Different Translational Activities of Inflammatory Regulators Associated with Hypervolemia in Haemodialysis Patients
Abstract
1. Introduction
2. Results
2.1. Demographic Data
2.2. Leucocyte Numbers
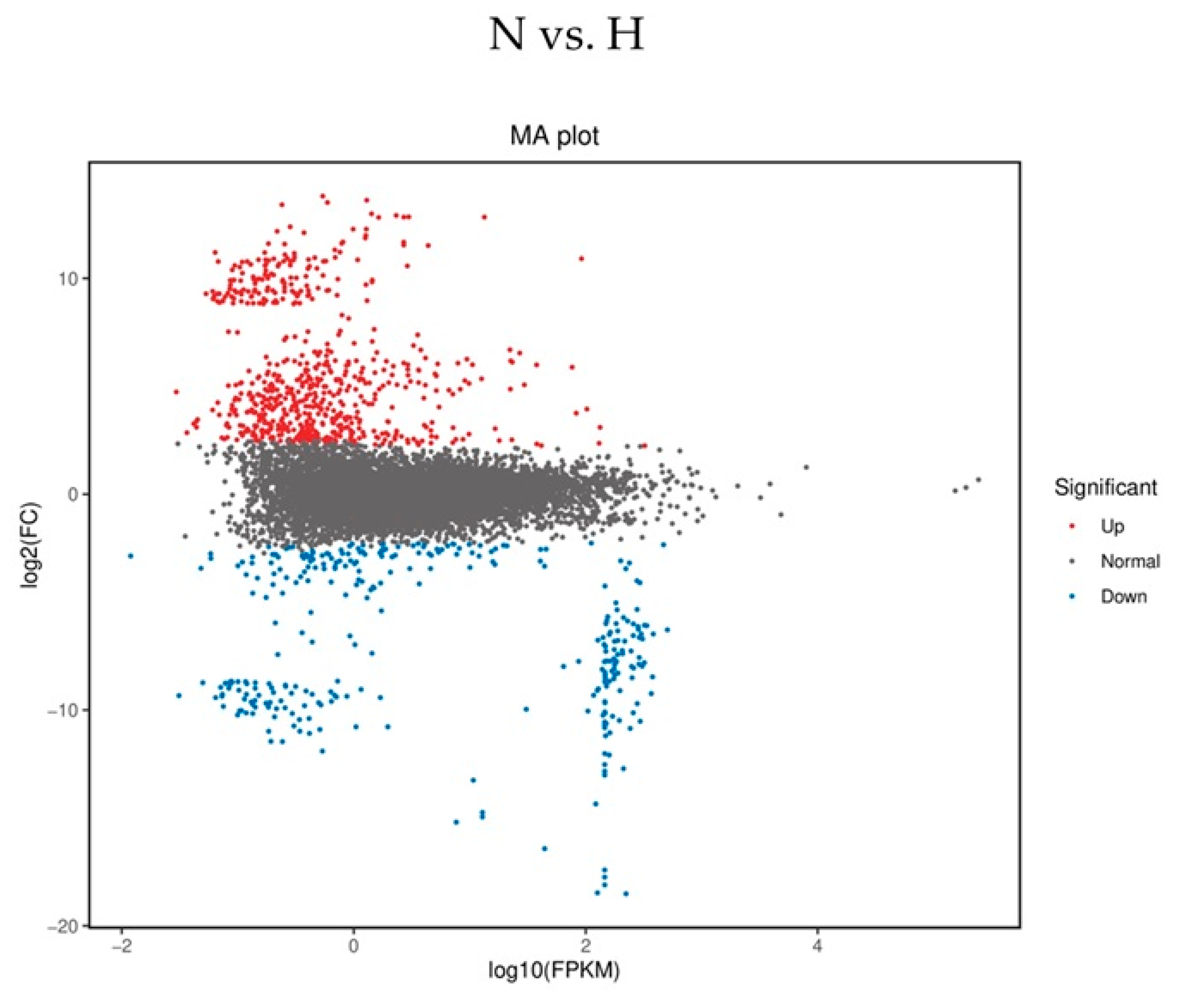
2.3. Sequence Analysis
2.4. mRNA Expression of TNFAIP and IL-10 Members and OTUD1 in Whole Blood
2.5. Polysome Profiling of IL-10, TIPE2 and OTUD1
2.6. Multivariate Linear Regression Model
3. Discussion
Study Limitations
4. Materials and Methods
4.1. Study Population
4.2. Absolute Cell Count Determination
4.3. RNA Analysis Isolated from TEMPUS Tubes
4.4. Sequence Analysis of Hyper- and Normovolemic Whole Blood Samples (mRNA-Seq)
4.5. PBMC Isolation
4.6. Treatment of PBMCs
4.7. Preparation of Cytosolic Lysates
4.8. Preparation of Linear Sucrose Gradients
4.9. RNA/cDNA/qPCR from Profiling Experiments
4.10. Cytokine Analysis
4.11. Statistics
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Palmer, B.F.; Clegg, D.J. Fluid overload as a therapeutic target for the preservative management of chronic kidney disease. Curr. Opin. Nephrol. Hypertens. 2020, 29, 22–28. [Google Scholar] [CrossRef]
- Hansen, B. Fluid Overload. Front. Vet. Sci. 2021, 18, 668688. [Google Scholar] [CrossRef]
- Banerjee, D.; Ma, J.Z.; Collins, A.J.; Herzog, C.A. Long-term survival of incident hemodialysis patients who are hospitalized for congestive heart failure, pulmonary edema, or fluid overload. Clin. J. Am. Soc. Nephrol. CJASN 2007, 2, 1186–1190. [Google Scholar] [CrossRef]
- Lopez, T.; Banerjee, D. Management of fluid overload in hemodialysis patients. Kidney Int. 2021, 100, 1170–1173. [Google Scholar] [CrossRef]
- Dekker, M.J.E.; Konings, C.; Canaud, B.; van der Sande, F.M.; Stuard, S.; Raimann, J.G.; Öztürk, E.; Usvyat, L.; Kotanko, P.; Kooman, J.P. Interactions Between Malnutrition, Inflammation, and Fluid Overload and Their Associations With Survival in Prevalent Hemodialysis Patients. J. Ren. Nutr. 2018, 28, 435–444. [Google Scholar] [CrossRef]
- Hung, S.-C.; Kuo, K.-L.; Peng, C.-H.; Wu, C.-H.; Lien, Y.-C.; Wang, Y.-C.; Tarng, D.-C. Volume overload correlates with cardiovascular risk factors in patients with chronic kidney disease. Kidney Int. 2014, 85, 703–709. [Google Scholar] [CrossRef]
- Ulrich, C.; Wilke, A.; Schleicher, N.; Girndt, M.; Fiedler, R. Hypervolemia-Induced Immune Disturbances Do Not Involve IL-1ß but IL-6 and IL-10 Activation in Haemodialysis Patients. Toxins 2020, 12, 159. [Google Scholar] [CrossRef]
- Guijarro, C.; Egido, J. Transcription factor-kappa B (NF-kappa B) and renal disease. Kidney Int. 2001, 59, 415–424. [Google Scholar] [CrossRef] [PubMed]
- Carlini, V.; Noonan, D.M.; Abdalalem, E.; Goletti, D.; Sansone, C.; Calabrone, L.; Albini, A. The multifaceted nature of IL-10: Regulation, role in immunological homeostasis and its relevance to cancer, COVID-19 and post-COVID conditions. Front. Immunol. 2023, 14, 1161067. [Google Scholar] [CrossRef] [PubMed]
- Bordoloi, D.; Banik, K.; Shabnam, B.; Padmavathi, G.; Monisha, J.; Arfuso, F.; Dharmarajan, A.; Mao, X.; Lim, L.H.K.; Wang, L.; et al. TIPE Family of Proteins and Its Implications in Different Chronic Diseases. Int. J. Mol. Sci. 2018, 19, 2974. [Google Scholar] [CrossRef] [PubMed]
- Oikawa, D.; Shimizu, K.; Tokunaga, F. Pleiotropic Roles of a KEAP1-Associated Deubiquitinase, OTUD1. Antioxidants 2023, 12, 350. [Google Scholar] [CrossRef]
- Nguyen, H.D.; Aljamaei, H.M.; Stadnyk, A.W. The Production and Function of Endogenous Interleukin-10 in Intestinal Epithelial Cells and Gut Homeostasis. Cell. Mol. Gastroenterol. Hepatol. 2021, 12, 1343–1352. [Google Scholar] [CrossRef]
- Singh, K.; Misra, D.P. Interleukin-10: Role in arterial wall homeostasis and dampening of inflammation in Takayasu arteritis. Int. J. Rheum. Dis. 2023, 26, 1663–1666. [Google Scholar] [CrossRef] [PubMed]
- Yeung, S.T.; Ovando, L.J.; Russo, A.J.; Rathinam, V.A.; Khanna, K.M. CD169+ macrophage intrinsic IL-10 production regulates immune homeostasis during sepsis. Cell Rep. 2023, 42, 112171. [Google Scholar] [CrossRef]
- Han, C.; Sun, L.; Pan, Q.; Sun, Y.; Wang, W.; Chen, Y. Polysome profiling followed by quantitative PCR for identifying potential micropeptide encoding long non-coding RNAs in suspension cell lines. STAR Protoc. 2022, 3, 101037. [Google Scholar] [CrossRef] [PubMed]
- Gandin, V.; Sikström, K.; Alain, T.; Morita, M.; McLaughlan, S.; Larsson, O.; Topisirovic, I. Polysome fractionation and analysis of mammalian translatomes on a genome-wide scale. J. Vis. Exp. JoVE 2014, e51455. [Google Scholar] [CrossRef]
- Ulrich, C.; Canim, Z.; Herberger, E.; Girndt, M.; Fiedler, R. Inflammation in Hypervolemic Hemodialysis Patients: The Roles of RelB and Caspase-4. Int. J. Mol. Sci. 2023, 24, 17550. [Google Scholar] [CrossRef]
- White, S.; Lin, L.; Hu, K. NF-κB and tPA Signaling in Kidney and Other Diseases. Cells 2020, 9, 1348. [Google Scholar] [CrossRef]
- Ghosh, S.; May, M.J.; Kopp, E.B. NF-kappa B and Rel proteins: Evolutionarily conserved mediators of immune responses. Annu. Rev. Immunol. 1998, 16, 225–260. [Google Scholar] [CrossRef] [PubMed]
- Harrigan, J.A.; Jacq, X.; Martin, N.M.; Jackson, S.P. Deubiquitylating enzymes and drug discovery: Emerging opportunities. Nature reviews. Drug Discov. 2018, 17, 57–78. [Google Scholar] [CrossRef]
- Liu, S.-Q.; Ren, C.; Yao, R.-Q.; Wu, Y.; Luan, Y.-Y.; Dong, N.; Yao, Y.-M. TNF-α-induced protein 8-like 2 negatively regulates the immune function of dendritic cells by suppressing autophagy via the TAK1/JNK pathway in septic mice. Cell Death Dis. 2021, 12, 1032. [Google Scholar] [CrossRef]
- Li, T.; Wang, W.; Gong, S.; Sun, H.; Zhang, H.; Yang, A.-G.; Chen, Y.H.; Li, X. Genome-wide analysis reveals TNFAIP8L2 as an immune checkpoint regulator of inflammation and metabolism. Mol. Immunol. 2018, 99, 154–162. [Google Scholar] [CrossRef]
- Sun, H.; Gong, S.; Carmody, R.J.; Hilliard, A.; Li, L.; Sun, J.; Kong, L.; Xu, L.; Hilliard, B.; Hu, S.; et al. TIPE2, a negative regulator of innate and adaptive immunity that maintains immune homeostasis. Cell 2008, 133, 415–426. [Google Scholar] [CrossRef]
- Oikawa, D.; Gi, M.; Kosako, H.; Shimizu, K.; Takahashi, H.; Shiota, M.; Hosomi, S.; Komakura, K.; Wanibuchi, H.; Tsuruta, D.; et al. OTUD1 deubiquitinase regulates NF-κB- and KEAP1-mediated inflammatory responses and reactive oxygen species-associated cell death pathways. Cell Death Dis. 2022, 13, 694. [Google Scholar] [CrossRef]
- Ming, T.; Liu, H.; Yuan, M.; Tian, J.; Fang, Q.; Liu, Y.; Kong, Q.; Wang, Q.; Song, X.; Xia, Z.; et al. The deubiquitinase OTUD1 deubiquitinates TIPE2 and plays a protective role in sepsis-induced lung injury by targeting TAK1-mediated MAPK and NF-κB signaling. Biochem. Pharmacol. 2024, 227, 116418. [Google Scholar] [CrossRef]
- Dawood, A.; Fiedler, R.; Markau, S.; Girndt, M.; Ulrich, C. Polysome Profiling Proves Impaired IL-10 and Caspase-8 Translation in PBMCs of Hemodialysis Patients. Biomolecules 2025, 15, 335. [Google Scholar] [CrossRef]
- Chassé, H.; Boulben, S.; Costache, V.; Cormier, P.; Morales, J. Analysis of translation using polysome profiling. Nucleic Acids Res. 2017, 45, e15. [Google Scholar] [CrossRef] [PubMed]
- Weir, M.R. Hypervolemia and blood pressure: Powerful indicators of increased mortality among hemodialysis patients. Hypertension 2010, 56, 341–343. [Google Scholar] [CrossRef] [PubMed]
- Antlanger, M.; Hecking, M.; Haidinger, M.; Werzowa, J.; Kovarik, J.J.; Paul, G.; Eigner, M.; Bonderman, D.; Hörl, W.H.; Säemann, M.D. Fluid overload in hemodialysis patients: A cross-sectional study to determine its association with cardiac biomarkers and nutritional status. BMC Nephrol. 2013, 14, 266. [Google Scholar] [CrossRef]
- Ulrich, C.; Fiedler, R.; Herberger, E.; Canim, Z.; Markau, S.; Girndt, M. Hypervolemia in Dialysis Patients Impairs STAT3 Signaling and Upregulates miR-142-3p: Effects on IL-10 and IL-6. Int. J. Mol. Sci. 2024, 25, 3719. [Google Scholar] [CrossRef] [PubMed]
- Lou, Y.; Liu, S. The TIPE (TNFAIP8) family in inflammation, immunity, and cancer. Mol. Immunol. 2011, 49, 4–7. [Google Scholar] [CrossRef] [PubMed]
- Liang, H.; Liu, X.; Guo, W.; Xiong, W.; Ren, D.; Liu, W. OTUD1 downregulates PD-L1 expression by deubiquitinating STAT3 and promotes the immune response in CcRCC. Cell. Oncol. 2025; ahead of print. [Google Scholar]
- Zhang, H.-G.; Wang, B.; Yang, Y.; Liu, X.; Wang, J.; Xin, N.; Li, S.; Miao, Y.; Wu, Q.; Guo, T.; et al. Depression compromises antiviral innate immunity via the AVP-AHI1-Tyk2 axis. Cell Res. 2022, 32, 897–913. [Google Scholar] [CrossRef]
- Zhang, Z.; Wang, D.; Wang, P.; Zhao, Y.; You, F. OTUD1 Negatively Regulates Type I IFN Induction by Disrupting Noncanonical Ubiquitination of IRF3. J. Immunol. 2020, 204, 1904–1918. [Google Scholar] [CrossRef]
- Lu, D.; Song, J.; Sun, Y.; Qi, F.; Liu, L.; Jin, Y.; McNutt, M.A.; Yin, Y. Mutations of deubiquitinase OTUD1 are associated with autoimmune disorders. J. Autoimmun. 2018, 94, 156–165. [Google Scholar] [CrossRef]
- Rysz, J.; Franczyk, B.; Ławiński, J.; Gluba-Brzózka, A. Oxidative Stress in ESRD Patients on Dialysis and the Risk of Cardiovascular Diseases. Antioxidants 2020, 9, 1079. [Google Scholar] [CrossRef]
- Karamouzis, I.; Sarafidis, P.A.; Karamouzis, M.; Iliadis, S.; Haidich, A.-B.; Sioulis, A.; Triantos, A.; Vavatsi-Christaki, N.; Grekas, D.M. Increase in oxidative stress but not in antioxidant capacity with advancing stages of chronic kidney disease. Am. J. Nephrol. 2008, 28, 397–404. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.R.; Kim, J.-K.; Lee, H.-S.; Kim, S.G.; Choi, E.-K. Serum levels of protein carbonyl, a marker of oxidative stress, are associated with overhydration, sarcopenia and mortality in hemodialysis patients. BMC Nephrol. 2020, 21, 281. [Google Scholar] [CrossRef] [PubMed]


| N (n = 25) | H (n = 17) | p-Value N vs. H | CO (n = 9) | p-Value CO vs. N vs. H | |
|---|---|---|---|---|---|
| Age (years) | 55.6 ± 13.9 | 65.6 ± 14.9 | 0.030 | 39.7 ± 18.4 | 0.005 H vs. CO |
| Gender (f, %) | 52.0 | 29.4 | 0.208 | 33.0 | 0.306 |
| BMI (kg/m2) | 26.0 ± 5.3 | 29.3 ± 9.0 | 0.143 | - | |
| Diabetes (%) | 20.0 | 41.2 | 0.174 | - | |
| Epo (I.U./week) | 8850 ± 4904 | 12,157 ± 7753 | 0.097 | - | |
| Phase angle (°) | 5.4 ± 1.0 | 4.1 ± 0.9 | 0.001 | - | |
| Resistance | 629.1 ± 111.2 | 449.7 ± 62.5 | 0.001 | - | |
| Reactance | 59.0 ± 13.6 | 32.4 ± 8.1 | 0.001 | - | |
| Total body water (L) | 37.4 ± 9.3 | 47.6 ± 8.7 | 0.001 | - | |
| BCM (Kg/m2) | 25.0 ± 7.8 | 26.5 ± 7.1 | 0.523 | - | |
| ICW (L) | 22.9 ± 4.4 | 26.1 ± 3.9 | 0.019 | - | |
| ECW (L) | 14.5 ± 5.0 | 21.4 ± 5.6 | 0.001 | - | |
| Sys. RR after HD (mmHg) | 131.9 ± 19.5 | 148.9 ± 25.9 | 0.020 | - | |
| Dia. RR after HD (mmHg) | 75.4 ± 15.5 | 70.8 ± 13.2 | 0.320 | - | |
| PP after HD (bpm) | 71.8 ± 10.9 | 66.5 ± 11.5 | 0.082 | - | |
| Kt/V | 1.5 ± 0.4 | 1.3 ± 0.2 | 0.026 | - | |
| Creatinine (µmol/L) | 921.9 ± 218.5 | 667.8 ± 206.8 | 0.001 | - | |
| Hb (mmol/L) | 6.9 ± 0.4 | 6.7 ± 0.8 | 0.288 | - | |
| CRP (mg/dL) | 18.3 ± 11.8 | 29.9 ± 17.4 | 0.024 | 0.9 ± 0.6 | 0.001 |
| Albumin (g/L) | 40.6 ± 3.3 | 38.9 ± 3.5 | 0.108 | - | |
| Bicarbonate (mmol/L) | 22.6 ± 1.5 | 24.2 ± 2.0 | 0.004 | - | |
| Sodium (mmol/L) | 138.1 ± 3.3 | 138.8 ± 3.5 | 0.527 | - | |
| Potassium (mmol/L) | 5.7 ± 0.7 | 5.5 ± 0.8 | 0.377 | - | |
| Calcium (mmol/L) | 2.2 ± 0.2 | 2.3 ± 0.2 | 0.615 | - | |
| Phosphate (mmol/L) | 2.0 ± 0.4 | 1.7 ± 0.4 | 0.019 | - |
| N (n = 25) | H (n = 17) | CO (n = 9) | p-Value N vs. H vs. CO | |
|---|---|---|---|---|
| CD45+ Leucocytes (106/mL) | 9.7 ± 2.8 | 9.5 ± 3.2 | 9.3 ± 2.4 | 0.089 |
| CD15+ Granulocytes (106/mL) | 5.5 ± 2.2 | 5.8 ± 2.2 | 4.2 ± 1.1 | 0.191 |
| CD3+ Lymphocytes (106/mL) | 1.3 ± 0.36 | 1.2 ± 0.5 | 1.6 ± 0.4 | 0.086 |
| CD14+ Monocytes (106/mL) | 0.7 ± 0.3 | 0.7 ± 0.2 | 0.6 ± 0.1 | 0.396 |
| CD4+ Lymphocytes (106/mL) | 0.8 ± 0.4 | 0.6 ± 0.2 | 0.9 ± 0.4 | 0.047 |
| CD8+ Lymphocytes (106/mL) | 0.4 ± 0.3 | 0.3 ± 0.2 | 0.5 ± 0.3 | 0.079 |
| CD19+ B-cells (106/mL) | 0.3 ± 0.2 | 0.2 ± 0.1 | 0.3 ± 0.3 | 0.553 |
| Downregulation in N | |||
|---|---|---|---|
| Symbol | FKPM (N) | FKPM (H) | p-Value |
| ASPRV1 | 2.94 × 106 | 1.01 × 107 | 1.00 × 10−2 |
| CTSE | 4.51 × 100 | 1.86 × 106 | 4.02 × 10−2 |
| EXOC4 | 4.78 × 107 | 1.36 × 107 | 1.37 × 10−2 |
| GPLD1 | 2.55 × 100 | 1.20 × 106 | 1.19 × 10−2 |
| HBG2 | 6.90 × 108 | 2.01 × 109 | 3.31 × 10−2 |
| HLA-DRB5 | 4.35 × 107 | 1.01 × 108 | 1.96 × 10−3 |
| IGKV2D-28 | 5.48 × 106 | 1.08 × 107 | 3.77 × 10−2 |
| MYBL2 | 4.64 × 100 | 1.54 × 106 | 1.62 × 10−2 |
| MYL6B | 1.32 × 105 | 2.43 × 106 | 5.59 × 10−3 |
| OTUD1 | 2.22 × 100 | 2.02 × 106 | 3.98 × 10−3 |
| Upregulation in N | |||
| ABCA3 | 4.15 × 108 | 1.02 × 100 | 1.70 × 10−4 |
| ANO5 | 2.03 × 100 | 1.27 × 10−1 | 1.25 × 10−4 |
| AXIN1 | 2.92 × 109 | 1.14 × 107 | 1.04 × 10−4 |
| CAPN15 | 2.00 × 109 | 8.41 × 106 | 1.15 × 10−4 |
| CCDC154 | 3.68 × 108 | 6.77 × 10−1 | 1.38 × 10−4 |
| CCNF | 9.31 × 107 | 1.00 × 100 | 2.05 × 10−4 |
| CIAO3 | 2.44 × 109 | 2.07 × 106 | 1.02 × 10−4 |
| FAM173A | 1.67 × 109 | 8.09 × 106 | 2.06 × 10−4 |
| MCRIP2 | 1.54 × 109 | 4.36 × 106 | 1.14 × 10−4 |
| SOX8 | 1.10 × 109 | 3.02 × 10−1 | 1.01 × 10−4 |
| TNFAIP8L2 | 1.05 × 108 | 3.07 × 107 | 4.92 × 10−2 |
| Symbol | FKPM (N) | FKPM (H) | p-Value |
|---|---|---|---|
| TNFAIP3 (A20) | 4.538 × 107 | 9.224 × 106 | 0.278 |
| TNFAIP5 (Pentraxin3) | 3.871 × 100 | 1.410 × 100 | 0.443 |
| TNFAIP8L2 (TIPE2) | 1.05 × 108 | 3.07 × 107 | 0.049 |
| IL-10 | n.d. | n.d. | |
| IL-22 | n.d. | n.d. | |
| IL-26 | n.d. | n.d. |
| Symbol | (N) | (H) | p-Value |
|---|---|---|---|
| TNFAIP3 (A20) | 1.7 ± 0.9 | 1.4 ± 0.6 | 0.397 |
| TNFAIP5 (Pentraxin3) | 1.5 ± 1.0 | 1.9 ± 1.2 | 0.451 |
| TNFAIP8L2 | 1.5 ± 0.7 | 1.0 ± 0.2 | 0.006 |
| IL-10 | 1.5 ± 0.9 | 2.0 ± 1.2 | 0.198 |
| IL-22 | 2.4 ± 1.4 | 2.9 ± 1.1 | 0.194 |
| IL-26 | 0.8 ± 0.6 | 1.3 ± 1.2 | 0.350 |
| OTUD1 | 1.5 ± 0.6 | 1.3 ± 0.2 | 0.295 |
| Dependent Variable | Parameter | Regression Coefficient B | Standard Error | T | Significance | 95% Confidence Interval | Partial Eta-Quadrat | |
|---|---|---|---|---|---|---|---|---|
| Lower Limit | Upper Limit | |||||||
| OTUD1 mRNA (F5—monosomal) | Constant | 7.367 | 2.469 | 2.983 | 0.005 | 2.337 | 12.397 | 0.218 |
| Kt/V | 0.850 | 1.297 | 0.655 | 0.517 | −1.791 | 3.491 | 0.013 | |
| CRP | −0.081 | 0.030 | −2.657 | 0.012 | −0.142 | −0.019 | 0.181 | |
| Age | −0.004 | 0.030 | −0.145 | 0.886 | −0.066 | 0.057 | 0.001 | |
| Phase angle (°) | Constant | 6.451 | 0.891 | 7.245 | 0.000 | 4.638 | 8.265 | 0.621 |
| Kt/V | 0.509 | 0.468 | 1.089 | 0.284 | −0.443 | 1.462 | 0.036 | |
| CRP | −0.023 | 0.011 | −2.127 | 0.041 | −0.046 | −0.001 | 0.124 | |
| Age | −0.029 | 0.011 | −2.630 | 0.013 | −0.051 | −0.006 | 0.178 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ulrich, C.; Dawood, A.; Fiedler, R.; Markau, S.; Girndt, M. Different Translational Activities of Inflammatory Regulators Associated with Hypervolemia in Haemodialysis Patients. Int. J. Mol. Sci. 2025, 26, 8922. https://doi.org/10.3390/ijms26188922
Ulrich C, Dawood A, Fiedler R, Markau S, Girndt M. Different Translational Activities of Inflammatory Regulators Associated with Hypervolemia in Haemodialysis Patients. International Journal of Molecular Sciences. 2025; 26(18):8922. https://doi.org/10.3390/ijms26188922
Chicago/Turabian StyleUlrich, Christof, Amanda Dawood, Roman Fiedler, Silke Markau, and Matthias Girndt. 2025. "Different Translational Activities of Inflammatory Regulators Associated with Hypervolemia in Haemodialysis Patients" International Journal of Molecular Sciences 26, no. 18: 8922. https://doi.org/10.3390/ijms26188922
APA StyleUlrich, C., Dawood, A., Fiedler, R., Markau, S., & Girndt, M. (2025). Different Translational Activities of Inflammatory Regulators Associated with Hypervolemia in Haemodialysis Patients. International Journal of Molecular Sciences, 26(18), 8922. https://doi.org/10.3390/ijms26188922






