Molecular Features and Actionable Gene Targets of Testicular Germ Cell Tumors in a Real-World Setting
Abstract
1. Introduction
2. Results
3. Discussion
4. Materials and Methods
4.1. Patient Cohort and Methods
4.2. Gene Variant Call Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cancer of the Testis-Cancer Stat Facts [Internet]. SEER. Available online: https://seer.cancer.gov/statfacts/html/testis.html (accessed on 10 April 2025).
- Singh, R.; Fazal, Z.; Freemantle, S.J.; Spinella, M.J. Between a Rock and a Hard Place: An Epigenetic-Centric View of Testicular Germ Cell Tumors. Cancers 2021, 13, 1506. [Google Scholar] [CrossRef]
- Facts About Testicular Cancer|Testicular Cancer Statistics [Internet]. Available online: https://www.cancer.org/cancer/types/testicular-cancer/about/key-statistics.html (accessed on 10 April 2025).
- Lobo, J.; Acosta, A.M.; Netto, G.J. Molecular Biomarkers with Potential Clinical Application in Testicular Cancer. Mod. Pathol. 2023, 36, 100307. [Google Scholar] [CrossRef] [PubMed]
- Lobo, J.; Jerónimo, C.; Henrique, R. Cisplatin Resistance in Testicular Germ Cell Tumors: Current Challenges from Various Perspectives. Cancers 2020, 12, 1601. [Google Scholar] [CrossRef] [PubMed]
- Mao, S.; Huang, S. Zinc and Copper Levels in Bladder Cancer: A Systematic Review and Meta-Analysis. Biol. Trace Elem. Res. 2013, 153, 5–10. [Google Scholar] [CrossRef] [PubMed]
- Nunes, S.C.; Serpa, J. Glutathione in Ovarian Cancer: A Double-Edged Sword. Int. J. Mol. Sci. 2018, 19, 1882. [Google Scholar] [CrossRef]
- Yan, F.; Pang, J.; Peng, Y.; Molina, J.R.; Yang, P.; Liu, S. Elevated Cellular PD1/PD-L1 Expression Confers Acquired Resistance to Cisplatin in Small Cell Lung Cancer Cells. PLoS ONE 2016, 11, e0162925. [Google Scholar] [CrossRef]
- Hsu, W.H.; Zhao, X.; Zhu, J.; Kim, I.K.; Rao, G.; McCutcheon, J.; Hsu, S.-T.; Teicher, B.; Kallakury, B.; Dowlati, A.; et al. Checkpoint Kinase 1 Inhibition Enhances Cisplatin Cytotoxicity and Overcomes Cisplatin Resistance in SCLC by Promoting Mitotic Cell Death. J. Thorac. Oncol. 2019, 14, 1032–1045. [Google Scholar] [CrossRef]
- de Vries, G.; Rosas-Plaza, X.; van Vugt, M.A.T.M.; Gietema, J.A.; de Jong, S. Testicular cancer: Determinants of cisplatin sensitivity and novel therapeutic opportunities. Cancer Treat. Rev. 2020, 88, 102054. [Google Scholar] [CrossRef]
- Elmorsy, E.A.; Saber, S.; Hamad, R.S.; Abdel-Reheim, M.A.; El-kott, A.F.; AlShehri, M.A.; Morsy, K.; Salama, S.A.; Youssef, M.E. Advances in understanding cisplatin-induced toxicity: Molecular mechanisms and protective strategies. Eur. J. Pharm. Sci. 2024, 203, 106939. [Google Scholar] [CrossRef]
- Baroni, T.; Arato, I.; Mancuso, F.; Calafiore, R.; Luca, G. On the Origin of Testicular Germ Cell Tumors: From Gonocytes to Testicular Cancer. Front. Endocrinol. 2019, 10, 343. [Google Scholar] [CrossRef]
- De Toni, L.; Šabovic, I.; Cosci, I.; Ghezzi, M.; Foresta, C.; Garolla, A. Testicular Cancer: Genes, Environment, Hormones. Front. Endocrinol. 2019, 10, 408. [Google Scholar] [CrossRef]
- Gilligan, T.; Lin, D.W.; Adra, N.; Bagrodia, A.; Feldman, D.R.; Yamoah, K.; Aggarwal, R.; Chandrasekar, T.; Costa, D.; Drakaki, A.; et al. NCCN Guidelines® Insights: Testicular Cancer, Version 2.2025: Featured Updates to the NCCN Guidelines. J. Natl. Compr. Cancer Netw. 2025, 23, e250018. [Google Scholar] [CrossRef] [PubMed]
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.; Gastier-Foster, J.; Grody, W.W.; Hegde, M.; Lyon, E.; Spector, E.; et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015, 17, 405–424. [Google Scholar] [CrossRef] [PubMed]
- Tate, J.G.; Bamford, S.; Jubb, H.C.; Sondka, Z.; Beare, D.M.; Bindal, N.; Boutselakis, H.; Cole, C.G.; Creatore, C.; Dawson, E.; et al. COSMIC: The Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res. 2019, 47, D941–D947. [Google Scholar] [CrossRef] [PubMed]
- Kopanos, C.; Tsiolkas, V.; Kouris, A.; Chapple, C.E.; Albarca Aguilera, M.; Meyer, R.; Massouras, A. VarSome: The human genomic variant search engine. Bioinformatics 2019, 35, 1978–1980. [Google Scholar] [CrossRef]
- Landrum, M.J.; Lee, J.M.; Benson, M.; Brown, G.R.; Chao, C.; Chitipiralla, S.; Gu, B.; Hart, J.; Hoffman, D.; Jang, W.; et al. ClinVar: Improving access to variant interpretations and supporting evidence. Nucleic Acids Res. 2018, 46, D1062–D1067. [Google Scholar] [CrossRef]
- Cabral, E.R.M.; Pacanhella, M.F.; Lengert, A.V.H.; Reis, M.B.D.; Leal, L.F.; Lima, M.A.D.; da Silva, A.L.V.; Pinto, I.A.; Reis, R.M.; Pinto, M.T.; et al. Somatic mutation detection and KRAS amplification in testicular germ cell tumors. Front. Oncol. 2023, 13, 1133363. [Google Scholar] [CrossRef]
- Cárcano, F.M.; Lengert, A.H.; Vidal, D.O.; Scapulatempo Neto, C.; Queiroz, L.; Marques, H.; Baltazar, F.; Berardinelli, G.; Martinelli, C.M.S.; da Silva, E.C.A.; et al. Absence of microsatellite instability and BRAF (V600E) mutation in testicular germ cell tumors. Andrology 2016, 4, 866–872. [Google Scholar] [CrossRef]
- Shen, H.; Shih, J.; Hollern, D.P.; Wang, L.; Bowlby, R.; Tickoo, S.K.; Mungall, A.J.; Newton, Y.; Hegde, A.M.; Armenia, J.; et al. Integrated Molecular Characterization of Testicular Germ Cell Tumors. Cell Rep. 2018, 23, 3392–3406. [Google Scholar] [CrossRef]
- Fankhauser, C.D.; Curioni-Fontecedro, A.; Allmann, V.; Beyer, J.; Tischler, V.; Sulser, T.; Moch, H.; Bode, P.K. Frequent PD-L1 expression in testicular germ cell tumors. Br. J. Cancer 2015, 113, 411–413. [Google Scholar] [CrossRef]
- Hacioglu, B.M.; Kodaz, H.; Erdogan, B.; Cinkaya, A.; Tastekin, E.; Hacibekiroglu, I.; Turkmen, E.; Kostek, O.; Genc, E.; Uzunoglu, S.; et al. K-RAS and N-RAS mutations in testicular germ cell tumors. Bosn. J. Basic Med. Sci. 2017, 17, 159–163. [Google Scholar] [CrossRef]
- Ní Leathlobhair, M.; Frangou, A.; Kinnersley, B.; Cornish, A.J.; Chubb, D.; Lakatos, E.; Arumugam, P.; Gruber, A.J.; Law, P.; Tapinos, A.; et al. Genomic landscape of adult testicular germ cell tumours in the 100,000 Genomes Project. Nat. Commun. 2024, 15, 9247. [Google Scholar] [CrossRef]
- Stephenson, A.; Eggener, S.E.; Bass, E.B.; Chelnick, D.M.; Daneshmand, S.; Feldman, D.; Gilligan, T.; Karam, J.A.; Leibovich, B.; Liauw, S.L.; et al. Diagnosis and Treatment of Early Stage Testicular Cancer: AUA Guideline. J. Urol. 2019, 202, 272–281. [Google Scholar] [CrossRef] [PubMed]
- Ou, X.; Gao, G.; Habaz, I.A.; Wang, Y. Mechanisms of resistance to tyrosine kinase inhibitor-targeted therapy and overcoming strategies. MedComm (2020) 2024, 5, e694. [Google Scholar] [CrossRef] [PubMed]
- De Roock, W.; Claes, B.; Bernasconi, D.; De Schutter, J.; Biesmans, B.; Fountzilas, G.; Kalogeras, K.T.; Kotoula, V.; Papamichael, D.; Laurent-Puig, P.; et al. Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: A retrospective consortium analysis. Lancet Oncol. 2010, 11, 753–762. [Google Scholar] [CrossRef] [PubMed]
- Meric-Bernstam, F.; Hurwitz, H.; Raghav, K.P.S.; McWilliams, R.R.; Fakih, M.; VanderWalde, A.; Swanton, C.; Kurzrock, R.; Burris, H.; Sweeney, C.; et al. Pertuzumab plus trastuzumab for HER2-amplified metastatic colorectal cancer (MyPathway): An updated report from a multicentre, open-label, phase 2a, multiple basket study. Lancet Oncol. 2019, 20, 518–530. [Google Scholar] [CrossRef]
- Khan, M.R.; Sheehan, P.K.; Bazin, A.; Leonard, C.; Aleem, U.; Corrigan, L.; McDermott, R. Late side effects of testicular cancer and treatment: A comprehensive review. Discov. Oncol. 2024, 15, 646. [Google Scholar] [CrossRef]
- Lubberts, S.; Meijer, C.; Demaria, M.; Gietema, J.A. Early ageing after cytotoxic treatment for testicular cancer and cellular senescence: Time to act. Crit. Rev. Oncol./Hematol. 2020, 151, 102963. [Google Scholar] [CrossRef]
- Hiester, A.; Che, Y.; Lusch, A.; Kuß, O.; Niegisch, G.; Lorch, A.; Arsov, C.; Albers, P. Phase 2 Single-arm Trial of Primary Retroperitoneal Lymph Node Dissection in Patients with Seminomatous Testicular Germ Cell Tumors with Clinical Stage IIA/B (PRIMETEST). Eur. Urol. 2023, 84, 25–31. [Google Scholar] [CrossRef]
- Daneshmand, S.; Cary, C.; Masterson, T.; Einhorn, L.; Adra, N.; Boorjian, S.A.; Kollmannsberger, C.; Schuckman, A.; So, A.; Black, P.; et al. Surgery in Early Metastatic Seminoma: A Phase II Trial of Retroperitoneal Lymph Node Dissection for Testicular Seminoma With Limited Retroperitoneal Lymphadenopathy. J. Clin. Oncol. 2023, 41, 3009–3018. [Google Scholar] [CrossRef]
- Heidenreich, A.; Paffenholz, P.; Hartmann, F.; Seelemeyer, F.; Pfister, D. Retroperitoneal Lymph Node Dissection in Clinical Stage IIA/B Metastatic Seminoma: Results of the COlogne Trial of Retroperitoneal Lymphadenectomy In Metastatic Seminoma (COTRIMS). Eur. Urol. Oncol. 2024, 7, 122–127. [Google Scholar] [CrossRef]
- Berney, D.M.; Cree, I.; Rao, V.; Moch, H.; Srigley, J.R.; Tsuzuki, T.; Amin, M.B.; Comperat, E.M.; Hartmann, A.; Menon, S. An introduction to the WHO 5th edition 2022 classification of testicular tumours. Histopathology 2022, 81, 459–466. [Google Scholar] [CrossRef]
- Beaubier, N.; Tell, R.; Lau, D.; Parsons, J.R.; Bush, S.; Perera, J.; Sorrells, S.; Baker, T.; Chang, A.; Michuda, J.; et al. Clinical validation of the tempus xT next-generation targeted oncology sequencing assay. Oncotarget 2019, 10, 2384–2396. [Google Scholar] [CrossRef] [PubMed]
- STROBE [Internet]. STROBE. Available online: https://www.strobe-statement.org/ (accessed on 13 July 2025).
- Agha, R.; Mathew, G.; Rashid, R.; Kerwan, A.; Al-Jabir, A.; Sohrabi, C.; Franchi, T.; Nicola, M.; Agha, M.; PROCESS Group. Revised Preferred Reporting of Case Series in Surgery (PROCESS) Guideline: An update for the age of Artificial Intelligence. Prem. J. Sci. 2025, 10, 100080. [Google Scholar] [CrossRef]
- Suehnholz, S.P.; Nissan, M.H.; Zhang, H.; Kundra, R.; Nandakumar, S.; Lu, C.; Carrero, S.; Dhaneshwar, A.; Fernandez, N.; Xu, B.W.; et al. Quantifying the Expanding Landscape of Clinical Actionability for Patients with Cancer. Cancer Discov. 2024, 14, 49–65. [Google Scholar] [CrossRef]
- Chakravarty, D.; Gao, J.; Phillips, S.; Kundra, R.; Zhang, H.; Wang, J.; Rudolph, J.E.; Yaeger, R.; Soumerai, T.; Nissan, M.H.; et al. OncoKB: A Precision Oncology Knowledge Base. JCO Precis. Oncol. 2017, 1, 1–16. [Google Scholar] [CrossRef]
- Sim, N.L.; Kumar, P.; Hu, J.; Henikoff, S.; Schneider, G.; Ng, P.C. SIFT web server: Predicting effects of amino acid substitutions on proteins. Nucleic Acids Res. 2012, 40, W452–W457. [Google Scholar] [CrossRef]
- Choi, Y.; Chan, A.P. PROVEAN web server: A tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics 2015, 31, 2745–2747. [Google Scholar] [CrossRef]
- Chengsheng, T.; Huacheng, L.; Bing, X. AdaBoost typical Algorithm and its application research. MATEC Web Conf. 2017, 139, 00222. [Google Scholar] [CrossRef]
- Cutler, A.; Cutler, D.R.; Stevens, J.R. Random Forests. In Ensemble Machine Learning; Zhang, C., Ma, Y., Eds.; Springer: New York, NY, USA, 2012; pp. 157–175. Available online: https://link.springer.com/10.1007/978-1-4419-9326-7_5 (accessed on 13 July 2025).
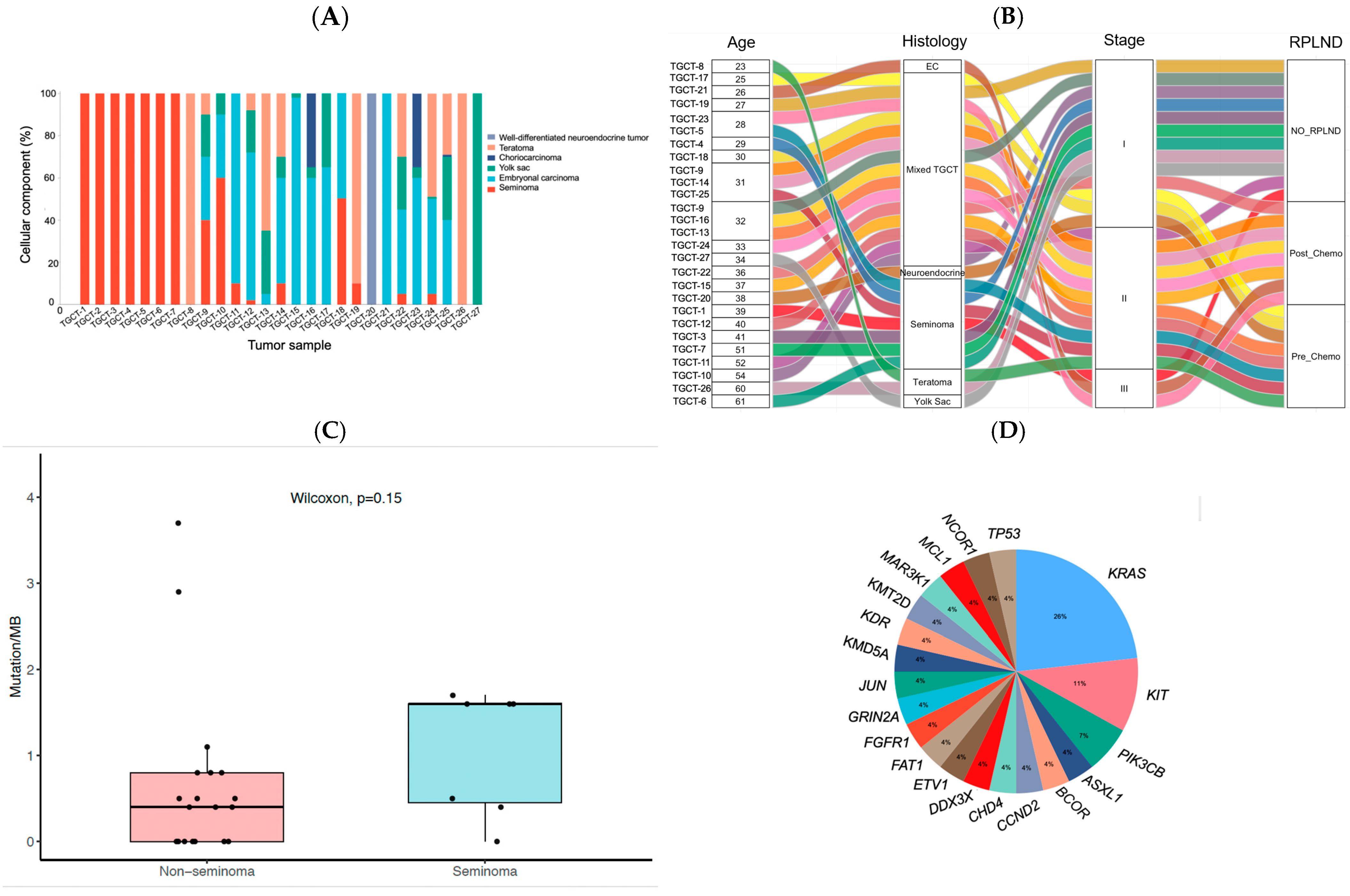
| Clinicopathological Variables | Number of Patients (%) | |
|---|---|---|
| Clinical disease stage | I | 13 (48) |
| II | 11 (41) | |
| III | 3 (11) | |
| Type of TGCT | Seminoma | 7 (26) |
| Non-Seminoma | 19 (70) | |
| Pre-pubertal teratoma | 1 (4) | |
| Surgeries Performed | Orchiectomy | 27 (100) |
| RPLND | 16 (59) | |
| Chemotherapy status prior RPLND | Chemotherapy-naïve | 8 (50) |
| Post-chemotherapy | 8 (50) | |
| Patient ID | Gene | AA Change | Alteration Effect | Function | Variant Allele Frequency | Mutations/MB |
|---|---|---|---|---|---|---|
| TGCT-1 | BCOR | p.S177fs | Frameshift | LOF | 10.50% | 1.6 |
| TGCT-2 | KIT | p.N822K # | Missense | GOF | 8.10% | 1.6 |
| PIK3CB | p.E1051K | Missense | GOF | 8.60% | ||
| TGCT-3 | KRAS | p.G12R | Missense | GOF | 4.00% | 1.6 |
| PIK3CB | p.E1051K | Missense | GOF | 8.60% | ||
| TGCT-6 | KIT | p.D816V # | Missense | GOF | 12.70% | 1.7 |
| TGCT-7 | KDR | -- | Copy number gain | -- | -- | 0.4 |
| KIT | -- | Copy number gain | -- | -- | ||
| p.N822K # | Missense | GOF | 39.36% | |||
| KRAS | -- | Copy number gain | -- | -- | ||
| TGCT-8 | KRAS | p.G12C | Missense | GOF | 59.70% | 3.7 |
| TGCT-10 | KRAS | p.G12V | Missense | GOF | 29.40% | 0.8 |
| TGCT-12 | GRIN2A | -- | Copy number loss | -- | -- | 0.0 |
| MCL1 | -- | Copy number gain | -- | -- | ||
| TGCT-14 | TP53 | p.L252_I254del | Inframe deletion | LOF | 16.60% | 0.5 |
| TGCT-18 | ETV1 | -- | Copy number gain | -- | -- | 0.8 |
| FGFR1 | -- | Copy number gain | -- | -- | ||
| JUN | -- | Copy number gain | -- | -- | ||
| KRAS | -- | Copy number gain | -- | -- | ||
| NCOR1 | p.Stgc43 | Stop codon | LOF | 2.12% | ||
| TGCT-21 | DDX3X | p.766-1G>T | Splice region | LOF | 4.90% | 0.0 |
| KMT2D | p.G5189 * | Stop codon | LOF | 2.90% | ||
| MAP3K1 | p.G331 * | Stop codon | LOF | 3.70% | ||
| TGCT-24 | CCND2 | -- | Copy number gain | -- | -- | 0.8 |
| CHD4 | -- | Copy number gain | -- | -- | ||
| KDM5A | -- | Copy number gain | -- | -- | ||
| KRAS | -- | Copy number gain | -- | -- | ||
| p.G12D | Missense | GOF | 59.30% | |||
| TGCT-25 | KRAS | -- | Copy number gain | -- | -- | 0.4 |
| TGCT-26 | ASXL1 | p.E565fs | Frameshift | LOF | 16.56% | 0.4 |
| TGCT-27 | FAT1 | p.L1889fs | Frameshift | LOF | 24.65% | 2.9 |
| Patient ID | Type of Variant | Gene | AA Alteration | Variant Effect | Variant Allele Frequency | ACMG Classification |
|---|---|---|---|---|---|---|
| TGCT-1 | Somatic | PRKN | p.L102F | Missense | 9.7% | VUS |
| Somatic | RANBP2 | p.P162A | Missense | 15.2% | VUS | |
| TGCT-2 | Somatic | C8orf34 | p.L523del | Inframe deletion | 6.9% | VUS |
| Germline | MSH6 | p.T999P | Missense | N.A. | Likely Pathogenic | |
| Germline | PMS2 | p.Q160H | Missense | N.A. | VUS | |
| TGCT-3 | Somatic | KEL | p.H581P | Missense | 6.2% | VUS |
| Germline | PALB2 | p.V836I | Missense | N.A. | Likely Pathogenic | |
| TGCT-4 | Somatic | FAM46C | p.N18S | Missense | 5.4% | N.A |
| TGCT-6 | Somatic | KMT2C | p.C3460G | Missense | 9.1% | Likely Pathogenic |
| Somatic | MAF | p.H187del | Inframe deletion | 6.0% | VUS | |
| Somatic | MTOR | p.K1981E | Missense | 12.8% | Likely Pathogenic | |
| TGCT-7 | Germline | MUTYH | p.L420M | Missense | N.A. | Pathogenic |
| TGCT-10 | Germline | RET | p.R180Q | Missense | N.A. | VUS |
| TGCT-11 | Germline | APOB | p.Y3295H | Missense | N.A. | VUS |
| Germline | MSH6 | p.P66L | Missense | N.A. | Likely Pathogenic | |
| Somatic | NF1 | p.L762M | Missense | 13.9% | Likely Pathogenic | |
| TGCT-13 | Somatic | CUX1 | p.R1374L | Missense | 10.4% | N.A |
| Somatic | KMT2D | p.R1918P | Missense | 12.1% | N.A | |
| TGCT-15 | Somatic | ERBB2 | p.E992K | Missense | 25.9% | Pathogenic |
| TGCT-16 | Germline | APOB | p.M1150K | Missense | N.A. | N.A |
| Germline | BRCA2 | p.F2058I | Missense | N.A. | N.A | |
| Germline | RB1 | p.E137D | Missense | N.A. | N.A | |
| TGCT-17 | Germline | BRCA2 | p.Q2858K | Missense | N.A. | N.A |
| TGCT-18 | Somatic | NF2 | p.T581I | Missense | 25.5% | Likely Pathogenic |
| TGCT-20 | Somatic | IL7R | p.G424V | Missense | 7.4% | N.A |
| TGCT-24 | Germline | APOB | p.G753E | Missense | N.A. | N.A |
| Germline | APOB | p.Y129C | Splice site | N.A. | N.A | |
| Somatic | JAK3 | p.R210W | Missense | 27.8% | Likely Pathogenic | |
| Germline | MSH2 | p.L119S | Missense | N.A. | N.A | |
| Germline | MSH6 | p.E277D | Missense | N.A. | N.A | |
| TGCT-25 | Somatic | GRM3 | p.C127Y | Missense | 15.3% | VUS |
| TGCT-26 | Germline | RET | p.E623K | Missense | N.A. | N.A |
| TGCT-27 | Somatic | HSP90AA1 | p.V266I | Missense | 17.4% | N.A |
| Somatic | RUNX1 | p.S167I | Missense | 20.5% | N.A | |
| Somatic | WRN | p.Q253 * | Stop codon | 28.3% | Pathogenic |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Morales-Grimany, R.; Giannikou, K.; Delgado, C.; Pandit, K.; Baky, F.; Amini, A.; Yuen, K.; Gerald, T.; Badia, R.; Taylor, J.; et al. Molecular Features and Actionable Gene Targets of Testicular Germ Cell Tumors in a Real-World Setting. Int. J. Mol. Sci. 2025, 26, 8963. https://doi.org/10.3390/ijms26188963
Morales-Grimany R, Giannikou K, Delgado C, Pandit K, Baky F, Amini A, Yuen K, Gerald T, Badia R, Taylor J, et al. Molecular Features and Actionable Gene Targets of Testicular Germ Cell Tumors in a Real-World Setting. International Journal of Molecular Sciences. 2025; 26(18):8963. https://doi.org/10.3390/ijms26188963
Chicago/Turabian StyleMorales-Grimany, Rafael, Krinio Giannikou, Cesar Delgado, Kshitij Pandit, Fady Baky, Armon Amini, Kit Yuen, Thomas Gerald, Rohit Badia, Jacob Taylor, and et al. 2025. "Molecular Features and Actionable Gene Targets of Testicular Germ Cell Tumors in a Real-World Setting" International Journal of Molecular Sciences 26, no. 18: 8963. https://doi.org/10.3390/ijms26188963
APA StyleMorales-Grimany, R., Giannikou, K., Delgado, C., Pandit, K., Baky, F., Amini, A., Yuen, K., Gerald, T., Badia, R., Taylor, J., Wang, L., Javier-Desloges, J., Margulis, V., Woldu, S., Salmasi, A., Millard, F., Mckay, R. R., & Bagrodia, A. (2025). Molecular Features and Actionable Gene Targets of Testicular Germ Cell Tumors in a Real-World Setting. International Journal of Molecular Sciences, 26(18), 8963. https://doi.org/10.3390/ijms26188963






