The Association Between Genetics and Response to Treatment with Biologics in Patients with Psoriasis
Abstract
1. Introduction
2. Results
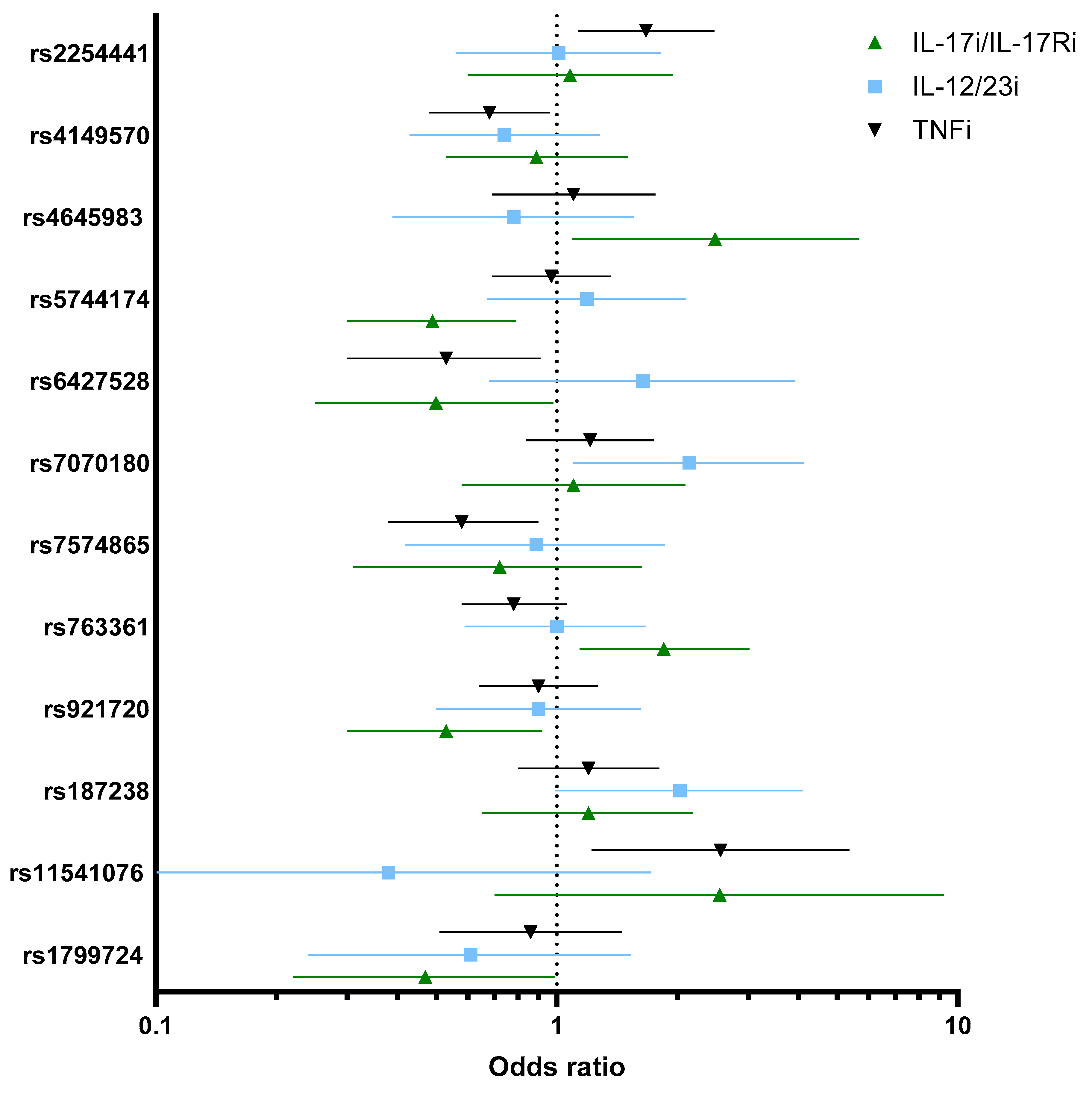
2.1. Association Between Genetics Variants and Response to TNFi
2.2. Association Between Genetic Variants and Response to IL-12/23i
2.3. Association Between Genetic Variants and Response to IL-17i/IL-17Ri
3. Discussion
4. Materials and Methods
4.1. Genetic Analyses
4.2. Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Reid, C.; Griffiths, C.E. Psoriasis and treatment: Past, present and future aspects. Acta Derm.-Venereol. 2020, 100, 69–79. [Google Scholar] [CrossRef]
- Nast, A.; Smith, C.; Spuls, P.I.; Avila Valle, G.; Bata-Csörgö, Z.; Boonen, H.; De Jong, E.; Garcia-Doval, I.; Gisondi, P.; Kaur-Knudsen, D. EuroGuiDerm Guideline on the systemic treatment of Psoriasis vulgaris—Part 1: Treatment and monitoring recommendations. J. Eur. Acad. Dermatol. Venereol. 2020, 34, 2461–2498. [Google Scholar] [CrossRef] [PubMed]
- Elmets, C.A.; Leonardi, C.L.; Davis, D.M.; Gelfand, J.M.; Lichten, J.; Mehta, N.N.; Armstrong, A.W.; Connor, C.; Cordoro, K.M.; Elewski, B.E. Joint AAD-NPF guidelines of care for the management and treatment of psoriasis with awareness and attention to comorbidities. J. Am. Acad. Dermatol. 2019, 80, 1073–1113. [Google Scholar] [CrossRef]
- Nikolai, L.; Egeberg, A.; Isufi, D.; Rasmussen, M.K.; Bryld, L.E.; Tomas, N.D.; Ajgeiy, K.K.; Bertelsen, T.; Lone, S. Response to interleukin-17A inhibitors according to prior biologic exposures: A Danish nationwide study. Acta Derm.-Venereol. 2023, 103, 12616. [Google Scholar]
- Hjort, G.; Schwarz, C.W.; Skov, L.; Loft, N. Clinical Characteristics Associated with Response to Biologics in the Treatment of Psoriasis: A Meta-analysis. JAMA Dermatol. 2024, 160, 830–837. [Google Scholar] [CrossRef]
- Nikolai, L.; Egeberg, A.; Rasmussen, M.K.; Bryld, L.E.; Nissen, C.V.; Dam, T.N.; Ajgeiy, K.K.; Iversen, L.; Lone, S. Response to biologics during the first six months of therapy in biologic-naïve patients with psoriasis predicts risk of disease flares: A Danish nationwide study. Acta Derm.-Venereol. 2021, 101, adv00357. [Google Scholar]
- Loft, N.; Skov, L.; Iversen, L.; Gniadecki, R.; Dam, T.; Brandslund, I.; Hoffmann, H.J.; Andersen, M.; Dessau, R.; Bergmann, A. Associations between functional polymorphisms and response to biological treatment in Danish patients with psoriasis. Pharmacogenom. J. 2018, 18, 494–500. [Google Scholar] [CrossRef]
- Bergmann, M.S.; Loft, N.; Schwarz, C.W.; Kaur-Knudsen, D.; Zachariae, C.; Skov, L. HLA-C* 06:02 in Danish patients with psoriasis and response to biological treatment. J. Dermatol. 2024, 52, 142–145. [Google Scholar] [CrossRef]
- Siewertsen, M.; Al-Sofi, R.; Dridi, H.; Ajenthen, G.D.; Zachariae, C.; Skov, L.; Loft, N. Association between HLA-Cw6 and response to treatment with biologics in patients with psoriasis: A systematic review and meta-analysis. J. Eur. Acad. Dermatol. Venereol. 2023, 37, e611–e614. [Google Scholar] [CrossRef]
- Dand, N.; Duckworth, M.; Baudry, D.; Russell, A.; Curtis, C.J.; Lee, S.H.; Evans, I.; Mason, K.J.; Alsharqi, A.; Becher, G. HLA-C* 06:02 genotype is a predictive biomarker of biologic treatment response in psoriasis. J. Allergy Clin. Immunol. 2019, 143, 2120–2130. [Google Scholar] [CrossRef]
- van Vugt, L.J.; van den Reek, J.M.; Hannink, G.; Coenen, M.J.; de Jong, E.M. Association of HLA-C* 06:02 status with differential response to ustekinumab in patients with psoriasis: A systematic review and meta-analysis. JAMA Dermatol. 2019, 155, 708–715. [Google Scholar] [CrossRef]
- Al-Sofi, R.F.; Bergmann, M.S.; Nielsen, C.H.; Andersen, V.; Skov, L.; Loft, N. The Association between Genetics and Response to Treatment with Biologics in Patients with Psoriasis, Psoriatic Arthritis, Rheumatoid Arthritis, and Inflammatory Bowel Diseases: A Systematic Review and Meta-Analysis. Int. J. Mol. Sci. 2024, 25, 5793. [Google Scholar] [CrossRef]
- Kobayashi, K.; Hernandez, L.D.; Galán, J.E.; Janeway, C.A.; Medzhitov, R.; Flavell, R.A. IRAK-M is a negative regulator of Toll-like receptor signaling. Cell 2002, 110, 191–202. [Google Scholar] [CrossRef]
- Potter, C.; Cordell, H.J.; Barton, A.; Daly, A.K.; Hyrich, K.L.; Mann, D.A.; Morgan, A.W.; Wilson, A.G.; Isaacs, J.D.; Genetics, B.i.R.A.; et al. Association between anti-tumour necrosis factor treatment response and genetic variants within the TLR and NFκB signalling pathways. Ann. Rheum. Dis. 2010, 69, 1315–1320. [Google Scholar] [CrossRef]
- Sode, J.; Vogel, U.; Bank, S.; Andersen, P.; Hetland, M.; Locht, H.; Heegaard, N.; Andersen, V. Confirmation of an IRAK3 polymorphism as a genetic marker predicting response to anti-TNF treatment in rheumatoid arthritis. Pharmacogenom. J. 2018, 18, 81–86. [Google Scholar] [CrossRef]
- Polo y La Borda, J.; Campos, J.; Sanz, J.; Andréu, J.L.; Mulero, J.; Sánchez, A. Predictive clinical-genetic model of long-term non-response to tumor necrosis factor-alpha inhibitor therapy in spondyloarthritis. Int. J. Rheum. Dis. 2019, 22, 1529–1537. [Google Scholar] [CrossRef]
- Baxi, S.; Greenblatt, R.; Bacchetti, P.; Cohen, M.; DeHovitz, J.; Anastos, K.; Gange, S.; Young, M.; Aouizerat, B. Evaluating the association of single-nucleotide polymorphisms with tenofovir exposure in a diverse prospective cohort of women living with HIV. Pharmacogenom. J. 2018, 18, 245–250. [Google Scholar] [CrossRef] [PubMed]
- del Fresno, C.; Soler-Rangel, L.; Soares-Schanoski, A.; Gómez-Piña, V.; González-León, M.C.; Gómez-García, L.; Mendoza-Barberá, E.; Rodríguez-Rojas, A.; García, F.; Fuentes-Prior, P. Inflammatory responses associated with acute coronary syndrome up-regulate IRAK-M and induce endotoxin tolerance in circulating monocytes. J. Endotoxin Res. 2007, 13, 39–52. [Google Scholar] [CrossRef]
- Martin, M.; Romero, X.; de la Fuente, M.A.; Tovar, V.; Zapater, N.; Esplugues, E.; Pizcueta, P.; Bosch, J.; Engel, P. CD84 functions as a homophilic adhesion molecule and enhances IFN-γ secretion: Adhesion is mediated by Ig-like domain 1. J. Immunol. 2001, 167, 3668–3676. [Google Scholar] [CrossRef] [PubMed]
- Cui, J.; Stahl, E.A.; Saevarsdottir, S.; Miceli, C.; Diogo, D.; Trynka, G.; Raj, T.; Mirkov, M.U.; Canhao, H.; Ikari, K. Genome-wide association study and gene expression analysis identifies CD84 as a predictor of response to etanercept therapy in rheumatoid arthritis. PLoS Genet. 2013, 9, e1003394. [Google Scholar] [CrossRef] [PubMed]
- Van den Reek, J.; Coenen, M.; Van De L’isle Arias, M.; Zweegers, J.; Rodijk-Olthuis, D.; Schalkwijk, J.; Vermeulen, S.; Joosten, I.; van de Kerkhof, P.; Seyger, M. Polymorphisms in CD84, IL12B and TNFAIP3 are associated with response to biologics in patients with psoriasis. Br. J. Dermatol. 2017, 176, 1288–1296. [Google Scholar] [CrossRef] [PubMed]
- Membrive-Jiménez, C.; Pérez-Ramírez, C.; Arias-Santiago, S.; Richetta, A.G.; Ottini, L.; Pineda-Lancheros, L.E.; Ramírez-Tortosa, M.d.C.; Jiménez-Morales, A. Impact of Functional Polymorphisms on Drug Survival of Biological Therapies in Patients with Moderate-to-Severe Psoriasis. Int. J. Mol. Sci. 2023, 24, 8703. [Google Scholar] [CrossRef] [PubMed]
- Ferreiro-Iglesias, A.; Montes, A.; Perez-Pampin, E.; Cañete, J.D.; Raya, E.; Magro-Checa, C.; Vasilopoulos, Y.; Caliz, R.; Ferrer, M.A.; Joven, B. Evaluation of 12 GWAS-drawn SNPs as biomarkers of rheumatoid arthritis response to TNF inhibitors. A potential SNP association with response to etanercept. PLoS ONE 2019, 14, e0213073. [Google Scholar] [CrossRef]
- Lopez-Rodriguez, R.; Perez-Pampin, E.; Marquez, A.; Blanco, F.J.; Joven, B.; Carreira, P.; Ferrer, M.A.; Caliz, R.; Valor, L.; Narvaez, J. Validation study of genetic biomarkers of response to TNF inhibitors in rheumatoid arthritis. PLoS ONE 2018, 13, e0196793. [Google Scholar] [CrossRef]
- Prieto-Pérez, R.; Solano-López, G.; Cabaleiro, T.; Román, M.; Ochoa, D.; Talegón, M.; Baniandrés, O.; López-Estebaranz, J.; De La Cueva, P.; Daudén, E. New polymorphisms associated with response to anti-TNF drugs in patients with moderate-to-severe plaque psoriasis. Pharmacogenom. J. 2018, 18, 70–75. [Google Scholar] [CrossRef]
- Ovejero-Benito, M.C.; Prieto-Perez, R.; Llamas-Velasco, M.; Belmonte, C.; Cabaleiro, T.; Roman, M.; Ochoa, D.; Talegon, M.; Saiz-Rodriguez, M.; Dauden, E. Polymorphisms associated with etanercept response in moderate-to-severe plaque psoriasis. Pharmacogenomics 2017, 18, 631–638. [Google Scholar] [CrossRef]
- Sode, J.; Vogel, U.; Bank, S.; Andersen, P.S.; Hetland, M.L.; Locht, H.; Heegaard, N.H.; Andersen, V. Genetic variations in pattern recognition receptor loci are associated with anti-TNF response in patients with rheumatoid arthritis. PLoS ONE 2015, 10, e0139781. [Google Scholar] [CrossRef]
- Bank, S.; Andersen, P.; Burisch, J.; Pedersen, N.; Roug, S.; Galsgaard, J.; Turino, S.; Brodersen, J.; Rashid, S.; Rasmussen, B. Genetically determined high activity of IL-12 and IL-18 in ulcerative colitis and TLR5 in Crohns disease were associated with non-response to anti-TNF therapy. Pharmacogenom. J. 2018, 18, 87–97. [Google Scholar] [CrossRef]
- Zhang, C.; Shestopaloff, K.; Hollis, B.; Kwok, C.H.; Hon, C.; Hartmann, N.; Tian, C.; Wozniak, M.; Santos, L.; West, D. Response to anti-IL17 therapy in inflammatory disease is not strongly impacted by genetic background. Am. J. Hum. Genet. 2023, 110, 1817–1824. [Google Scholar] [CrossRef]
- Loft, N.; Egeberg, A.; Rasmussen, M.; Bryld, L.; Nissen, C.; Dam, T.; Ajgeiy, K.; Iversen, L.; Skov, L. Prevalence and characterization of treatment-refractory psoriasis and super-responders to biologic treatment: A nationwide study. J. Eur. Acad. Dermatol. Venereol. 2022, 36, 1284–1291. [Google Scholar] [CrossRef] [PubMed]
- Mahil, S.; Wilson, N.; Dand, N.; Reynolds, N.; Griffiths, C.; Emsley, R.; Marsden, A.; Evans, I.; Warren, R.; Stocken, D. Psoriasis treat to target: Defining outcomes in psoriasis using data from a real-world, population-based cohort study (the British Association of Dermatologists Biologics and Immunomodulators Register, BADBIR). Br. J. Dermatol. 2020, 182, 1158–1166. [Google Scholar] [CrossRef] [PubMed]
- Puig, L. PASI 90 response: The new standard in therapeutic efficacy for psoriasis. J. Eur. Acad. Dermatol. Venereol. 2015, 29, 645–648. [Google Scholar] [CrossRef]
- Enevold, C.; Oturai, A.B.; Sørensen, P.S.; Ryder, L.P.; Koch-Henriksen, N.; Bendtzen, K. Multiple sclerosis and polymorphisms of innate pattern recognition receptors TLR1-10, NOD1-2, DDX58, and IFIH1. J. Neuroimmunol. 2009, 212, 125–131. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, K.M.; Mannucci, A.; Kimpton, C.P.; Gill, P. A rapid and quantitative DNA sex test: Fluorescence-based PCR analysis of XY homologous gene amelogenin. Biotechniques 1993, 15, 636–638, 640–641. [Google Scholar] [PubMed]
| TNFi N = 319 | IL-12/23i N = 109 | IL-17i N = 146 | |
|---|---|---|---|
| Age, years, mean (SD) | 43.9 (15.4) | 40.7 (14.9) | 46.4 (15.3) |
| Sex, male, n (%) | 211 (66.1%) | 78 (71.6%) | 95 (65.2%) |
| BMI, mean (SD) | 29.3 (6.8) | 28.8 (6.3) | 29.7 (8.5) |
| Baseline PASI, mean (SD) | 10.2 (6.2) | 10.0 (5.7) | 9.4 (5.1) |
| PsA, n (%) | 73 (22.9%) | 17 (15.7%) | 34 (23.2%) |
| Biologic naïve | 228 (71.5%) | 59 (53.9%) | 39 (26.7%) |
| SNP (rs Number Major/Minor Allele) | TNFi (n = 319) ORcrude (95% CI), p-Value | TNFi (n = 319) ORadj (95% CI), p-Value | IL-12/23i (n = 109) ORcrude (95% CI), p-Value | IL-12/23i (n = 109) ORadj (95% CI), p-Value | IL-17i/IL-17Ri (n = 146) ORcrude (95% CI), p-Value | IL-17i/IL-17Ri (n = 146) ORadj (95% CI), p-Value |
|---|---|---|---|---|---|---|
| PSTPIP1 (rs2254441 G/A) MAF = 0.21 | 1.67 (1.13–2.47), 0.0096 | 1.70 (1.14–2.53), 0.0081 | 1.01 (0.56–1.82), 0.96 | 0.92 (0.50–1.71), 0.80 | 1.08 (0.60–1.94), 0.79 | 0.94 (0.50–1.75), 0.85 |
| TNFRSF1A (rs4149570 C/A) MAF = 0.38 | 0.68 (0.48–0.96), 0.032 | 0.66 (0.46–0.94), 0.023 | 0.74 (0.43–1.28), 0.29 | 0.74 (0.42–1.31), 0.30 | 0.89 (0.53–1.50), 0.67 | 0.92 (0.53–1.61), 0.78 |
| CASP9 (rs4645983 G/A) MAF = 0.23 | 1.10 (0.69–1.76), 0.66 (n = 224) | 1.11 (0.69–1.79), 0.64 (n = 224) | 0.78 (0.39–1.56), 0.49 (n = 78) | 0.68 (0.32–1.43), 0.31 (n = 78) | 2.48 (1.09–5.68), 0.030 (n = 101) | 2.21 (0.90–5.40), 0.081 (n = 101) |
| TLR5 (rs5744174 A/G) MAF = 0.45 | 0.92 (0.66–1.28), 0.63 | 0.92 (0.66–1.28), 0.63 | 1.08 (0.64–1.77), 0.77 | 0.91 (0.53–1.56), 0.74 | 2.00 (1.14–3.50), 0.015 | 1.99 (1.095–3.64), 0.024 |
| CD84 (rs6427528 G/A) MAF = 0.099 | 0.53 (0.30–0.91), 0.023 | 0.53 (0.30–0.92), 0.026 | 1.64 (0.68–3.93), 0.26 | 1.53 (0.60–3.90), 0.36 | 0.50 (0.25–0.98), 0.045 | 0.51 (0.25–1.05), 0.066 |
| GFRA1 (rs7070180 C/T) MAF = 0.25 | 1.21 (0.84–1.75), 0.28 (n = 315) | 1.23 (0.85–1.78), 0.26 (n = 315) | 2.14 (1.10–4.14), 0.024 (n = 108) | 2.17 (1.057–4.48), 0.034 (n = 108) | 1.10 (0.58–2.09), 0.76 (n = 144) | 1.08 (0.54–2.13), 0.82 (n = 144) |
| STAT4 (rs7574865 G/T) MAF = 0.19 | 0.58 (0.38–0.90), 0.014 (n = 264) | 0.60 (0.39–0.92), 0.020 (n = 264) | 0.89 (0.42–1.86), 0.76 (n = 83) | 0.72 (0.31–1.63), 0.43 (n = 83) | 1.12 (0.62–2.05), 0.69 (n = 127) | 1.27 (0.66–2.43), 0.45 (n = 127) |
| CD226 (rs763361 C/T) MAF = 0.48 | 0.78 (0.58–1.06), 0.11 | 0.-79 (0.58–1.07), 0.13 | 1.00 (0.59–1.67), 0.10 | 0.94 (0.54–1.63), 0.83 | 1.85 (1.14–3.02), 0.012 | 1.80 (1.090–2.99), 0.021 |
| LINC02964 (rs921720 G/A) MAF = 0.39 | 0.90 (0.64–1.27), 0.56 (n = 317) | 0.92 (0.65–1.29), 0.63 (n = 317) | 0.90 (0.50–1.62), 0.73 (n = 108) | 0.87 (0.45–1.66), 0.67 (n = 108) | 0.53 (0.30–0.92), 0.025 (n = 144) | 0.45 (0.25–0.83), 0.0099 (n = 144) |
| IL-18 (rs187238 C/G) MAF = 0.27 | 1.20 (0.80–1.80), 0.36 (n = 264) | 1.17 (0.78–1.78), 0.43 (n = 264) | 2.03 (0.99–4.14), 0.051 (n = 87) | 2.59 (1.11–6.02), 0.026 (n = 87) | 1.20 (0.65–2.18), 0.55 (n = 129) | 1.22 (0.65–2.30), 0.53 (n = 129) |
| IRAK3 (rs11541076 A/T) MAF = 0.15 | 2.56 (1.22–5.37), 0.012 (n = 127) | 2.47 (1.13–5.38), 0.022 (n = 127) | 0.38 (0.084–1.72), 0.21 (n = 44) | 0.23 (0.044–1.28), 0.094 (n = 44) | 2.55 (0.70–9.22), 0.15 (n = 56) | 2.21 (0.57–8.55), 0.24 (n = 56) |
| TNF (rs1799724 C/T) MAF = 0.10 | 0.86 (0.51–1.45), 0.57 | 0.84 (0.50–1.43), 0.54 | 0.61 (0.24–1.53), 0.29 | 0.52 (0.19–1.38), 0.19 | 0.47 (0.22–0.99), 0.049 | 0.61 (0.27 (0.27–1.37), 0.23 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Loft, N.; Altintas, S.; Al-Sofi, R.; Isufi, D.; Zachariae, C.; Kaur-Knudsen, D.; Nielsen, C.H.; Skov, L. The Association Between Genetics and Response to Treatment with Biologics in Patients with Psoriasis. Int. J. Mol. Sci. 2025, 26, 8998. https://doi.org/10.3390/ijms26188998
Loft N, Altintas S, Al-Sofi R, Isufi D, Zachariae C, Kaur-Knudsen D, Nielsen CH, Skov L. The Association Between Genetics and Response to Treatment with Biologics in Patients with Psoriasis. International Journal of Molecular Sciences. 2025; 26(18):8998. https://doi.org/10.3390/ijms26188998
Chicago/Turabian StyleLoft, Nikolai, Sule Altintas, Rownaq Al-Sofi, Daniel Isufi, Claus Zachariae, Diljit Kaur-Knudsen, Claus Henrik Nielsen, and Lone Skov. 2025. "The Association Between Genetics and Response to Treatment with Biologics in Patients with Psoriasis" International Journal of Molecular Sciences 26, no. 18: 8998. https://doi.org/10.3390/ijms26188998
APA StyleLoft, N., Altintas, S., Al-Sofi, R., Isufi, D., Zachariae, C., Kaur-Knudsen, D., Nielsen, C. H., & Skov, L. (2025). The Association Between Genetics and Response to Treatment with Biologics in Patients with Psoriasis. International Journal of Molecular Sciences, 26(18), 8998. https://doi.org/10.3390/ijms26188998






