Quercetin and Its Structural Analogs as NUDT5 Inhibitors: A Preliminary In Silico Study
Abstract
1. Introduction
2. Results
2.1. Molecular Docking
2.2. Hydrogen Bond Analysis
2.3. MM/GBSA Binding Free Energy Calculation
2.4. Lipinski’s Rule of Five and ADMET Profile
2.5. Toxicity Profile
2.6. Binding Mode Interactions
2.7. Correlation Between Binding Affinity Scores and Physicochemical Parameters
3. Discussion
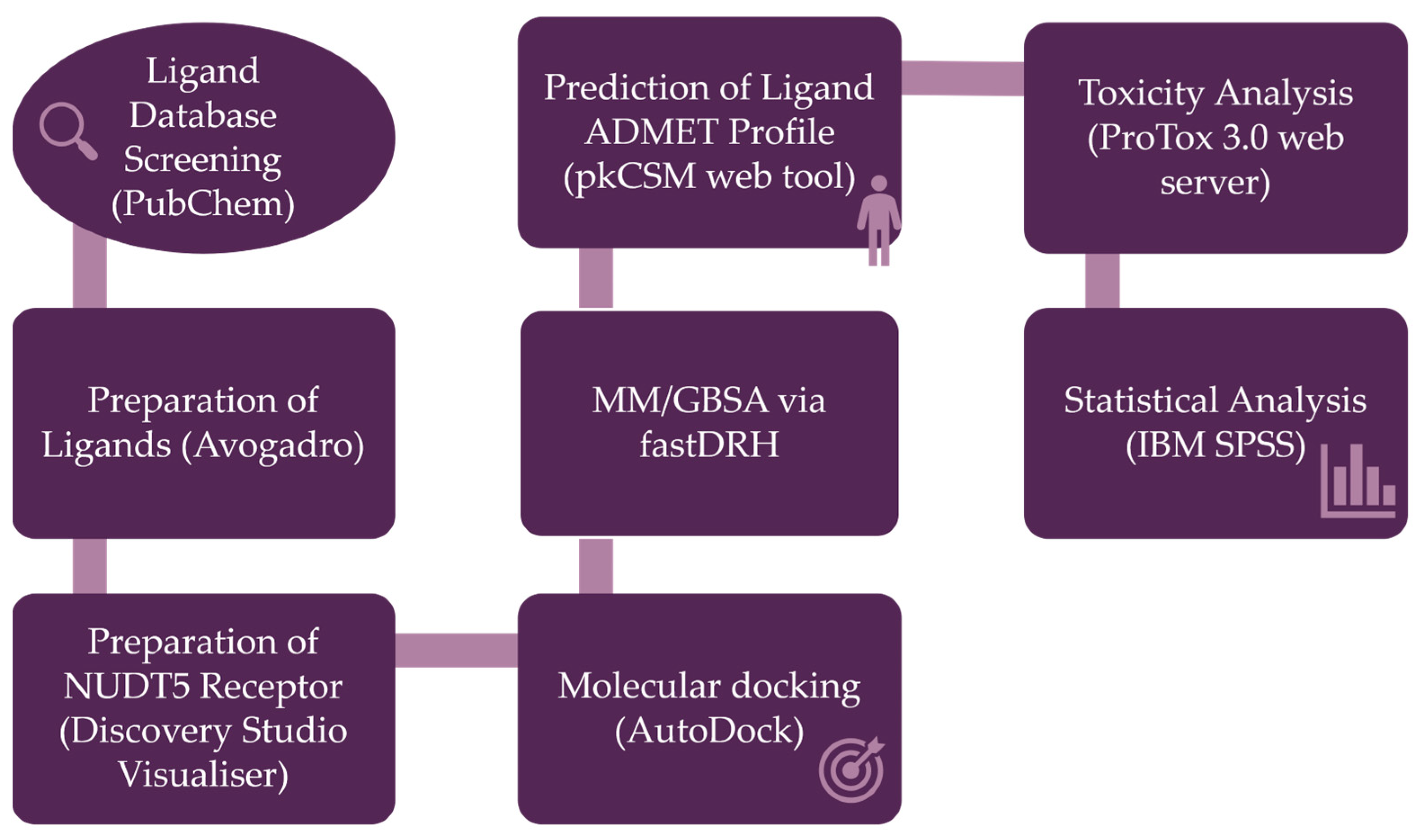
4. Materials and Methods
4.1. Preparation of Ligands for Molecular Docking
4.2. Preparation of the NUDT5 Receptor for Docking
4.3. Molecular Docking Procedure
4.4. Binding Free Energy Calculations
4.5. Determination of the Key Parameters of Lipinski’s Rule of Five
4.6. Prediction of ADMET Profile
4.7. Toxicity Analysis
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Klar, N.; Rosenzweig, M.; Diergaarde, B.; Brufsky, A. Features associated with long-term survival in patients with metastatic breast cancer. Clin. Breast Cancer 2019, 19, 304–310. [Google Scholar] [CrossRef]
- Cavallaro, P.A.; De Santo, M.; Belsito, E.L.; Longobucco, C.; Curcio, M.; Morelli, C.; Pasqua, L.; Leggio, A. Peptides Targeting HER2-Positive Breast Cancer Cells and Applications in Tumor Imaging and Delivery of Chemotherapeutics. Nanomaterials 2023, 13, 2476. [Google Scholar] [CrossRef]
- Zaimy, M.A.; Saffarzadeh, N.; Mohammadi, A.; Pourghadamyari, H.; Izadi, P.; Sarli, A.; Moghaddam, L.K.; Paschepari, S.R.; Azizi, H.; Torkamandi, S.; et al. New methods in the diagnosis of cancer and gene therapy of cancer based on nanoparticles. Cancer Gene Ther. 2017, 24, 233–243. [Google Scholar] [CrossRef] [PubMed]
- Dwivedi, P.S.R.; Shastry, C.S. Anti-tumor potential and mode of action of karanjin against breast cancer; an in-silico approach. Arab. J. Chem. 2023, 16, 104778. [Google Scholar] [CrossRef]
- Islam, R.; Yan, M.P.; Yen, K.P.; Rasol, N.E.; Meng, C.K.; Wai, L.K. Synthesis and biological evaluation of chromone derivatives against triple-negative breast cancer cells. Med. Chem. Res. 2023, 32, 884–898. [Google Scholar] [CrossRef]
- Shaker, B.; Ahmad, S.; Lee, J.; Jung, C.; Na, D. In silico methods and tools for drug discovery. Comput. Biol. Med. 2021, 137, 104851. [Google Scholar] [CrossRef]
- Iwaloye, O.; Ottu, V.; Olawale, F.; Babalola, O.O.; Elekofehinti, O.O.; Kikiowo, B.; Adegboyega, A.E.; Ogbonna, H.N.; Adeboboye, C.F.; Folorunso, I.M.; et al. Computer-aided drug design in anti-cancer drug discovery: What have we learnt and what is the way forward? Inform. Med. Unlocked 2023, 41, 101332. [Google Scholar] [CrossRef]
- Cui, W.; Aouidate, A.; Wang, S.; Yu, Q.; Li, Y.; Yuan, S. Discovering Anti-Cancer Drugs via Computational Methods. Front. Pharmacol. 2020, 11, 733. [Google Scholar] [CrossRef]
- Niranjan, V.; Jayaprasad, S.; Uttarkar, A.; Kusanur, R.; Kumar, J. Design of Novel Coumarin Derivatives as NUDT5 Antagonists That Act by Restricting ATP Synthesis in Breast Cancer Cells. Molecules 2023, 28, 89. [Google Scholar] [CrossRef]
- Pickup, K.E.; Pardow, F.; Carbonell-Caballero, J.; Lioutas, A.; Villanueva-Cañas, J.L.; Wright, R.H.G.; Beato, M. Expression of Oncogenic Drivers in 3D Cell Culture Depends on Nuclear ATP Synthesis by NUDT5. Cancers 2019, 11, 1337. [Google Scholar] [CrossRef]
- Qian, J.; Ma, Y.; Tahaney, W.M.; Moyer, C.L.; Lanier, A.; Hill, J.; Coleman, D.; Koupaei, N.; Hilsenbeck, S.G.; Savage, M.I.; et al. The Novel Phosphatase NUDT5 Is a Critical Regulator of Triple-Negative Breast Cancer Growth. Breast Cancer Res. 2024, 26, 23. [Google Scholar] [CrossRef] [PubMed]
- Monkkonen, T.; Traustadóttir, G.Á.; Koledova, Z. Unraveling the Breast: Advances in Mammary Biology and Cancer Methods. J. Mammary Gland Biol. Neoplasia. 2021, 25, 233–236. [Google Scholar] [CrossRef] [PubMed]
- Odunitan, T.T.; Saibu, O.A.; Apanisile, B.T.; Omoboyowa, D.A.; Balogun, T.A.; Awe, A.V.; Ajayi, M.T.; Olagunju, G.V.; Muhammad, F.M.; Akinboade, M.W.; et al. Integrating Biocomputational Techniques for Breast Cancer Drug Discovery via the HER-2, BCRA, VEGF and ER Protein Targets. Comput. Biol. Med. 2024, 168, 107737. [Google Scholar] [CrossRef]
- Tong, X.-Y.; Liao, X.; Gao, M.; Lv, B.-M.; Chen, X.-H.; Chu, X.-Y.; Zhang, Q.-Y.; Zhang, H.-Y. Identification of NUDT5 Inhibitors from Approved Drugs. Front. Mol. Biosci. 2020, 7, 44. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Yao, J.; Han, C.; Yang, J.; Chaudhry, M.T.; Wang, S.; Liu, H.; Yin, Y. Quercetin, Inflammation and Immunity. Nutrients 2016, 8, 167. [Google Scholar] [CrossRef]
- Di Petrillo, A.; Orrù, G.; Fais, A.; Fantini, M.C. Quercetin and Its Derivates as Antiviral Potentials: A Comprehensive Review. Phytother. Res. 2021, 36, 266–278. [Google Scholar] [CrossRef]
- Tang, S.-M.; Deng, X.-T.; Zhou, J.; Li, Q.-P.; Ge, X.-X.; Miao, L. Pharmacological basis and new insights of quercetin action in respect to its anti-cancer effects. Biomed. Pharmacother. 2020, 121, 109604. [Google Scholar] [CrossRef]
- Liu, L.; Barber, E.; Kellow, N.J.; Williamson, G. Improving Quercetin Bioavailability: A Systematic Review and Meta-Analysis of Human Intervention Studies. Food Chem. 2025, 477, 143630. [Google Scholar] [CrossRef]
- Kandemir, K.; Tomas, M.; McClements, D.J.; Capanoglu, E. Recent Advances on the Improvement of Quercetin Bioavailability. Trends Food Sci. Technol. 2022, 119, 192–200. [Google Scholar] [CrossRef]
- Frenț, O.-D.; Stefan, L.; Morgovan, C.M.; Duteanu, N.; Dejeu, I.L.; Marian, E.; Vicaș, L.; Manole, F. A Systematic Review: Quercetin—Secondary Metabolite of the Flavonol Class, with Multiple Health Benefits and Low Bioavailability. Int. J. Mol. Sci. 2024, 25, 12091. [Google Scholar] [CrossRef]
- Septembre-Malaterre, A.; Boumendjel, A.; Seteyen, A.-L.S.; Boina, C.; Gasque, P.; Guiraud, P.; Sélambarom, J. Focus on the high therapeutic potentials of quercetin and its derivatives. Phytomed. Plus. 2022, 2, 100220. [Google Scholar] [CrossRef]
- Alizadeh, S.R.; Ebrahimzadeh, M.A. Quercetin derivatives: Drug design, development, and biological activities, a review. Eur. J. Med. Chem. 2022, 229, 114068. [Google Scholar] [CrossRef]
- Magar, R.T.; Sohng, J.K. A Review on Structure, Modifications and Structure-Activity Relation of Quercetin and Its Derivatives. J. Microbiol. Biotechnol. 2020, 30, 11–20. [Google Scholar] [CrossRef]
- Page, B.D.G.; Valerie, N.C.K.; Wright, R.H.G.; Wallner, O.; Isaksson, R.; Carter, M.; Rudd, S.G.; Loseva, O.; Jemth, A.-S.; Almlöf, I.; et al. Targeted NUDT5 Inhibitors Block Hormone Signaling in Breast Cancer Cells. Nat. Commun. 2018, 9, 250. [Google Scholar] [CrossRef]
- Jeffrey, G.A. An Introduction to Hydrogen Bonding; Oxford University Press: New York, NY, USA, 1997. [Google Scholar]
- Domínguez-Villa, F.X.; Durán-Iturbide, N.A.; Ávila-Zárraga, J.G. Synthesis, molecular docking, and in silico ADME/Tox profiling studies of new 1-aryl-5-(3-azidopropyl) indol-4-ones: Potential inhibitors of SARS CoV-2 main protease. Bioorg. Chem. 2021, 106, 104497. [Google Scholar] [CrossRef]
- Zackria, A.A.; Pattabiraman, R.; Murthy, T.P.K.; Kumar, S.B.; Mathew, B.B.; Biju, V.G. Computational Screening of Natural Compounds from Salvia Plebeia R. Br. For Inhibition of SARS-CoV-2 Main Protease. Vegetos 2021, 35, 345–359. [Google Scholar] [CrossRef] [PubMed]
- Sergazy, S.; Shulgau, Z.; Zhulikeyeva, A.; Ramankulov, Y.; Palamarchuk, I.V.; Kulakov, I.V. Cytoprotective Activity of Newly Synthesized 3-(Arylmethylamino)-6-Methyl-4-Phenylpyridin-2 (1 H)-Ones Derivatives. Molecules 2022, 27, 5362. [Google Scholar] [CrossRef]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Dubey, A.; Alanazi, A.M.; Bhardwaj, R.; Ragusa, A. Identification of Potential NUDT5 Inhibitors from Marine Bacterial Natural Compounds via Molecular Dynamics and Free Energy Landscape Analysis. Mol. Divers. 2025, 29, 1929–1944. [Google Scholar] [CrossRef]
- Hasan, M.M.; Khan, Z.; Chowdhury, M.S.; Khan, M.A.; Moni, M.A.; Rahman, M.H. In Silico Molecular Docking and ADME/T Analysis of Quercetin Compound with Its Evaluation of Broad-Spectrum Therapeutic Potential against Particular Diseases. Inform. Med. Unlocked 2022, 29, 100894. [Google Scholar] [CrossRef]
- Ivanova, L.; Karelson, M. The Impact of Software Used and the Type of Target Protein on Molecular Docking Accuracy. Molecules 2022, 27, 9041. [Google Scholar] [CrossRef]
- Gligorić, E.; Igić, R.; Teofilović, B.; Grujić-Letić, N. Phytochemical Screening of Ultrasonic Extracts of Salix Species and Molecular Docking Study of Salix-Derived Bioactive Compounds Targeting Pro-Inflammatory Cytokines. Int. J. Mol. Sci. 2023, 24, 11848. [Google Scholar] [CrossRef]
- Ruswanto, R.; Nofianti, T.; Mardianingrum, R.; Kesuma, D.; Siswandono, N. Design, Molecular Docking, and Molecular Dynamics of Thiourea-Iron (III) Metal Complexes as NUDT5 Inhibitors for Breast Cancer Treatment. Heliyon 2022, 8, e10694. [Google Scholar] [CrossRef]
- Rashid, M.; Maqbool, A.; Shafiq, N.; Jardan, Y.A.B.; Parveen, S.; Bourhia, M.; Nafidi, H.-A.; Khan, R.A. The Combination of Multi-Approach Studies to Explore the Potential Therapeutic Mechanisms of Imidazole Derivatives as an MCF-7 Inhibitor in Therapeutic Strategies. Front. Chem. 2023, 11, 1197665. [Google Scholar] [CrossRef]
- Bujalowski, W.; Jezewska, M.J.; Bujalowski, P.J. Signal and binding. I. Physico-chemical response to macromolecule–ligand interactions. Biophys. Chem. 2017, 222, 7–24. [Google Scholar] [CrossRef]
- Grabowski, S.J. Intramolecular hydrogen bond energy and its decomposition—O–H∙∙∙O interactions. Crystals 2020, 11, 5. [Google Scholar] [CrossRef]
- Bulusu, G.; Desiraju, G.R. Strong and weak hydrogen bonds in protein-ligand recognition. J. Indian Inst. Sci. 2020, 100, 31–41. [Google Scholar] [CrossRef]
- Karami, T.K.; Hailu, S.; Feng, S.; Graham, R.; Gukasyan, H.J. Eyes on Lipinski’s Rule of Five: A New “Rule of Thumb” for Physicochemical Design Space of Ophthalmic Drugs. J. Ocul. Pharmacol. Ther. 2022, 38, 43–55. [Google Scholar] [CrossRef] [PubMed]
- Elhady, S.S.; Abdelhameed, R.F.A.; Malatani, R.T.; Alahdal, A.M.; Bogari, H.A.; Almalki, A.J.; Mohammad, K.A.; Ahmed, S.A.; Khedr, A.I.M.; Darwish, K.M. Molecular Docking and Dynamics Simulation Study of Hyrtios erectus Isolated Scalarane Sesterterpenes as Potential SARS-CoV-2 Dual Target Inhibitors. Biology 2021, 10, 389. [Google Scholar] [CrossRef] [PubMed]
- Pollastri, M.P. Overview on the Rule of Five. Curr. Protoc. Pharmacol. 2010, 49, 9.12.1–9.12.8. [Google Scholar] [CrossRef]
- Singh, P.; Kumar, V.; Lee, G.; Jung, T.S.; Ha, M.W.; Hong, J.C.; Lee, K.W. Pharmacophore-Oriented Identification of Potential Leads as CCR5 Inhibitors to Block HIV Cellular Entry. Int. J. Mol. Sci. 2022, 23, 16122. [Google Scholar] [CrossRef] [PubMed]
- Pires, D.E.; Blundell, T.L.; Ascher, D.B. pkCSM: Predicting small-molecule pharmacokinetic and toxicity properties using graph-based signatures. J. Med. Chem. 2015, 58, 4066–4072. [Google Scholar] [CrossRef]
- Wang, Y.; Xing, J.; Xu, Y.; Zhou, N.; Peng, J.; Xiong, Z.; Liu, X.; Luo, X.; Luo, C.; Chen, K.; et al. In Silico ADME/T Modelling for Rational Drug Design. Q. Rev. Biophys. 2015, 48, 488–515. [Google Scholar] [CrossRef] [PubMed]
- Yusuf, M. Insights into the in-silico research: Current scenario, advantages, limits, and future perspectives. Life Silico 2023, 1, 13–25. [Google Scholar]
- Zhou, S.F. Drugs behave as substrates, inhibitors and inducers of human cytochrome P450 3A4. Curr. Drug Metab. 2008, 9, 310–322. [Google Scholar] [CrossRef]
- Zhao, M.; Ma, J.; Li, M.; Zhang, Y.; Jiang, B.; Zhao, X.; Huai, C.; Shen, L.; Zhang, N.; He, L.; et al. Cytochrome P450 Enzymes and Drug Metabolism in Humans. Int. J. Mol. Sci. 2021, 22, 12808. [Google Scholar] [CrossRef]
- Hu, B.; Zhou, X.; Mohutsky, M.A.; Desai, P.V. Structure–Property Relationships and Machine Learning Models for Addressing CYP3A4-Mediated Victim Drug–Drug Interaction Risk in Drug Discovery. Mol. Pharm. 2020, 17, 3600–3608. [Google Scholar] [CrossRef]
- Pang, X.; Fu, W.; Wang, J.; Kang, D.; Xu, L.; Zhao, Y.; Liu, A.-L.; Du, G.-H. Identification of Estrogen Receptor α Antagonists from Natural Products via In Vitro and In Silico Approaches. Oxid. Med. Cell Longev. 2018, 2018, 6040149. [Google Scholar] [CrossRef]
- Taghizadeh, M.S.; Niazi, A.; Moghadam, A.; Alireza, A. Experimental, Molecular Docking and Molecular Dynamic Studies of Natural Products Targeting Overexpressed Receptors in Breast Cancer. PLoS ONE 2022, 17, e0267961. [Google Scholar] [CrossRef]
- Elhady, S.S.; Eltamany, E.E.; Shaaban, A.E.; Bagalagel, A.A.; Muhammad, Y.A.; El-Sayed, N.M.; Ayyad, S.N.; Ahmed, A.A.M.; Elgawish, M.S.; Ahmed, S.A. Jaceidin Flavonoid Isolated from Chiliadenus montanus Attenuates Tumor Progression in Mice via VEGF Inhibition: In Vivo and In Silico Studies. Plants 2020, 9, 1031. [Google Scholar] [CrossRef] [PubMed]
- Maugeri, A.; Calderaro, A.; Patanè, G.T.; Navarra, M.; Barreca, D.; Cirmi, S.; Felice, M.R. Targets Involved in the Anti-Cancer Activity of Quercetin in Breast, Colorectal and Liver Neoplasms. Int. J. Mol. Sci. 2023, 24, 2952. [Google Scholar] [CrossRef]
- Li, S.; Zhao, Q.; Wang, B.; Yuan, S.; Wang, X.; Li, K. Quercetin reversed MDR in breast cancer cells through down-regulating P-gp expression and eliminating cancer stem cells mediated by YB-1 nuclear translocation. Phytother. Res. 2018, 32, 1530–1536. [Google Scholar] [CrossRef]
- Prieto-Vila, M.; Shimomura, I.; Kogure, A.; Usuba, W.; Takahashi, R.U.; Ochiya, T.; Yamamoto, Y. Quercetin Inhibits Lef1 and Resensitizes Docetaxel-Resistant Breast Cancer Cells. Molecules 2020, 25, 2576. [Google Scholar] [CrossRef]
- Safi, A.; Heidarian, E.; Ahmadi, R. Quercetin Synergistically Enhances the Anticancer Efficacy of Docetaxel through Induction of Apoptosis and Modulation of PI3K/AKT, MAPK/ERK, and JAK/STAT3 Signaling Pathways in MDA-MB-231 Breast Cancer Cell Line. Int. J. Mol. Cell Med. 2021, 10, 11–22. [Google Scholar] [CrossRef] [PubMed]
- Roshanazadeh, M.; Rezaei, H.B.; Rashidi, M. Quercetin synergistically potentiates the anti-metastatic effect of 5-fluorouracil on the MDA-MB-231 breast cancer cell line. Iran. J. Basic Med. Sci. 2021, 24, 928–934. [Google Scholar] [CrossRef]
- Mawalizadeh, F.; Mohammadzadeh, G.; Khedri, A.; Rashidi, M. Quercetin potentiates the chemosensitivity of MCF-7 breast cancer cells to 5-fluorouracil. Mol. Biol. Rep. 2021, 48, 7733–7742. [Google Scholar] [CrossRef]
- Xu, Z.; Zhao, D.; Zheng, X.; Huang, B.; Xia, X.; Pan, X. Quercetin Exerts Bidirectional Regulation Effects on the Efficacy of Tamoxifen in Estrogen Receptor-positive Breast Cancer Therapy: An in Vitro Study. Environ. Toxicol. 2020, 35, 1179–1193. [Google Scholar] [CrossRef]
- Grinter, S.Z.; Zou, X. Challenges, Applications, and Recent Advances of Protein-Ligand Docking in Structure-Based Drug Design. Molecules 2014, 19, 10150–10176. [Google Scholar] [CrossRef]
- Kim, S.; Chen, J.; Cheng, T.; Gindulyte, A.; He, J.; He, S.; Li, Q.; Shoemaker, B.A.; Thiessen, P.A.; Yu, B.; et al. PubChem 2023 Update. Nucleic Acids Res. 2022, 51, D1373–D1380. [Google Scholar] [CrossRef]
- Hanwell, M.D.; Curtis, D.E.; Lonie, D.C.; Vandermeersch, T.; Zurek, E.; Hutchison, G.R. Avogadro: An Advanced Semantic Chemical Editor, Visualization, and Analysis Platform. J. Cheminform. 2012, 4, 17. [Google Scholar] [CrossRef] [PubMed]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated Docking with Selective Receptor Flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef] [PubMed]
- Elsaman, T.; Awadalla, M.K.A.; Mohamed, M.S.; Eltayib, E.M.; Mohamed, M.A. Identification of Microbial-Based Natural Products as Potential CYP51 Inhibitors for Eumycetoma Treatment: Insights from Molecular Docking, MM-GBSA Calculations, ADMET Analysis, and Molecular Dynamics Simulations. Pharmaceuticals 2025, 18, 598. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Pan, H.; Sun, H.; Kang, Y.; Liu, H.; Cao, D.; Hou, T. FastDRH: A Webserver to Predict and Analyze Protein–Ligand Complexes Based on Molecular Docking and MM/PB(GB)SA Computation. Brief. Bioinform. 2022, 23, bbac201. [Google Scholar] [CrossRef]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef]
- Banerjee, P.; Kemmler, E.; Dunkel, M.; Preissner, R. ProTox 3.0: A Webserver for the Prediction of Toxicity of Chemicals. Nucleic Acids Res. 2024, 52, W513–W520. [Google Scholar] [CrossRef] [PubMed]
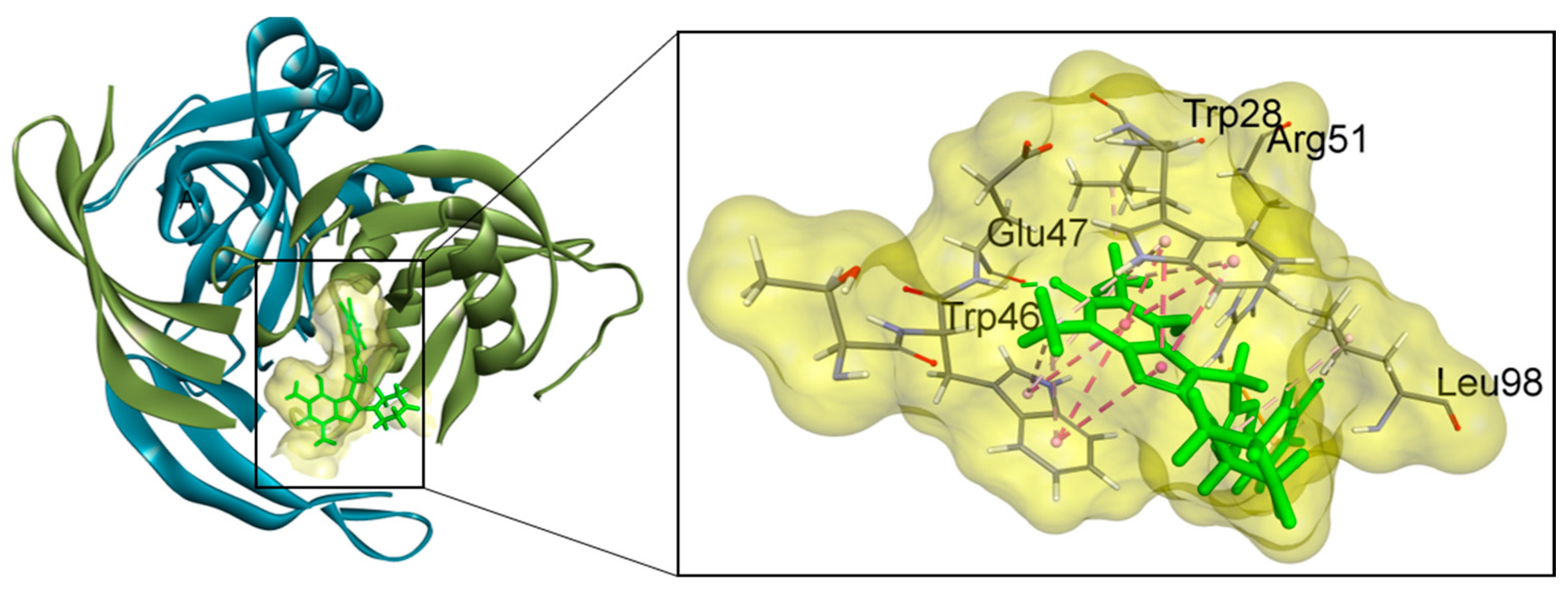
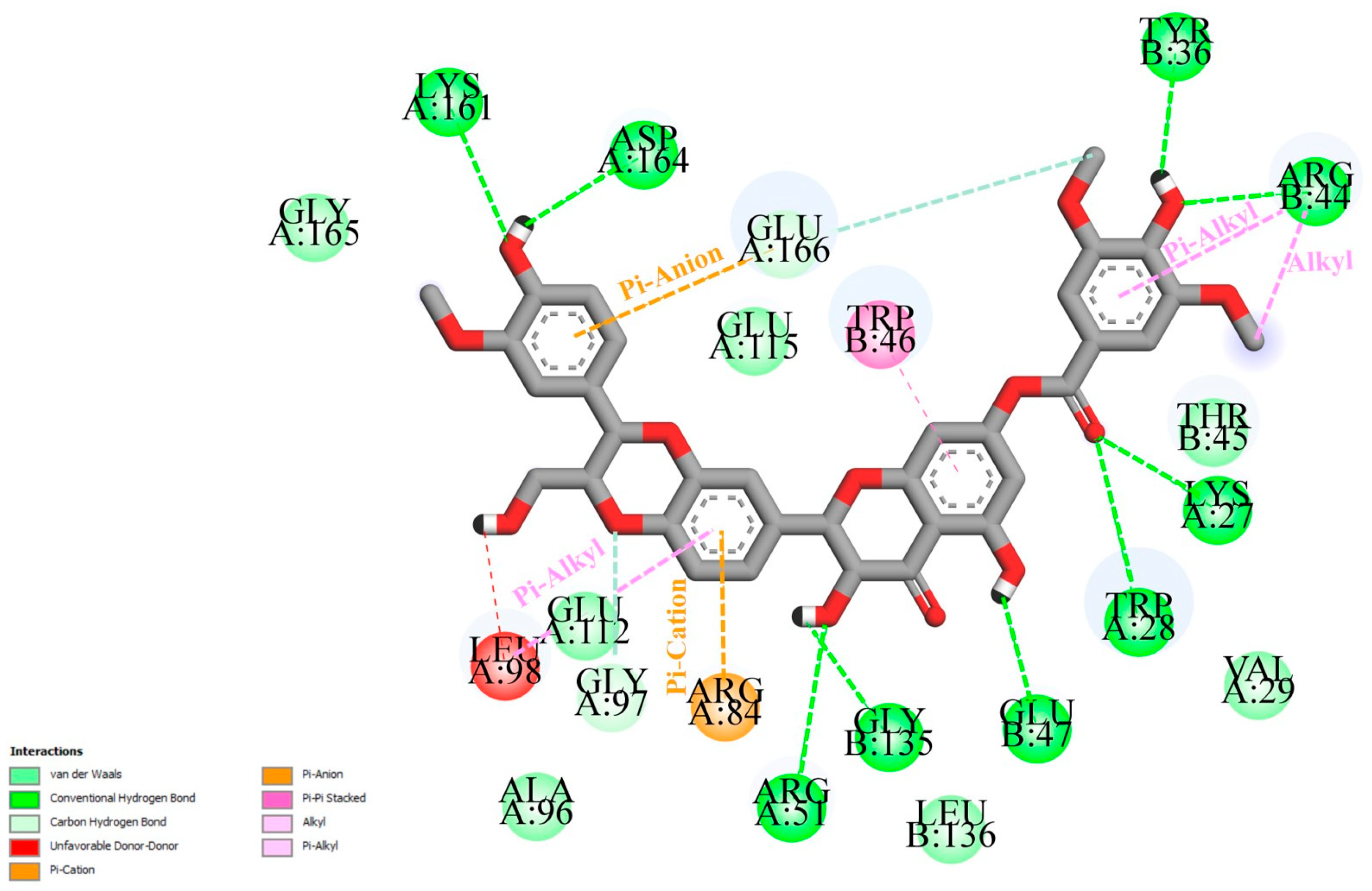
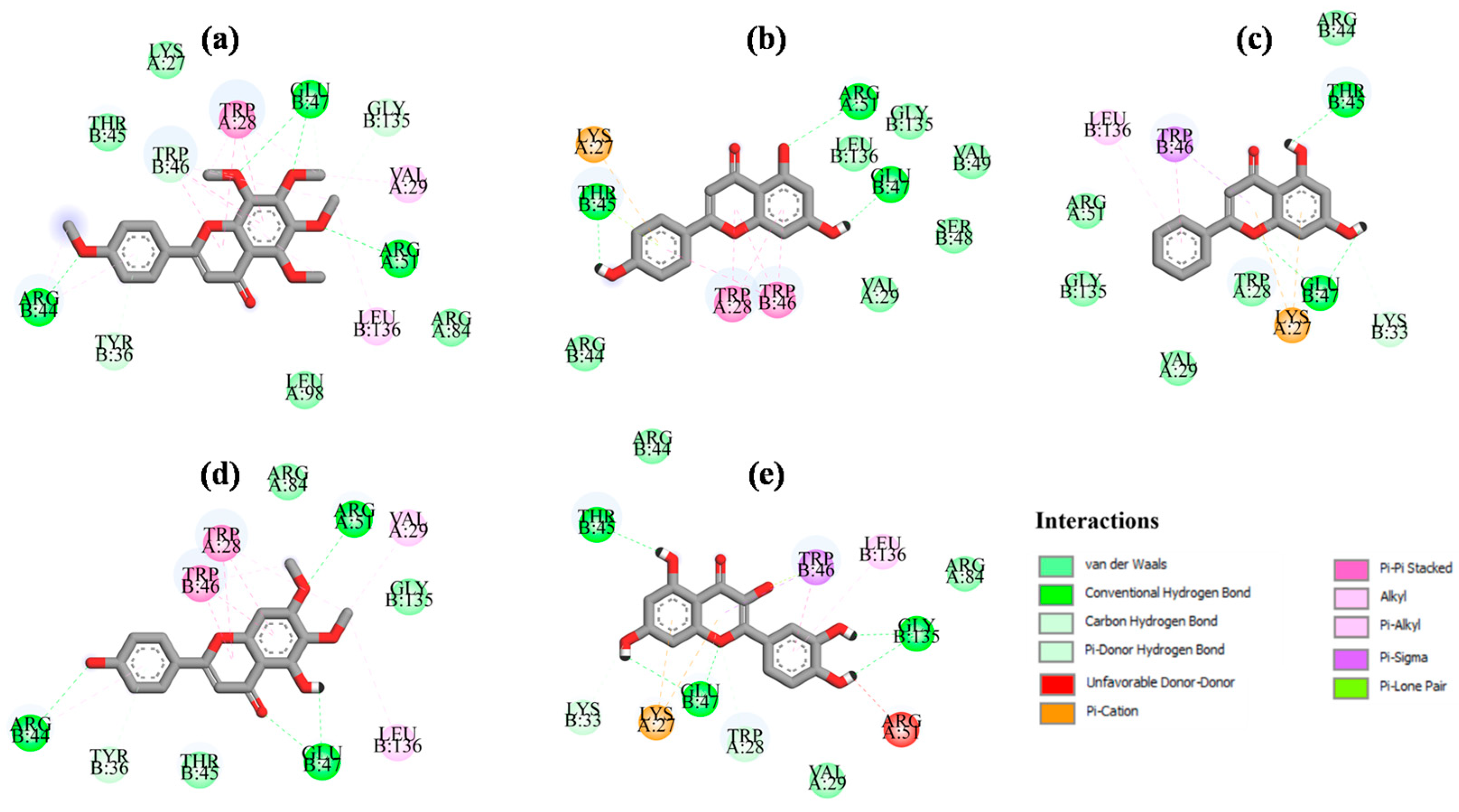
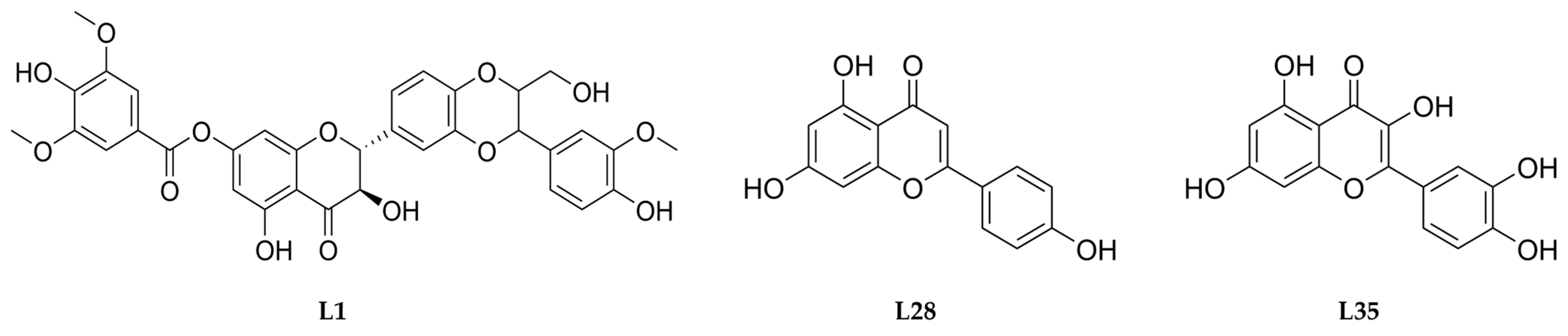
| Ligand | ΔGb (kcal/mol) | Interactions | |
|---|---|---|---|
| Hydrogen Bond | Van Der Waals Interactions | ||
| L1 | −11.24 | LysA:161, AspA:164, TyrB:36, ArgB:44, LysA:27, TrpA:28, GluB:47, GlyB:135, ArgA:51 | GlyA:165, GluA:115, ThrB:45, GluA:112, ValA:29, AlaA:96, LeuB:136 |
| L2 | −10.92 | ArgA:196, TyrB:36, ArgA:51 | LysA:161, AspA:164, GlyA:165, AspA:194, AlaA:63, AspB:133, GluA:166, ArgB:44, ThrB:45, GlnA:82, LeuB:136, GluB:47 |
| L3 | −10.76 | AspA:194, GlyA:165, TyrB:36, ArgA:51 | PheA:94, ArgA:196, ArgB:44, GluA:166, GlyA:97, ArgA:84, GluB:47, LeuB:136, ValA:29 |
| L4 | −10.42 | ArgA:51, LeuA:98, GluA:112, LysA:161, AspA:164, GluB:47 | LeuB:136, GlyB:135, GlyA:97, ArgA:111, ThrB:45, GluA:116 |
| L5 | −10.28 | TyrB:36, GluB:47 | ArgA:196, AspA:164, GlyA:165, ValA:62, GluA:166, GluA:93, GlyA:97, ArgA:84, LeuB:136, ValA:29 |
| L6 | −10.09 | GlyA:165, TrpA:28, PheA:167, LysA:28, GluB:47, GlnB:15, PheA:83 | ArgB:44, GluA:166, GlnA:82, LysB:33, ArgA:84 |
| L7 | −9.73 | AlaA:96, ArgA:51, GlyB:135, PheA167, TrpA:28, GluB:47 | GlyA:97, ArgA:84, GlnA:82, TrpB:46, ValA:168, GluA:166, LeuB:136, LysA:27, ValA:29, GlnB:15, AspB:38, ThrB:45, ProB:39 |
| L8 | −9.69 | LeuA:98, GlyB:135, GluA:112, GluB:47, AspA:164, TrpB:46, ThrB:45 | ArgA:84, LeuB:136, ArgA:111, ArgA:51, GluA:166, TrpA:28, GlyA:165, LysA:27 |
| L9 | −9.69 | GluA:112, GluB:47, AspA:164 | ArgA:111, LysA:161, ThrB:45 |
| L10 | −9.68 | ArgB:44, LysA:27, TrpA:28, LeuA:98, TrpB:46, ArgA:51, GlyB:135, GluB:47 | LysA:161, AspA:164, GlyA:165, ThrB:45, GluA:166, GluA:116, ArgA:111, GlyA:97, LeuB:136 |
| L11 | −9.65 | ArgA:84, ArgA:51, GluA:115, GluB:47, TyrB:36, ThrB:45 | GlnA:82, ArgA:111, GlyB:135, AlaA:96, GlyA:97, GluA:112, ValA:29, LeuB:136, LysA:161, LeuA:98, GluA:166, LysA:27, ArgB:44 |
| L12 | −9.64 | ArgA:51, ThrB:45, AlaA:96, ArgA:84 | ValA:29, GlyB:135, LeuB:136, GluB:47, TrpA:28, AspA:60, GlyA:61, GlyA:97, GluA:116, GlnA:82 |
| L13 | −9.53 | GluA:166, ThrB:45, GlyB:135, GluB:47 | ArgB:44, ArgA:84, TrpA:28, ValA:29 |
| L14 | −9.43 | LeuA:98, GluB:47 | ValA:62, CysA:139, LeuB:136, GlyA:97, GlyB:135, ArgA:196, AspB:133 |
| L15 | −9.42 | GluB:47, ArgA:196, AspA:194 | ValA:62, GlyA:97, IleA:141, LeuB:136, PheA:94, AspB:133, ArgB:44 |
| L16 | −9.42 | GluA:112, GluB:47, AspA:164, ThrB:45 | ArgA:111, GlyB:135, LeuB:136, TyrB:36, GluA:166 |
| L17 | −9.38 | ArgA:51, GlyB:135, LeuA:98, ArgA:111, TrpB:46, GluB:47 | GluA:115, ArgA:84, GluA:112, LeuB:136, GlyA:97, AspA:101, ThrA:53, LysA:27 |
| L18 | −9.34 | LeuA:98, GluB:47, ArgA:51, AspA:164 | ArgA:111, GlyA:97, GluA:112, ThrB:45, LysA:161, GluA:116, ValA:29, LeuB:136 |
| L19 | −9.19 | GluB:47, GluA:115, GluA:112, LeuA:98, AlaA:96, ArgA:84 | ValA:29, LeuB:136, TyrB:36, GlyB:135, IleA:99, GlyA:97, LysA:27, GlnA:82, GluA:116 |
| L20 | −8.87 | AlaA:96, GlnA:82, ArgA:84, GluB:47, ThrB:45, ArgA:51, TyrB:36 | GluA:116, SerB:48, ValA:29, LeuB:136, GluA:112, TrpA:28, TrpB:46, ArgB:44, GluA:166 |
| L21 | −8.82 | GluA:166, TyrB:36, ArgA:51, ThrB:45, GluB:47 | ArgA:84, GlyB:135, ArgB:44, LysA:27, LeuB:136, ValA:29, ValB:49, SerB:48, LysB:33 |
| L22 | −8.64 | ThrB:45, GluB:47, ArgA:51 | LysA:27, TyrB:36, ArgB:44, ValA:29, GluA:166, LeuB:136, SerB:48, GlyB:135 |
| L23 | −8.42 | ThrB:45, GlyB:135, ArgA:51, GluB:47 | ArgA:84, ArgB:44, ValA:29 |
| L24 | −8.41 | GluB:47, ArgB:44, ArgA:51 | LysA:27, ThrB:45, ArgA:84, LeuA:98 |
| L25 | −8.27 | ArgA:51, GluB:47, ArgB:44 | LeuA:98, ValA:29, LeuB:136, ThrB:45 |
| L26 | −8.21 | GluB:47, ThrB:45, GlyB:135, ArgA:51 | LeuA:98, ArgB:44, LeuB:136 |
| L27 | −8.15 | ArgA:51, GluB:47, ThrB:45 | LeuA:98, TyrB:36, GylB:135, SerB:48, LeuB:136, ValA:29, LysA:27 |
| L28 | −8.13 | ArgA:51, ThrB:45, GluB:47 | GlyB:135, LeuB:136, ValB:49, SerB:48, ValA:29, ArgB:44 |
| L29 | −8.08 | ThrB:45, ArgA:51, GlyB:135 | GluA:166, ArgA:84, GlyA:97, LysB:33, TrpA:28, GluB:47, LeuB:136 |
| L30 | −8.08 | ThrB:45, GluB:47 | ArgB:44, ArgA:51, GlyB:135, TrpA:28, VaA:29 |
| L31 | −8.06 | GluB:47, ThrB:45, ArgA:51 | ValA:29, LysA:27, SerB:48, LeuB:136, GlyB:135, ArgB:44 |
| L32 | −8.06 | ThrB:45, GlyB:135, ArgA:51, gluB:47 | ArgA:84, ArgB:44; TrpA:28, ValA:29 |
| L33 | −8.04 | ArgA:51, ArgB:44, GluB:47 | ArgA:84, GlyB:135, ThrB:45 |
| L34 | −8.01 | ArgA:51, GluB:47, ThrB:45 | TyrB:36, GlyB:135, LeuB:136, SerB:48, ValA:29 |
| L35 | −8.0 | ThrB:45, GlyB:135, GluB:47 | ArgB:44, ArgA:84, ValA:29 |
| C | −11.02 | GluB:47, ArgA:51 | GluA:166, GlyB:135, LeuA:98, LeuB:136, GlyA:97, GlyA:61, ValA:62 |
| T | −9.36 | LeuA98 | ArgA:84, GlyA:97, GluA:112, AlaA:96, GluB:47, ThrB:45, GlyB:135 |
| R | −10.83 | ArgA:84, ArgB:44, GluB:47 | TyrB:36, LeuB:136, ValA:29, ArgA:51, GlyA:97, LeuA:98, ArgA:111, GluA:115, GluA:112, AlaA:96, GluA:116 |
| Complex | Name | Types | Distance | Angle DHA |
|---|---|---|---|---|
| NUDT5-L1 | A:LYS27:NZ—:L1:O11 | Conventional Hydrogen Bond | 2.9052 | |
| A:TRP28:NE1—:L1:O11 | Conventional Hydrogen Bond | 3.0263 | ||
| A:ARG51:NH2—:L1:O4 | Conventional Hydrogen Bond | 2.6999 | ||
| A:LYS161:NZ—:L1:O10 | Conventional Hydrogen Bond | 2.7842 | ||
| B:ARG44:NH1—:L1:O14 | Conventional Hydrogen Bond | 3.0866 | ||
| :L1:H66—A:ASP164:O | Conventional Hydrogen Bond | 2.0592 | 146.072 | |
| :L1:H61—B:GLY135:O | Conventional Hydrogen Bond | 2.1563 | 152.734 | |
| :L1:H65—B:GLU47:O | Conventional Hydrogen Bond | 2.3719 | 112.251 | |
| :L1:H72—B:TYR36:OH | Conventional Hydrogen Bond | 3.0413 | 94.802 | |
| NUDT5-L2 | A:ARG51:NH1—:L2:O4 | Conventional Hydrogen Bond | 3.2128 | |
| A:ARG196:NH2—:L2:O12 | Conventional Hydrogen Bond | 2.8131 | ||
| B:TYR36:OH—:L2:O13 | Conventional Hydrogen Bond | 2.4423 | ||
| NUDT5-L3 | A:ARG51:NH1—:L3:O4 | Conventional Hydrogen Bond | 3.0429 | |
| B:TYR36:OH—:L3:O11 | Conventional Hydrogen Bond | 2.6881 | ||
| :L3:H64—A:ASP194:OD2 | Conventional Hydrogen Bond | 1.8865 | 116.528 | |
| :L3:H71—A:GLY165:O | Conventional Hydrogen Bond | 2.3005 | 153.343 | |
| NUDT5-L4 | A:ARG51:NH1—:L4:O4 | Conventional Hydrogen Bond | 2.9590 | |
| A:LYS161:NZ—:L4:O8 | Conventional Hydrogen Bond | 2.7955 | ||
| A:ASP164:N—:L4:O10 | Conventional Hydrogen Bond | 3.1628 | ||
| B:GLU47:N—:L4:O7 | Conventional Hydrogen Bond | 3.1725 | ||
| :L4:H54—A:LEU98:O | Conventional Hydrogen Bond | 2.7405 | 95.676 | |
| :L4:H54—A:GLU112:OE1 | Conventional Hydrogen Bond | 2.2999 | 94.878 | |
| NUDT5-L5 | B:TYR36:OH—:L5:O11 | Conventional Hydrogen Bond | 2.7707 | |
| B:GLU47:N—:L5:O6 | Conventional Hydrogen Bond | 3.3795 | ||
| B:GLU47:N—:L5:O7 | Conventional Hydrogen Bond | 2.6243 | ||
| NUDT5-L24 | A:ARG51:NH1—:L24:O4 | Conventional Hydrogen Bond | 3.0627 | |
| B:ARG44:NH1—:L24:O7 | Conventional Hydrogen Bond | 3.0560 | ||
| B:GLU47:N—:L24:O3 | Conventional Hydrogen Bond | 3.0000 | ||
| B:GLU47:N—:L24:O5 | Conventional Hydrogen Bond | 3.2257 | ||
| NUDT5-L28 | A:ARG51:NH1—:L28:O2 | Conventional Hydrogen Bond | 2.8998 | |
| A:ARG51:NH2—:L28:O2 | Conventional Hydrogen Bond | 3.1644 | ||
| :L28:H30—B:THR45:OG1 | Conventional Hydrogen Bond | 1.9621 | 154.122 | |
| :L28:H29—B:GLU47:O | Conventional Hydrogen Bond | 1.7022 | 158.33 | |
| NUDT5-L30 | B:GLU47:N—:L30:O1 | Conventional Hydrogen Bond | 3.2047 | |
| :L30:H29—B:GLU47:OE2 | Conventional Hydrogen Bond | 2.1768 | 148.949 | |
| :L30:H28—B:THR45:O | Conventional Hydrogen Bond | 2.0593 | 126.724 | |
| NUDT5-L33 | A:ARG51:NH1—:L33:O3 | Conventional Hydrogen Bond | 2.9274 | |
| A:ARG51:NH2—:L33:O3 | Conventional Hydrogen Bond | 3.1588 | ||
| B:ARG44:NH1—:L33:O6 | Conventional Hydrogen Bond | 3.0908 | ||
| B:GLU47:N—:L33:O5 | Conventional Hydrogen Bond | 2.5814 | ||
| :L33:H30—B:GLU47:O | Conventional Hydrogen Bond | 2.4846 | 133.572 | |
| NUDT5-L35 | B:GLU47:N—:Quercetin:O1 | Conventional Hydrogen Bond | 3.2024 | |
| :Quercetin:H31—B:GLY135:O | Conventional Hydrogen Bond | 2.1313 | 131.074 | |
| :Quercetin:H32—B:GLY135:O | Conventional Hydrogen Bond | 2.1999 | 113.465 | |
| :Quercetin:H30—B:GLU47:OE2 | Conventional Hydrogen Bond | 2.3163 | 146.64 | |
| :Quercetin:H29—B:THR45:O | Conventional Hydrogen Bond | 2.2237 | 129.379 | |
| NUDT5-C | A:ARG51:NH1—A:Control:O32 | Conventional Hydrogen Bond | 2.8128 | |
| B:GLU47:N—A:Control:O33 | Conventional Hydrogen Bond | 2.6402 | ||
| NUDT5-T | A:LEU98:N—:Tamoxifen:O1 | Conventional Hydrogen Bond | 3.0922 | |
| NUDT5-R | A:ARG84:HH22—:Raloxifen:O2 | Conventional Hydrogen Bond | 2.4916 | 128.533 |
| B:ARG44:HH12—:Raloxifen:O5 | Conventional Hydrogen Bond | 2.0731 | 161.962 | |
| :Raloxifen:H60—B:GLU47:O | Conventional Hydrogen Bond | 2.0734 | 156.171 |
| Ligand | ELE | VDW | Non-Polar SASA | Polar | Total Binding Free Energy |
|---|---|---|---|---|---|
| L5 | 0 | −75.66 | −6.38 | 34.2 | −47.83 |
| L10 | 0 | −83.53 | −7.18 | 43.77 | −46.94 |
| L1 | 0 | −74 | −5.97 | 34.76 | −45.21 |
| L3 | 0 | −71.18 | −5.91 | 36.16 | −40.93 |
| L2 | 0 | −76.37 | −6.34 | 41.85 | −40.86 |
| L8 | 0 | −59.17 | −4.88 | 25.92 | −38.13 |
| L9 | 0 | −61.5 | −5.07 | 30.15 | −36.41 |
| L15 | 0 | −61.47 | −5.06 | 32.07 | −34.46 |
| L14 | 0 | −54.43 | −4.28 | 27.18 | −31.53 |
| L4 | 0 | −51.3 | −5.13 | 29.18 | −27.24 |
| L25 | 0 | −41.7 | −3.27 | 18.1 | −26.86 |
| L34 | 0 | −41.88 | −3.76 | 18.8 | −26.84 |
| L19 | 0 | −53.12 | −4.88 | 31.57 | −26.43 |
| L22 | 0 | −43.53 | −3.7 | 21.71 | −25.51 |
| L20 | 0 | −58.42 | −5.59 | 39.01 | −25.01 |
| L32 | 0 | −34.7 | −2.66 | 12.8 | −24.55 |
| L13 | 0 | −49.16 | −4.24 | 29.53 | −23.86 |
| L6 | 0 | −45.23 | −3.62 | 25.55 | −23.3 |
| L16 | 0 | −51.38 | −5.2 | 33.33 | −23.26 |
| L17 | 0 | −49.22 | −4.88 | 30.88 | −23.23 |
| L18 | 0 | −55.68 | −5.35 | 37.85 | −23.18 |
| L27 | 0 | −36.55 | −2.46 | 16.18 | −22.84 |
| L24 | 0 | −41.24 | −3.08 | 21.93 | −22.38 |
| L7 | 0 | −58.32 | −5.1 | 41.15 | −22.27 |
| L26 | 0 | −38.29 | −2.95 | 19.08 | −22.16 |
| L12 | 0 | −53.7 | −5.12 | 36.87 | −21.96 |
| L31 | 0 | −34.57 | −2.37 | 15.35 | −21.57 |
| L23 | 0 | −38.45 | −3.11 | 20.09 | −21.45 |
| L35 | 0 | −33.63 | −2.81 | 15.87 | −20.58 |
| L11 | 0 | −58.5 | −5.92 | 45.36 | −19.06 |
| L28 | 0 | −39.05 | −3.66 | 26.67 | −16.04 |
| L30 | 0 | −37.1 | −3.57 | 27.48 | −13.2 |
| L21 | 0 | −52.77 | −4.75 | 45.35 | −12.16 |
| L29 | 0 | −39.67 | −3.97 | 32.34 | −11.31 |
| L33 | 0 | −34.9 | −3.77 | 32.8 | −5.88 |
| C | 0 | −59.1 | −3.96 | 16.91 | −46.16 |
| R | 0 | −55.19 | −4.86 | 28.56 | −31.51 |
| T | 0 | −45.13 | −3.98 | 21.87 | −27.24 |
| Ligand | Mr | nHBA | nHBD | logP | Number of Violations of Lipinski’s Rules | Solubility in Water (log mol/L) | Caco-2 Permeability [log cm/s] |
|---|---|---|---|---|---|---|---|
| (g/mol) | |||||||
| L1 | 662.59 | 14 | 5 | 2.82 | 2 | −3.146 | 0.126 |
| L2 | 684.64 | 13 | 4 | 3.95 | 2 | −3.144 | 0.301 |
| L3 | 634.54 | 14 | 7 | 2.05 | 3 | −2.906 | −0.93 |
| L4 | 524.52 | 10 | 2 | 2.63 | 1 | −4.104 | 0.848 |
| L5 | 612.58 | 11 | 4 | 3.57 | 2 | −3.394 | 0.246 |
| L6 | 482.44 | 10 | 5 | 1.59 | 0 | −3.204 | 0.435 |
| L7 | 608.54 | 15 | 8 | −0.44 | 3 | −2.929 | 0.305 |
| L8 | 482.44 | 10 | 5 | 1.59 | 0 | −3.204 | 0.435 |
| L9 | 496.46 | 10 | 4 | 1.97 | 0 | −3.375 | 0.222 |
| L10 | 658.6 | 13 | 5 | 3.18 | 2 | −3.125 | 0.097 |
| L11 | 580.53 | 14 | 8 | −0.79 | 3 | −2.919 | −0.658 |
| L12 | 466.44 | 9 | 4 | 1.84 | 0 | −3.639 | 0.242 |
| L13 | 448.38 | 11 | 7 | 0.16 | 2 | −2.903 | 0.048 |
| L14 | 524.52 | 10 | 2 | 2.91 | 1 | −4.468 | 0.559 |
| L15 | 496.46 | 10 | 4 | 1.97 | 0 | −3.735 | 0.22 |
| L16 | 480.42 | 10 | 5 | 2.33 | 0 | −2.971 | 0.457 |
| L17 | 538.54 | 10 | 1 | 3.12 | 1 | −4.128 | 0.91 |
| L18 | 482.44 | 10 | 5 | 1.59 | 0 | −3.204 | 0.435 |
| L19 | 612.53 | 16 | 10 | −1.76 | 3 | −2.897 | −1.302 |
| L20 | 652.6 | 18 | 8 | −0.65 | 3 | −2.978 | 0.291 |
| L21 | 610.52 | 16 | 10 | −1.29 | 3 | −2.892 | −0.949 |
| L22 | 370.35 | 7 | 5 | 2.69 | 0 | −2.918 | −0.107 |
| L23 | 330.29 | 7 | 3 | 2.2 | 0 | −3.241 | 0.387 |
| L24 | 372.37 | 7 | 0 | 3.02 | 0 | −4.792 | 1.245 |
| L25 | 368.38 | 6 | 3 | 3.53 | 0 | −3.92 | 0.352 |
| L26 | 330.29 | 7 | 3 | 2.02 | 0 | −3.399 | −0.002 |
| L27 | 300.26 | 6 | 3 | 2.19 | 0 | −3.238 | 0.326 |
| L28 | 270.24 | 5 | 3 | 2.11 | 0 | −3.329 | 1.007 |
| L29 | 354.35 | 6 | 4 | 3.22 | 0 | −3.303 | −0.185 |
| L30 | 254.24 | 4 | 2 | 2.55 | 0 | −3.538 | 0.945 |
| L31 | 286.24 | 6 | 4 | 1.73 | 0 | −3.094 | 0.096 |
| L32 | 316.26 | 7 | 4 | 1.65 | 0 | −3.000 | −0.003 |
| L33 | 314.29 | 6 | 2 | 2.46 | 0 | −3.481 | 1.022 |
| L34 | 302.28 | 6 | 3 | 1.91 | 0 | −3.047 | 0.294 |
| L35 | 302.24 | 7 | 5 | 1.23 | 0 | −2.925 | −0.229 |
| C | 491.33 | 7 | 1 | 1.99 | 0 | −2.856 | 0.253 |
| T | 371.51 | 2 | 0 | 5.77 | 1 | −5.929 | 1.065 |
| R | 459.56 | 5 | 2 | 5 | 0 | −3.716 | 0.77 |
| Ligand | LD50 (mg/kg) | Toxicity Class | Hepatotoxicity | Neurotoxicity | Nephrotoxicity | Cardiotoxicity | Carcinogenicity | Immunotoxicity | Mutagenicity | Cytotoxicity |
|---|---|---|---|---|---|---|---|---|---|---|
| L1 | 2000 | 4 | Inactive | Inactive | Active | Active | Inactive | Active | Inactive | Inactive |
| L2 | 25 | 2 | Inactive | Inactive | Active | Active | Inactive | Active | Inactive | Inactive |
| L3 | 2000 | 4 | Inactive | Inactive | Active | Active | Inactive | Active | Inactive | Inactive |
| L4 | 1000 | 4 | Inactive | Inactive | Active | Active | Inactive | Active | Inactive | Inactive |
| L5 | 25 | 2 | Inactive | Inactive | Active | Active | Inactive | Active | Inactive | Inactive |
| L8 | 2000 | 4 | Inactive | Inactive | Active | Active | Inactive | Active | Inactive | Inactive |
| L9 | 2000 | 4 | Inactive | Inactive | Active | Active | Inactive | Active | Inactive | Inactive |
| L10 | 25 | 2 | Inactive | Inactive | Active | Active | Inactive | Active | Inactive | Inactive |
| L14 | 2000 | 4 | Inactive | Inactive | Active | Active | Inactive | Active | Inactive | Inactive |
| L15 | 2000 | 4 | Inactive | Inactive | Active | Active | Inactive | Active | Inactive | Inactive |
| L17 | 1000 | 4 | Inactive | Inactive | Active | Active | Inactive | Inactive | Inactive | Inactive |
| L24 | 5000 | 5 | Inactive | Inactive | Active | Inactive | Inactive | Inactive | Inactive | Inactive |
| L28 | 2500 | 5 | Inactive | Inactive | Active | Inactive | Inactive | Inactive | Inactive | Inactive |
| L30 | 3919 | 5 | Inactive | Inactive | Active | Inactive | Inactive | Inactive | Inactive | Inactive |
| L33 | 4000 | 5 | Inactive | Inactive | Active | Inactive | Inactive | Inactive | Inactive | Inactive |
| L35 | 159 | 3 | Inactive | Inactive | Active | Inactive | Active | Inactive | Active | Inactive |
| C | 684 | 4 | Inactive | Active | Inactive | Inactive | Inactive | Inactive | Active | Inactive |
| T | 1190 | 4 | Active | Active | Inactive | Inactive | Inactive | Active | Inactive | Inactive |
| R | 1000 | 4 | Inactive | Active | Active | Inactive | Inactive | Inactive | Inactive | Inactive |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gligorić, E.; Vidić, M.; Teofilović, B.; Grujić-Letić, N. Quercetin and Its Structural Analogs as NUDT5 Inhibitors: A Preliminary In Silico Study. Int. J. Mol. Sci. 2025, 26, 8843. https://doi.org/10.3390/ijms26188843
Gligorić E, Vidić M, Teofilović B, Grujić-Letić N. Quercetin and Its Structural Analogs as NUDT5 Inhibitors: A Preliminary In Silico Study. International Journal of Molecular Sciences. 2025; 26(18):8843. https://doi.org/10.3390/ijms26188843
Chicago/Turabian StyleGligorić, Emilia, Milica Vidić, Branislava Teofilović, and Nevena Grujić-Letić. 2025. "Quercetin and Its Structural Analogs as NUDT5 Inhibitors: A Preliminary In Silico Study" International Journal of Molecular Sciences 26, no. 18: 8843. https://doi.org/10.3390/ijms26188843
APA StyleGligorić, E., Vidić, M., Teofilović, B., & Grujić-Letić, N. (2025). Quercetin and Its Structural Analogs as NUDT5 Inhibitors: A Preliminary In Silico Study. International Journal of Molecular Sciences, 26(18), 8843. https://doi.org/10.3390/ijms26188843





