resistant Mutant Topoisomerase 1 Is Dependent on SUMOylation" International Journal of Molecular Sciences 26, no. 18: 9017. https://doi.org/10.3390/ijms26189017
APA StyleSingh, S., Zhu, X., & Kim, N. (2025). Mutations and Recombination at G4 DNA-Forming Sequences Exacerbated by CPT-Resistant Mutant Topoisomerase 1 Is Dependent on SUMOylation. International Journal of Molecular Sciences, 26(18), 9017. https://doi.org/10.3390/ijms26189017




