Effective Reduction in Nuclear DNA Contamination Allows Sensitive Mitochondrial DNA Methylation Determination by LC-MS/MS
Abstract
1. Introduction
2. Results
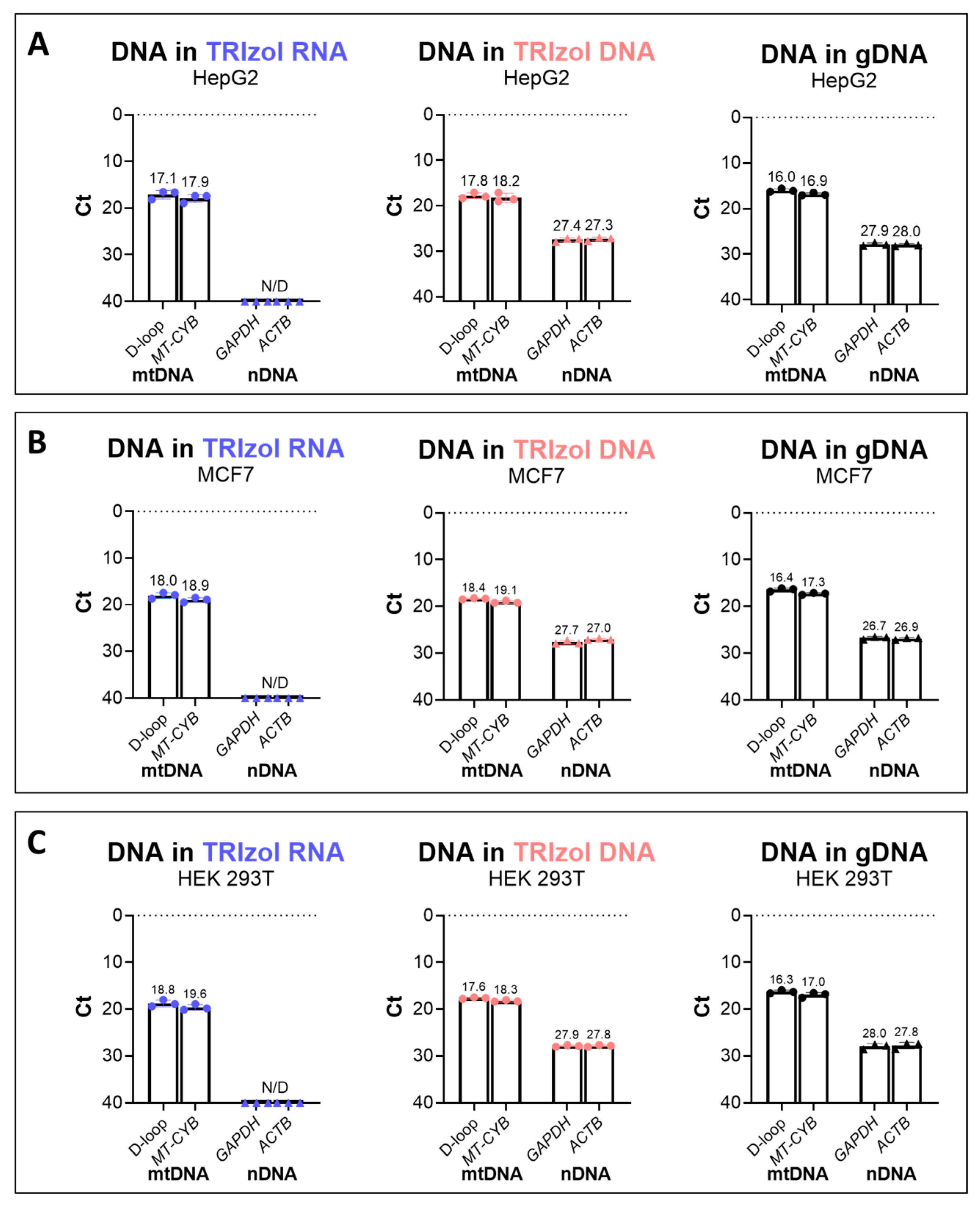
2.1. Isolation of mtDNA from the TRIzol RNA Phase to Mitigate nDNA Contamination
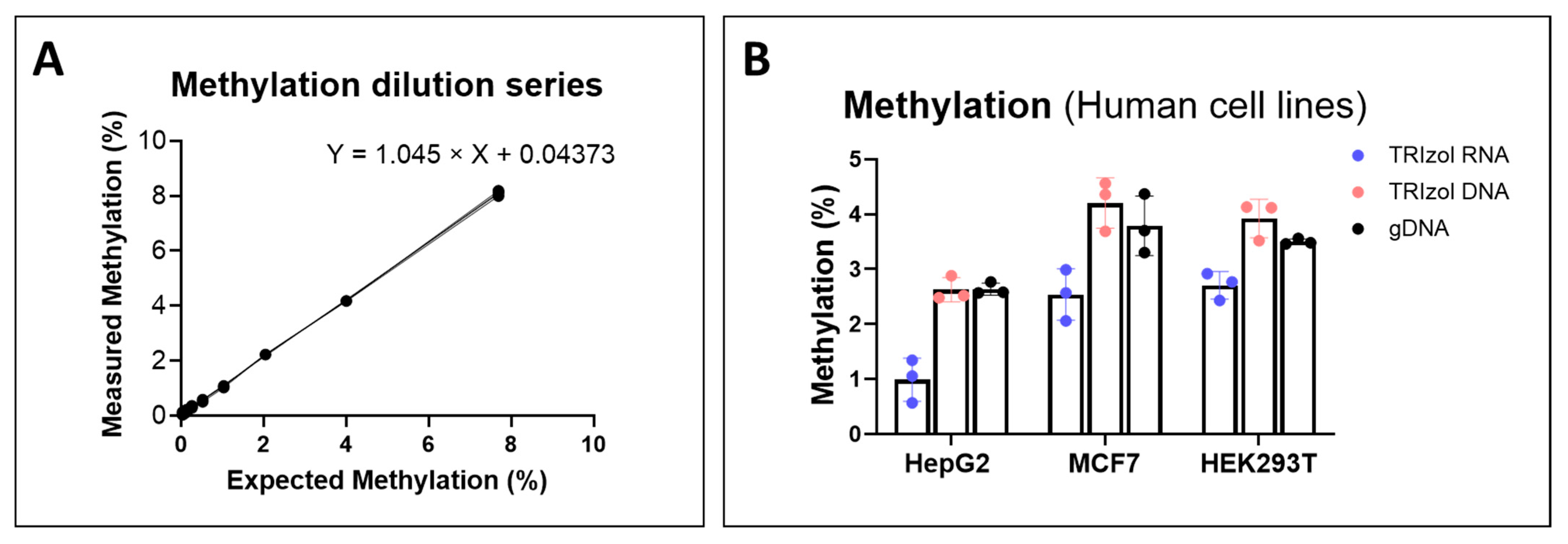
2.2. Methylated mtDNA Is Not Preferentially Enriched over Unmethylated mtDNA in the TRIzol RNA Phase (Or Vice Versa)
2.3. Assessment of mtDNA Methylation by LC-MS/MS
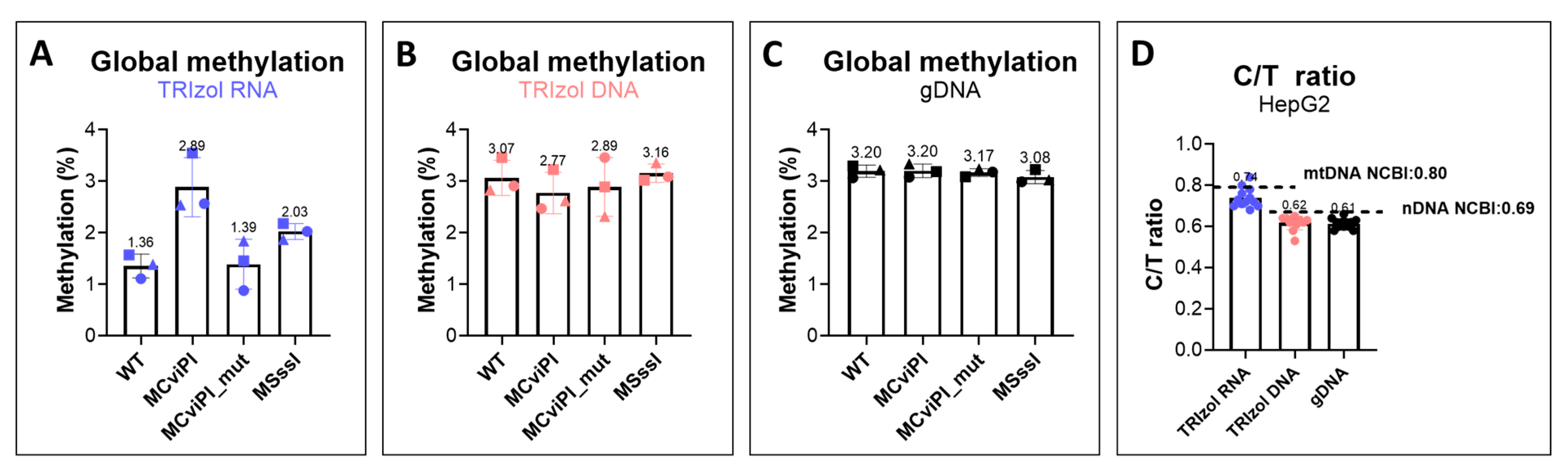
2.4. LC-MS/MS Analysis Confirmed the Increased mtDNA Methylation in HepG2 Transgenic Cell Lines
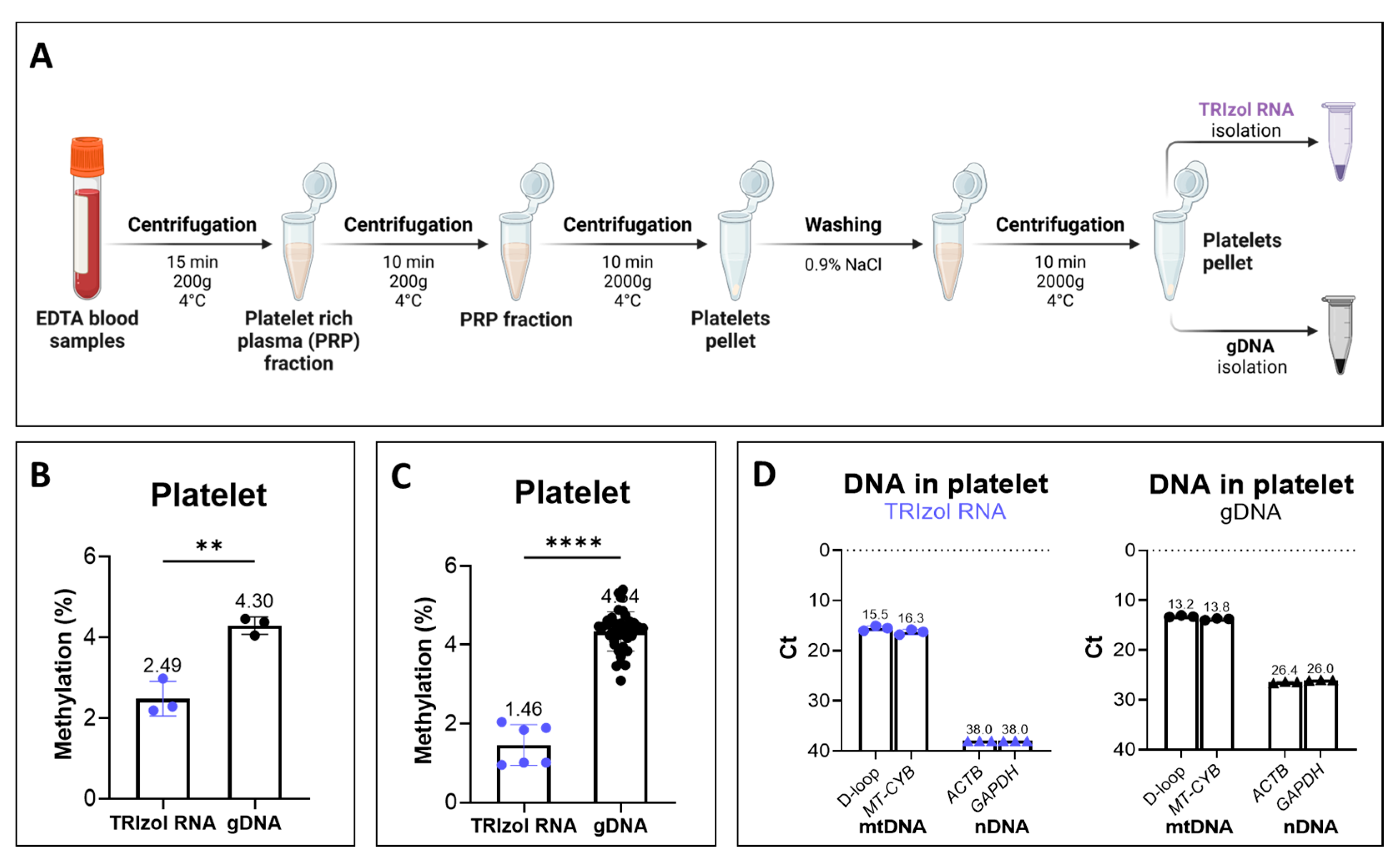
2.5. LC-MS/MS Demonstrated Lower Methylation Level in DNA Isolated from the TRIzol RNA Phase in Clinically Relevant Samples Compared to TRIzol DNA and gDNA Isolations
2.6. LC-MS/MS Confirmed the Low Methylation of mtDNA Isolated by TRIzol RNA Isolation for Mouse Brain Tissue
3. Discussion
4. Material and Methods
4.1. Cell Culture and Mouse Brain Tissues
4.2. Isolation of mtDNA and Total Genomic DNA
4.3. Quantitative Real-Time PCR
4.4. Pyrosequencing
4.5. Isolation of Platelets from Blood Samples
4.6. Crude Mitochondrial Fraction Isolation
4.7. LC-MS/MS
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ng, Y.S.; Bindoff, L.A.; Gorman, G.S.; Klopstock, T.; Kornblum, C.; Mancuso, M.; McFarland, R.; Sue, C.M.; Suomalainen, A.; Taylor, R.W.; et al. Mitochondrial disease in adults: Recent advances and future promise. Lancet Neurol. 2021, 20, 573–584. [Google Scholar] [CrossRef] [PubMed]
- Tabarki, B.; Hakami, W.; Alkhuraish, N.; Tlili-Graies, K.; Alfadhel, M. Spinal Cord Involvement in Pediatric-Onset Metabolic Disorders With Mendelian and Mitochondrial Inheritance. Front. Pediatr. 2021, 8, 599861. [Google Scholar] [CrossRef]
- Helgudóttir, S.S.; Mørkholt, A.S.; Lichota, J.; Bruun-Nyzell, P.; Andersen, M.C.; Kristensen, N.M.J.; Johansen, A.K.; Zinn, M.R.; Jensdóttir, H.M.; Nieland, J.D.V. Rethinking neurodegenerative diseases: Neurometabolic concept linking lipid oxidation to diseases in the central nervous system. Neural Regen. Res. 2023, 19, 1437–1445. [Google Scholar] [CrossRef] [PubMed]
- Russell, O.M.; Gorman, G.S.; Lightowlers, R.N.; Turnbull, D.M. Mitochondrial Diseases: Hope for the Future. Cell 2020, 181, 168–188. [Google Scholar] [CrossRef]
- Pham, L.; Arroum, T.; Wan, J.; Pavelich, L.; Bell, J.; Morse, P.T.; Lee, I.; Grossman, L.I.; Sanderson, T.H.; Malek, M.H.; et al. Regulation of mitochondrial oxidative phosphorylation through tight control of cytochrome c oxidase in health and disease—Implications for ischemia/reperfusion injury, inflammatory diseases, diabetes, and cancer. Redox Biol. 2024, 78, 103426. [Google Scholar] [CrossRef]
- Zong, Y.; Li, H.; Liao, P.; Chen, L.; Pan, Y.; Zheng, Y.; Zhang, C.; Liu, D.; Zheng, M.; Gao, J. Mitochondrial dysfunction: Mechanisms and advances in therapy. Signal Transduct. Target. Ther. 2024, 9, 124. [Google Scholar] [CrossRef]
- van der Wijst, M.G.; Rots, M.G. Mitochondrial epigenetics: An overlooked layer of regulation? Trends Genet. 2015, 31, 353–356. [Google Scholar] [CrossRef]
- Stoccoro, A.; Coppedè, F. Mitochondrial DNA Methylation and Human Diseases. Int. J. Mol. Sci. 2021, 22, 4594. [Google Scholar] [CrossRef]
- Coppedè, F. Mitochondrial DNA methylation and mitochondria-related epigenetics in neurodegeneration. Neural Regen. Res. 2023, 19, 405–406. [Google Scholar] [CrossRef] [PubMed]
- Mposhi, A.; Cortés-Mancera, F.; Heegsma, J.; de Meijer, V.E.; van de Sluis, B.; Sydor, S.; Bechmann, L.P.; Theys, C.; de Rijk, P.; De Pooter, T.; et al. Mitochondrial DNA methylation in metabolic associated fatty liver disease. Front. Nutr. 2023, 10, 964337. [Google Scholar] [CrossRef]
- Theys, C.; Ibrahim, J.; Mateiu, L.; Mposhi, A.; García-Pupo, L.; De Pooter, T.; De Rijk, P.; Strazisar, M.; İnce, I.A.; Vintea, I.; et al. Mitochondrial GpC and CpG DNA Hypermethylation Cause Metabolic Stress-Induced Mitophagy and Cholestophagy. Int. J. Mol. Sci. 2023, 24, 16412. [Google Scholar] [CrossRef]
- Park, S.H.; Lee, S.Y.; Kim, S.A. Mitochondrial DNA Methylation Is Higher in Acute Coronary Syndrome Than in Stable Coronary Artery Disease. In Vivo 2021, 35, 181–189. [Google Scholar] [CrossRef] [PubMed]
- Mongelli, A.; Mengozzi, A.; Geiger, M.; Gorica, E.; Mohammed, S.A.; Paneni, F.; Ruschitzka, F.; Costantino, S. Mitochondrial epigenetics in aging and cardiovascular diseases. Front. Cardiovasc. Med. 2023, 10, 1204483. [Google Scholar] [CrossRef]
- Owa, C.; Poulin, M.; Yan, L.; Shioda, T.; Albertini, E. Technical adequacy of bisulfite sequencing and pyrosequencing for detection of mitochondrial DNA methylation: Sources and avoidance of false-positive detection. PLoS ONE 2018, 13, e0192722. [Google Scholar] [CrossRef]
- Guitton, R.; Nido, G.S.; Tzoulis, C. No evidence of extensive non-CpG methylation in mtDNA. Nucleic Acids Res. 2022, 50, 9190–9194. [Google Scholar] [CrossRef]
- Shao, Z.; Han, Y.; Zhou, D. Optimized bisulfite sequencing analysis reveals the lack of 5-methylcytosine in mammalian mitochondrial DNA. BMC Genom. 2023, 24, 439. [Google Scholar] [CrossRef]
- Matsuda, S.; Yasukawa, T.; Sakaguchi, Y.; Ichiyanagi, K.; Unoki, M.; Gotoh, K.; Fukuda, K.; Sasaki, H.; Suzuki, T.; Kang, D. Accurate estimation of 5-methylcytosine in mammalian mitochondrial DNA. Sci. Rep. 2018, 8, 5801. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Du, Q.; Chen, L.; Fu, G.; Li, S.; Fu, L.; Zhang, X.; Ma, C.; Bin, C. CpG methylation patterns of human mitochondrial DNA. Sci. Rep. 2016, 6, 23421. [Google Scholar] [CrossRef] [PubMed]
- Mechta, M.; Ingerslev, L.R.; Fabre, O.; Picard, M.; Barrès, R. Evidence Suggesting Absence of Mitochondrial DNA Methylation. Front. Genet. 2017, 8, 166. [Google Scholar] [CrossRef]
- Lüth, T.; Wasner, K.; Klein, C.; Schaake, S.; Tse, R.; Pereira, S.L.; Laß, J.; Sinkkonen, L.; Grünewald, A.; Trinh, J. Nanopore Single-Molecule Sequencing for Mitochondrial DNA Methylation Analysis: Investigating Parkin-Associated Parkinsonism as a Proof of Concept. Front. Aging Neurosci. 2021, 13, 713084. [Google Scholar] [CrossRef]
- Goldsmith, C.; Rodríguez-Aguilera, J.R.; El-Rifai, I.; Jarretier-Yuste, A.; Hervieu, V.; Raineteau, O.; Saintigny, P.; de Sánchez, V.C.; Dante, R.; Ichim, G.; et al. Low biological fluctuation of mitochondrial CpG and non-CpG methylation at the single-molecule level. Sci. Rep. 2021, 11, 8032. [Google Scholar] [CrossRef] [PubMed]
- Bicci, I.; Calabrese, C.; Golder, Z.J.; Gomez-Duran, A.; Chinnery, P.F. Single-molecule mitochondrial DNA sequencing shows no evidence of CpG methylation in human cells and tissues. Nucleic Acids Res. 2021, 49, 12757–12768. [Google Scholar] [CrossRef]
- Dou, X.; Boyd-Kirkup, J.D.; McDermott, J.; Zhang, X.; Li, F.; Rong, B.; Zhang, R.; Miao, B.; Chen, P.; Cheng, H.; et al. The strand-biased mitochondrial DNA methylome and its regulation by DNMT3A. Genome Res. 2019, 29, 1622–1634. [Google Scholar] [CrossRef] [PubMed]
- Infantino, V.; Castegna, A.; Iacobazzi, F.; Spera, I.; Scala, I.; Andria, G.; Iacobazzi, V. Impairment of methyl cycle affects mitochondrial methyl availability and glutathione level in Down’s syndrome. Mol. Genet. Metab. 2011, 102, 378–382. [Google Scholar] [CrossRef]
- Liu, H.; Zhang, H.; Zhang, Y.; Xu, S.; Zhao, H.; He, H.; Liu, X. Modeling mtDNA hypermethylation vicious circle mediating Aβ-induced endothelial damage memory in HCMEC/D3 cell. Aging 2020, 12, 18343–18362. [Google Scholar] [CrossRef]
- Castellani, C.A.; Longchamps, R.J.; Sun, J.; Guallar, E.; Arking, D.E. Thinking outside the nucleus: Mitochondrial DNA copy number in health and disease. Mitochondrion 2020, 53, 214–223. [Google Scholar] [CrossRef]
- Lister, R.; Pelizzola, M.; Dowen, R.H.; Hawkins, R.D.; Hon, G.; Tonti-Filippini, J.; Nery, J.R.; Lee, L.; Ye, Z.; Ngo, Q.-M.; et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 2009, 462, 315–322. [Google Scholar] [CrossRef]
- Globisch, D.; Münzel, M.; Müller, M.; Michalakis, S.; Wagner, M.; Koch, S.; Brückl, T.; Biel, M.; Carell, T.; Croft, A.K. Tissue Distribution of 5-Hydroxymethylcytosine and Search for Active Demethylation Intermediates. PLoS ONE 2010, 5, e15367. [Google Scholar] [CrossRef] [PubMed]
- Breiling, A.; Lyko, F. Epigenetic regulatory functions of DNA modifications: 5-methylcytosine and beyond. Epigenetics Chromatin 2015, 8, 24. [Google Scholar] [CrossRef]
- Devall, M.; Burrage, J.; Caswell, R.; Johnson, M.; Troakes, C.; Al-Sarraj, S.; Jeffries, A.R.; Mill, J.; Lunnon, K. A Comparison of Mitochondrial DNA Isolation Methods in Frozen Post-Mortem Human Brain Tissue—Applications for Studies of Mitochondrial Genetics in Brain Disorders. BioTechniques 2015, 59, 241–246. [Google Scholar] [CrossRef]
- Zheng, J.; Liu, Y.; Liang, Z.; Wang, Y.A.; Yu, W.; Ding, B.; Wang, H. A Rigorous Nuclear DNA-Excluding Mass Spectrometry Assay for Accurate Identification of 5-Methylcytosine in Mitochondrial DNA. Anal. Chem. 2025, 97, 5914–5918. [Google Scholar] [CrossRef]
- Nakhle, J.; Özkan, T.; Lněničková, K.; Briolotti, P.; Vignais, M.-L. Methods for Simultaneous and Quantitative Isolation of Mitochondrial DNA, Nuclear DNA and RNA from Mammalian Cells. BioTechniques 2020, 69, 436–442. [Google Scholar] [CrossRef]
- Mposhi, A.; Liang, L.; Mennega, K.P.; Yildiz, D.; Kampert, C.; Hof, I.H.; Jellema, P.G.; de Koning, T.J.; Faber, K.N.; Ruiters, M.H.J.; et al. The Mitochondrial Epigenome: An Unexplored Avenue to Explain Unexplained Myopathies? Int. J. Mol. Sci. 2022, 23, 2197. [Google Scholar] [CrossRef]
- Andrews, R.M.; Kubacka, I.; Chinnery, P.F.; Lightowlers, R.N.; Turnbull, D.M.; Howell, N. Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat. Genet. 1999, 23, 147. [Google Scholar] [CrossRef]
- Piovesan, A.; Pelleri, M.C.; Antonaros, F.; Strippoli, P.; Caracausi, M.; Vitale, L. On the length, weight and GC content of the human genome. BMC Res. Notes 2019, 12, 106. [Google Scholar] [CrossRef]
- Freedman, J.E.; Larson, M.G.; Tanriverdi, K.; O’Donnell, C.J.; Morin, K.; Hakanson, A.S.; Vasan, R.S.; Johnson, A.D.; Iafrati, M.D.; Benjamin, E.J. Relation of Platelet and Leukocyte Inflammatory Transcripts to Body Mass Index in the Framingham Heart Study. Circulation 2010, 122, 119–129. [Google Scholar] [CrossRef]
- Chebbo, M.; Assou, S.; Pantesco, V.; Duez, C.; Alessi, M.C.; Chanez, P.; Gras, D. Platelets Purification Is a Crucial Step for Transcriptomic Analysis. Int. J. Mol. Sci. 2022, 23, 3100. [Google Scholar] [CrossRef] [PubMed]
- Devall, M.; Soanes, D.M.; Smith, A.R.; Dempster, E.L.; Smith, R.G.; Burrage, J.; Iatrou, A.; Hannon, E.; Troakes, C.; Moore, K.; et al. Genome-wide characterization of mitochondrial DNA methylation in human brain. Front. Endocrinol. 2023, 13, 1059120. [Google Scholar] [CrossRef]
- Tatarkina, M.A.; Lobanova, V.V.; Kozenkov, I.I.I.I.; Efimenko, B.E.; Dzhigkaev, A.K. Isolation of highly purified genomic material from mitochondria of muscle tissue cells. bioRxiv 2022, 1–12. [Google Scholar] [CrossRef]
- Hao, Z.; Wu, T.; Cui, X.; Zhu, P.; Tan, C.; Dou, X.; Hsu, K.-W.; Lin, Y.-T.; Peng, P.-H.; Zhang, L.-S.; et al. N6-Deoxyadenosine Methylation in Mammalian Mitochondrial DNA. Mol. Cell 2020, 78, 382–395.e8. [Google Scholar] [CrossRef] [PubMed]
- Coppedè, F.; Stoccoro, A. Mitoepigenetics and neurodegenerative diseases. Front. Endocrinol. 2019, 10, 86. [Google Scholar] [CrossRef]
- Koh, C.W.Q.; Goh, Y.T.; Toh, J.D.W.; Neo, S.P.; Ng, S.B.; Gunaratne, J.; Gao, Y.-G.; Quake, S.R.; Burkholder, W.F.; Goh, W.S.S. Single-nucleotide-resolution sequencing of human N6-methyldeoxyadenosine reveals strand-asymmetric clusters associated with SSBP1 on the mitochondrial genome. Nucleic Acids Res. 2018, 46, 11659–11670. [Google Scholar] [CrossRef]
- Zheng, L.; Chen, X.; He, X.; Wei, H.; Li, X.; Tan, Y.; Min, J.; Chen, M.; Zhang, Y.; Dong, M.; et al. METTL4-Mediated Mitochondrial DNA N6-Methyldeoxyadenosine Promoting Macrophage Inflammation and Atherosclerosis. Circulation 2025, 151, 946–965. [Google Scholar] [CrossRef] [PubMed]
- Sturm, Á.; Sharma, H.; Bodnár, F.; Aslam, M.; Kovács, T.; Németh, Á.; Hotzi, B.; Billes, V.; Sigmond, T.; Tátrai, K.; et al. N6-Methyladenine Progressively Accumulates in Mitochondrial DNA during Aging. Int. J. Mol. Sci. 2023, 24, 14858. [Google Scholar] [CrossRef]
- Hahn, A.; Hung, G.C.C.; Ahier, A.; Dai, C.-Y.; Kirmes, I.; Forde, B.M.; Campbell, D.; Lee, R.S.Y.; Sucic, J.; Onraet, T.; et al. Misregulation of mitochondrial 6mA promotes the propagation of mutant mtDNA and causes aging in C. elegans. Cell Metab. 2024, 36, 2528–2541.e11. [Google Scholar] [CrossRef] [PubMed]
- van Doormaal, J.J.; Idema, I.G.; Muskiet, F.A.J.; Martini, I.A.; Doorenbos, H. Effects of short-term high dose intake of evening primrose oil on plasma and cellular fatty acid compositions, α-tocopherol levels, and erythropoiesis in normal and Type 1 (insulin-dependent) diabetic men. Diabetologia 1988, 31, 576–584. [Google Scholar] [CrossRef] [PubMed]
- Wieckowski, M.R.; Giorgi, C.; Lebiedzinska, M.; Duszynski, J.; Pinton, P. Isolation of mitochondria-associated membranes and mitochondria from animal tissues and cells. Nat. Protoc. 2009, 4, 1582–1590. [Google Scholar] [CrossRef]
- Zhou, G.; Pang, H.; Tang, Y.; Yao, X.; Ding, Y.; Zhu, S.; Guo, S.; Qian, D.; Shen, J.; Qian, Y.; et al. Hydrophilic interaction ultra-performance liquid chromatography coupled with triple-quadrupole tandem mass spectrometry (HILIC-UPLC–TQ-MS/MS) in multiple-reaction monitoring (MRM) for the determination of nucleobases and nucleosides in ginkgo seeds. Food Chem. 2014, 150, 260–266. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liang, L.; González Molina, L.A.; Jellema, P.G.; van Faassen, M.; Otten, L.T.A.; Mennega, K.P.; Hof, I.H.; Dijck-Brouwer, D.A.J.; Dolga, A.M.; Rots, M.G.; et al. Effective Reduction in Nuclear DNA Contamination Allows Sensitive Mitochondrial DNA Methylation Determination by LC-MS/MS. Int. J. Mol. Sci. 2025, 26, 8864. https://doi.org/10.3390/ijms26188864
Liang L, González Molina LA, Jellema PG, van Faassen M, Otten LTA, Mennega KP, Hof IH, Dijck-Brouwer DAJ, Dolga AM, Rots MG, et al. Effective Reduction in Nuclear DNA Contamination Allows Sensitive Mitochondrial DNA Methylation Determination by LC-MS/MS. International Journal of Molecular Sciences. 2025; 26(18):8864. https://doi.org/10.3390/ijms26188864
Chicago/Turabian StyleLiang, Lin, Luis Alfonso González Molina, Pytrick G. Jellema, Martijn van Faassen, Laura T. A. Otten, Kevin P. Mennega, Ingrid H. Hof, D. A. Janneke Dijck-Brouwer, Amalia M. Dolga, Marianne G. Rots, and et al. 2025. "Effective Reduction in Nuclear DNA Contamination Allows Sensitive Mitochondrial DNA Methylation Determination by LC-MS/MS" International Journal of Molecular Sciences 26, no. 18: 8864. https://doi.org/10.3390/ijms26188864
APA StyleLiang, L., González Molina, L. A., Jellema, P. G., van Faassen, M., Otten, L. T. A., Mennega, K. P., Hof, I. H., Dijck-Brouwer, D. A. J., Dolga, A. M., Rots, M. G., & Niezen-Koning, K. E. (2025). Effective Reduction in Nuclear DNA Contamination Allows Sensitive Mitochondrial DNA Methylation Determination by LC-MS/MS. International Journal of Molecular Sciences, 26(18), 8864. https://doi.org/10.3390/ijms26188864






