The Influence of Cohesin on the Short-Scale Dynamics of Intact and Damaged Chromatin in Different Phases of the Cell Cycle
Abstract
1. Introduction
2. Results
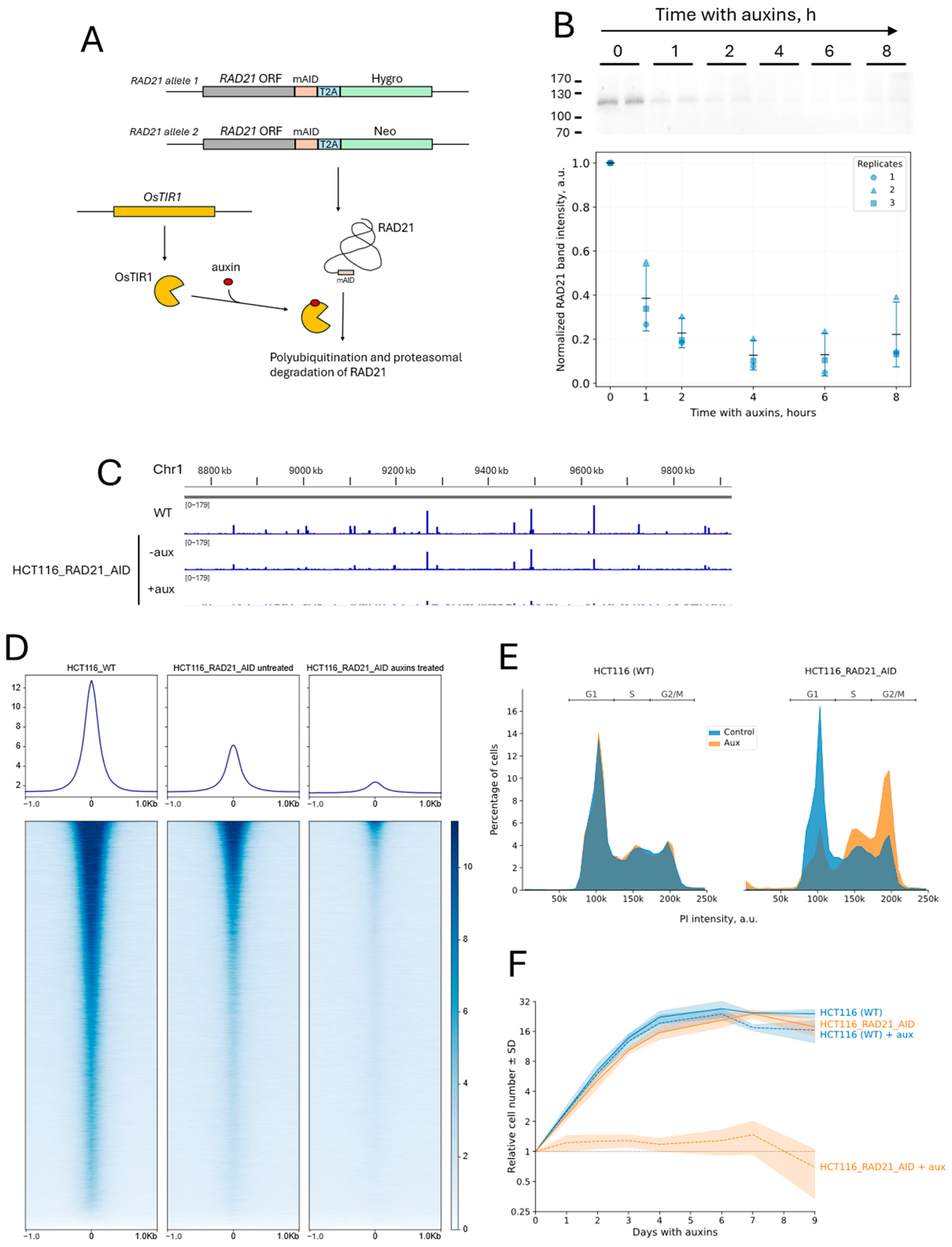
2.1. Cell Line with Auxin-Induced Degradation of RAD21 and with the CRISPR-Sirius Imaging System
2.2. Cohesin Depletion Increases the Spatial Dynamics of the Visualized Locus
2.3. Cohesin Constrains the Dynamics of Both Replicated and Unreplicated Chromatin
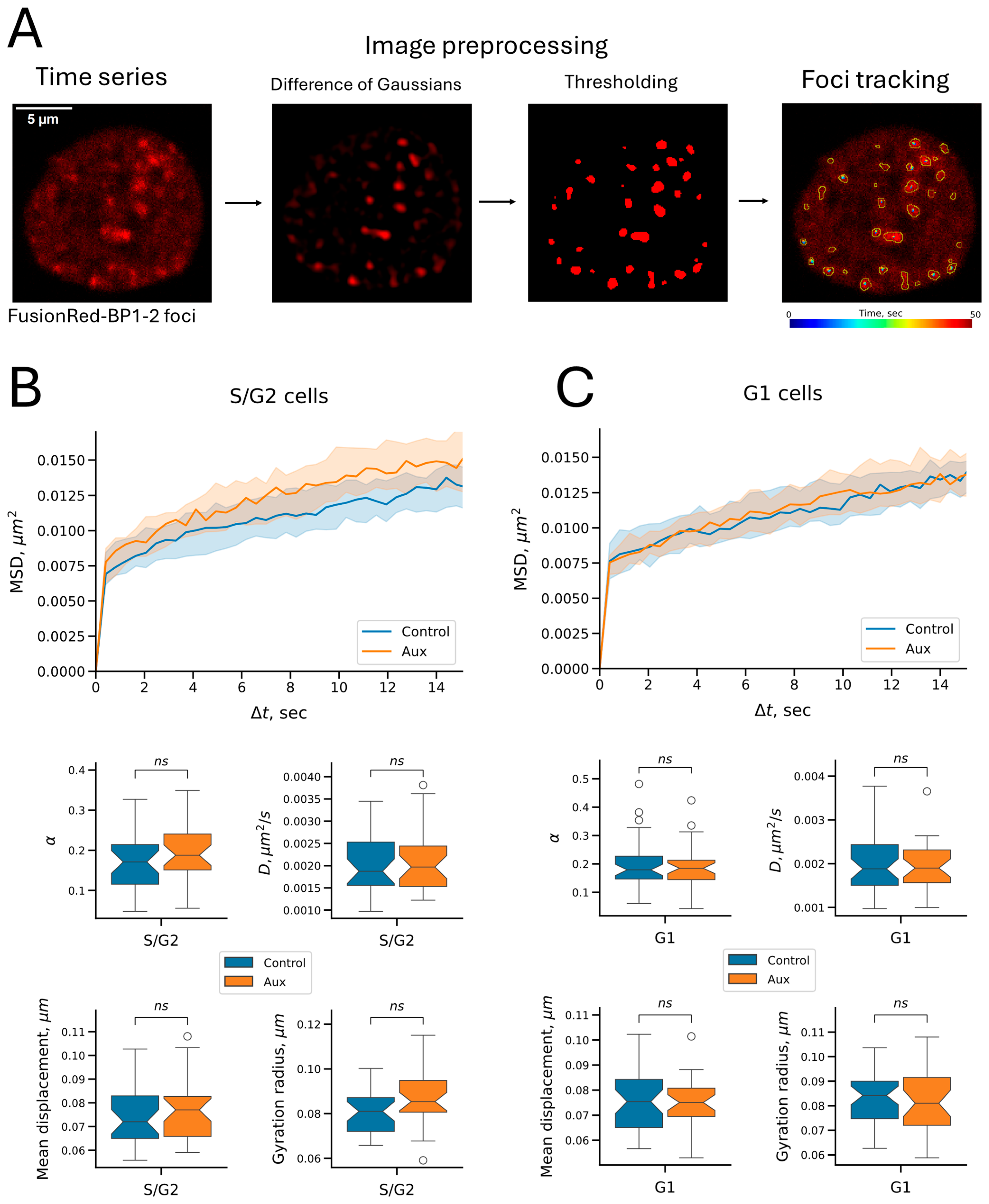
2.4. Cohesin Does Not Affect the Mobility of Repair Foci on the Time Scale Studied
3. Discussion
4. Materials and Methods
4.1. Plasmid Construction
4.2. Lentiviral Particles Production
4.3. Generation of HCT116 Cells with the Auxin-Inducible RAD21 Depletion and the CRISPR-Sirius System
4.4. Integration of the FusionRed-BP1-2 Gene
4.5. Western Blotting
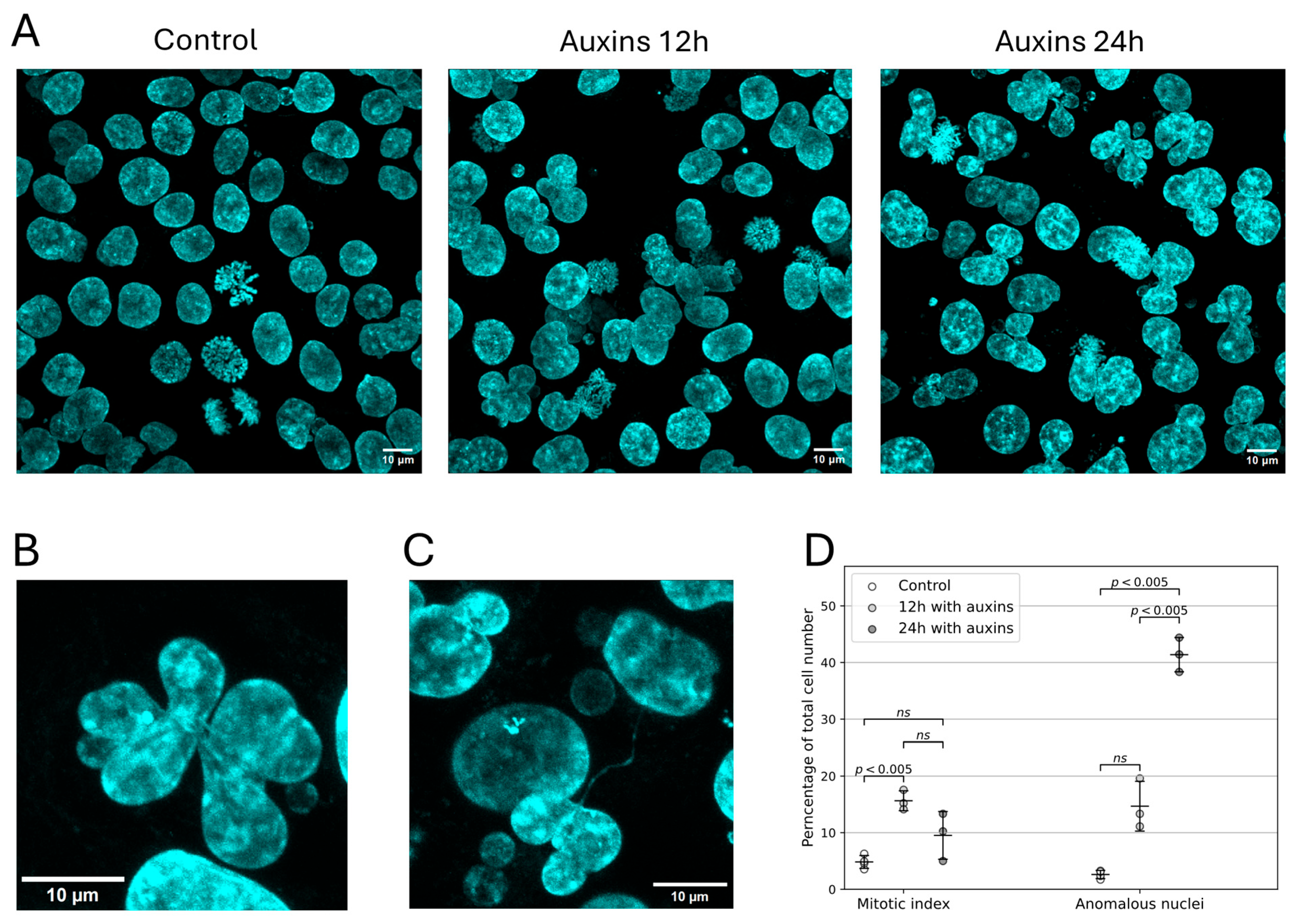
4.6. Analysis of Cell Cycle, Mitotic Index and Growth Curves
4.7. ChIP-Seq
4.8. PRO-Seq Data Analysis
4.9. C6 Locus Visualization and Tracking
4.10. FusionRed-BP1-2 Foci Visualization and Tracking
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AID | Auxin-inducible degron |
| DSB | DNA double-strand break |
| MSD | Mean square displacement |
| NHEJ | Non-homologous end joining |
References
- Oldenkamp, R.; Rowland, B.D. A Walk through the SMC Cycle: From Catching DNAs to Shaping the Genome. Mol. Cell 2022, 82, 1616–1630. [Google Scholar] [CrossRef] [PubMed]
- Hoencamp, C.; Rowland, B.D. Genome Control by SMC Complexes. Nat. Rev. Mol. Cell Biol. 2023, 24, 633–650. [Google Scholar] [CrossRef]
- Golov, A.K.; Gavrilov, A.A. Cohesin Complex: Structure and Principles of Interaction with DNA. Biochem. Mosc. 2024, 89, 585–600. [Google Scholar] [CrossRef]
- Haering, C.H.; Farcas, A.-M.; Arumugam, P.; Metson, J.; Nasmyth, K. The Cohesin Ring Concatenates Sister DNA Molecules. Nature 2008, 454, 297–301. [Google Scholar] [CrossRef]
- Gligoris, T.G.; Scheinost, J.C.; Bürmann, F.; Petela, N.; Chan, K.-L.; Uluocak, P.; Beckouët, F.; Gruber, S.; Nasmyth, K.; Löwe, J. Closing the Cohesin Ring: Structure and Function of Its Smc3-Kleisin Interface. Science 2014, 346, 963–967. [Google Scholar] [CrossRef]
- Srinivasan, M.; Scheinost, J.C.; Petela, N.J.; Gligoris, T.G.; Wissler, M.; Ogushi, S.; Collier, J.E.; Voulgaris, M.; Kurze, A.; Chan, K.-L.; et al. The Cohesin Ring Uses Its Hinge to Organize DNA Using Non-Topological as Well as Topological Mechanisms. Cell 2018, 173, 1508–1519.e18. [Google Scholar] [CrossRef]
- Rao, S.S.P.; Huang, S.-C.; Glenn St Hilaire, B.; Engreitz, J.M.; Perez, E.M.; Kieffer-Kwon, K.-R.; Sanborn, A.L.; Johnstone, S.E.; Bascom, G.D.; Bochkov, I.D.; et al. Cohesin Loss Eliminates All Loop Domains. Cell 2017, 171, 305–320.e24. [Google Scholar] [CrossRef]
- Wutz, G.; Várnai, C.; Nagasaka, K.; Cisneros, D.A.; Stocsits, R.R.; Tang, W.; Schoenfelder, S.; Jessberger, G.; Muhar, M.; Hossain, M.J.; et al. Topologically Associating Domains and Chromatin Loops Depend on Cohesin and Are Regulated by CTCF, WAPL, and PDS5 Proteins. EMBO J. 2017, 36, 3573–3599. [Google Scholar] [CrossRef]
- Gibcus, J.H.; Samejima, K.; Goloborodko, A.; Samejima, I.; Naumova, N.; Nuebler, J.; Kanemaki, M.T.; Xie, L.; Paulson, J.R.; Earnshaw, W.C.; et al. A Pathway for Mitotic Chromosome Formation. Science 2018, 359, eaao6135. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Emerson, D.J.; Gilgenast, T.G.; Titus, K.R.; Lan, Y.; Huang, P.; Zhang, D.; Wang, H.; Keller, C.A.; Giardine, B.; et al. Chromatin Structure Dynamics during the Mitosis-to-G1 Phase Transition. Nature 2019, 576, 158–162. [Google Scholar] [CrossRef] [PubMed]
- Golov, A.K.; Gavrilov, A.A. Cohesin-Dependent Loop Extrusion: Molecular Mechanics and Role in Cell Physiology. Biochem. Mosc. 2024, 89, 601–625. [Google Scholar] [CrossRef]
- Sanborn, A.L.; Rao, S.S.P.; Huang, S.-C.; Durand, N.C.; Huntley, M.H.; Jewett, A.I.; Bochkov, I.D.; Chinnappan, D.; Cutkosky, A.; Li, J.; et al. Chromatin Extrusion Explains Key Features of Loop and Domain Formation in Wild-Type and Engineered Genomes. Proc. Natl. Acad. Sci. USA 2015, 112, E6456–E6465. [Google Scholar] [CrossRef]
- Fudenberg, G.; Imakaev, M.; Lu, C.; Goloborodko, A.; Abdennur, N.; Mirny, L.A. Formation of Chromosomal Domains by Loop Extrusion. Cell Rep. 2016, 15, 2038–2049. [Google Scholar] [CrossRef] [PubMed]
- Nuebler, J.; Fudenberg, G.; Imakaev, M.; Abdennur, N.; Mirny, L.A. Chromatin Organization by an Interplay of Loop Extrusion and Compartmental Segregation. Proc. Natl. Acad. Sci. USA 2018, 115, E6697–E6706. [Google Scholar] [CrossRef]
- Mach, P.; Kos, P.I.; Zhan, Y.; Cramard, J.; Gaudin, S.; Tünnermann, J.; Marchi, E.; Eglinger, J.; Zuin, J.; Kryzhanovska, M.; et al. Cohesin and CTCF Control the Dynamics of Chromosome Folding. Nat. Genet. 2022, 54, 1907–1918. [Google Scholar] [CrossRef] [PubMed]
- Gabriele, M.; Brandão, H.B.; Grosse-Holz, S.; Jha, A.; Dailey, G.M.; Cattoglio, C.; Hsieh, T.-H.S.; Mirny, L.; Zechner, C.; Hansen, A.S. Dynamics of CTCF- and Cohesin-Mediated Chromatin Looping Revealed by Live-Cell Imaging. Science 2022, 376, 496–501. [Google Scholar] [CrossRef]
- Sonoda, E.; Matsusaka, T.; Morrison, C.; Vagnarelli, P.; Hoshi, O.; Ushiki, T.; Nojima, K.; Fukagawa, T.; Waizenegger, I.C.; Peters, J.-M.; et al. Scc1/Rad21/Mcd1 Is Required for Sister Chromatid Cohesion and Kinetochore Function in Vertebrate Cells. Dev. Cell 2001, 1, 759–770. [Google Scholar] [CrossRef]
- Bauerschmidt, C.; Arrichiello, C.; Burdak-Rothkamm, S.; Woodcock, M.; Hill, M.A.; Stevens, D.L.; Rothkamm, K. Cohesin Promotes the Repair of Ionizing Radiation-Induced DNA Double-Strand Breaks in Replicated Chromatin. Nucleic Acids Res. 2010, 38, 477–487. [Google Scholar] [CrossRef]
- Meisenberg, C.; Pinder, S.I.; Hopkins, S.R.; Wooller, S.K.; Benstead-Hume, G.; Pearl, F.M.G.; Jeggo, P.A.; Downs, J.A. Repression of Transcription at DNA Breaks Requires Cohesin throughout Interphase and Prevents Genome Instability. Mol. Cell 2019, 73, 212–223.e7. [Google Scholar] [CrossRef] [PubMed]
- Fedkenheuer, M.; Shang, Y.; Jung, S.; Fedkenheuer, K.; Park, S.; Mazza, D.; Sebastian, R.; Nagashima, H.; Zong, D.; Tan, H.; et al. A Dual Role of Cohesin in DNA DSB Repair. Nat. Commun. 2025, 16, 843. [Google Scholar] [CrossRef]
- Gelot, C.; Guirouilh-Barbat, J.; Le Guen, T.; Dardillac, E.; Chailleux, C.; Canitrot, Y.; Lopez, B.S. The Cohesin Complex Prevents the End Joining of Distant DNA Double-Strand Ends. Mol. Cell 2016, 61, 15–26. [Google Scholar] [CrossRef]
- Barber, T.D.; McManus, K.; Yuen, K.W.Y.; Reis, M.; Parmigiani, G.; Shen, D.; Barrett, I.; Nouhi, Y.; Spencer, F.; Markowitz, S.; et al. Chromatid Cohesion Defects May Underlie Chromosome Instability in Human Colorectal Cancers. Proc. Natl. Acad. Sci. USA 2008, 105, 3443–3448. [Google Scholar] [CrossRef] [PubMed]
- Losada, A. Cohesin in Cancer: Chromosome Segregation and Beyond. Nat. Rev. Cancer 2014, 14, 389–393. [Google Scholar] [CrossRef]
- Horsfield, J.A. Full Circle: A Brief History of Cohesin and the Regulation of Gene Expression. FEBS J. 2023, 290, 1670–1687. [Google Scholar] [CrossRef] [PubMed]
- Dorsett, D.; Ström, L. The Ancient and Evolving Roles of Cohesin in Gene Expression and DNA Repair. Curr. Biol. 2012, 22, R240–R250. [Google Scholar] [CrossRef]
- Arnould, C.; Rocher, V.; Finoux, A.-L.; Clouaire, T.; Li, K.; Zhou, F.; Caron, P.; Mangeot, P.E.; Ricci, E.P.; Mourad, R.; et al. Loop Extrusion as a Mechanism for Formation of DNA Damage Repair Foci. Nature 2021, 590, 660–665. [Google Scholar] [CrossRef]
- Kim, S.-T.; Xu, B.; Kastan, M.B. Involvement of the Cohesin Protein, Smc1, in Atm-Dependent and Independent Responses to DNA Damage. Genes. Dev. 2002, 16, 560–570. [Google Scholar] [CrossRef]
- Kitagawa, R.; Bakkenist, C.J.; McKinnon, P.J.; Kastan, M.B. Phosphorylation of SMC1 Is a Critical Downstream Event in the ATM–NBS1–BRCA1 Pathway. Genes. Dev. 2004, 18, 1423–1438. [Google Scholar] [CrossRef]
- Watrin, E.; Peters, J.-M. The Cohesin Complex Is Required for the DNA Damage-Induced G2/M Checkpoint in Mammalian Cells. EMBO J. 2009, 28, 2625–2635. [Google Scholar] [CrossRef]
- Ma, H.; Tu, L.-C.; Naseri, A.; Chung, Y.-C.; Grunwald, D.; Zhang, S.; Pederson, T. CRISPR-Sirius: RNA Scaffolds for Signal Amplification in Genome Imaging. Nat. Methods 2018, 15, 928–931. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.; Tu, L.-C.; Chung, Y.-C.; Naseri, A.; Grunwald, D.; Zhang, S.; Pederson, T. Cell Cycle– and Genomic Distance–Dependent Dynamics of a Discrete Chromosomal Region. J. Cell Biol. 2019, 218, 1467–1477. [Google Scholar] [CrossRef] [PubMed]
- Chung, Y.-C.; Bisht, M.; Thuma, J.; Tu, L.-C. Single-Chromosome Dynamics Reveals Locus-Dependent Dynamics and Chromosome Territory Orientation. J. Cell Sci. 2023, 136, jcs260137. [Google Scholar] [CrossRef] [PubMed]
- Viushkov, V.S.; Lomov, N.A.; Rubtsov, M.A. A Comparison of Two Versions of the CRISPR-Sirius System for the Live-Cell Visualization of the Borders of Topologically Associating Domains. Cells 2024, 13, 1440. [Google Scholar] [CrossRef]
- Michaelis, C.; Ciosk, R.; Nasmyth, K. Cohesins: Chromosomal Proteins That Prevent Premature Separation of Sister Chromatids. Cell 1997, 91, 35–45. [Google Scholar] [CrossRef]
- Robinett, C.C.; Straight, A.; Li, G.; Willhelm, C.; Sudlow, G.; Murray, A.; Belmont, A.S. In Vivo Localization of DNA Sequences and Visualization of Large-Scale Chromatin Organization Using Lac Operator/Repressor Recognition. J. Cell Biol. 1996, 135, 1685–1700. [Google Scholar] [CrossRef]
- Dimitrova, N.; Chen, Y.-C.M.; Spector, D.L.; De Lange, T. 53BP1 Promotes Non-Homologous End Joining of Telomeres by Increasing Chromatin Mobility. Nature 2008, 456, 524–528. [Google Scholar] [CrossRef]
- Xu, H.; Balakrishnan, K.; Malaterre, J.; Beasley, M.; Yan, Y.; Essers, J.; Appeldoorn, E.; Thomaszewski, J.M.; Vazquez, M.; Verschoor, S.; et al. Rad21-Cohesin Haploinsufficiency Impedes DNA Repair and Enhances Gastrointestinal Radiosensitivity in Mice. PLoS ONE 2010, 5, e12112, Correction in PLoS ONE 2010, 5. https://doi.org/10.1371/annotation/12224797-353c-4e9c-92f3-a0de9b527415. [Google Scholar] [CrossRef]
- White, J.K.; Gerdin, A.-K.; Karp, N.A.; Ryder, E.; Buljan, M.; Bussell, J.N.; Salisbury, J.; Clare, S.; Ingham, N.J.; Podrini, C.; et al. Genome-Wide Generation and Systematic Phenotyping of Knockout Mice Reveals New Roles for Many Genes. Cell 2013, 154, 452–464. [Google Scholar] [CrossRef]
- Natsume, T.; Kiyomitsu, T.; Saga, Y.; Kanemaki, M.T. Rapid Protein Depletion in Human Cells by Auxin-Inducible Degron Tagging with Short Homology Donors. Cell Rep. 2016, 15, 210–218. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, K.; Fukagawa, T.; Takisawa, H.; Kakimoto, T.; Kanemaki, M. An Auxin-Based Degron System for the Rapid Depletion of Proteins in Nonplant Cells. Nat. Methods 2009, 6, 917–922. [Google Scholar] [CrossRef]
- Cremer, M.; Brandstetter, K.; Maiser, A.; Rao, S.S.P.; Schmid, V.J.; Guirao-Ortiz, M.; Mitra, N.; Mamberti, S.; Klein, K.N.; Gilbert, D.M.; et al. Cohesin Depleted Cells Rebuild Functional Nuclear Compartments after Endomitosis. Nat. Commun. 2020, 11, 6146. [Google Scholar] [CrossRef]
- Yesbolatova, A.; Natsume, T.; Hayashi, K.; Kanemaki, M.T. Generation of Conditional Auxin-Inducible Degron (AID) Cells and Tight Control of Degron-Fused Proteins Using the Degradation Inhibitor Auxinole. Methods 2019, 164–165, 73–80. [Google Scholar] [CrossRef]
- Yesbolatova, A.; Saito, Y.; Kitamoto, N.; Makino-Itou, H.; Ajima, R.; Nakano, R.; Nakaoka, H.; Fukui, K.; Gamo, K.; Tominari, Y.; et al. The Auxin-Inducible Degron 2 Technology Provides Sharp Degradation Control in Yeast, Mammalian Cells, and Mice. Nat. Commun. 2020, 11, 5701. [Google Scholar] [CrossRef]
- Li, S.; Prasanna, X.; Salo, V.T.; Vattulainen, I.; Ikonen, E. An Efficient Auxin-Inducible Degron System with Low Basal Degradation in Human Cells. Nat. Methods 2019, 16, 866–869. [Google Scholar] [CrossRef] [PubMed]
- Yunusova, A.M.; Smirnov, A.V.; Pristyazhnuk, I.E.; Shnaider, T.A.; Maltseva, E.K.; Afonnikova, S.D.; Gusev, O.A.; Battulin, N.R. Assessing Cell Lines with Inducible Depletion of Cohesin and Condensins Components through Analysis of Metaphase Chromosome Morphology. Vavilov J. Genet. Breed. 2024, 28, 138–147. [Google Scholar] [CrossRef]
- Liu, Y.; Dekker, J. CTCF–CTCF Loops and Intra-TAD Interactions Show Differential Dependence on Cohesin Ring Integrity. Nat. Cell Biol. 2022, 24, 1516–1527. [Google Scholar] [CrossRef]
- Carroll, J.; He, J.; Ding, S.; Fearnley, I.M.; Walker, J.E. TMEM70 and TMEM242 Help to Assemble the Rotor Ring of Human ATP Synthase and Interact with Assembly Factors for Complex I. Proc. Natl. Acad. Sci. USA 2021, 118, e2100558118. [Google Scholar] [CrossRef] [PubMed]
- Amitai, A.; Seeber, A.; Gasser, S.M.; Holcman, D. Visualization of Chromatin Decompaction and Break Site Extrusion as Predicted by Statistical Polymer Modeling of Single-Locus Trajectories. Cell Rep. 2017, 18, 1200–1214. [Google Scholar] [CrossRef]
- Shukron, O.; Seeber, A.; Amitai, A.; Holcman, D. Advances Using Single-Particle Trajectories to Reconstruct Chromatin Organization and Dynamics. Trends Genet. 2019, 35, 685–705. [Google Scholar] [CrossRef] [PubMed]
- Seeber, A.; Hauer, M.H.; Gasser, S.M. Chromosome Dynamics in Response to DNA Damage. Annu. Rev. Genet. 2018, 52, 295–319. [Google Scholar] [CrossRef]
- Fradet-Turcotte, A.; Canny, M.D.; Escribano-Díaz, C.; Orthwein, A.; Leung, C.C.Y.; Huang, H.; Landry, M.-C.; Kitevski-LeBlanc, J.; Noordermeer, S.M.; Sicheri, F.; et al. 53BP1 Is a Reader of the DNA-Damage-Induced H2A Lys 15 Ubiquitin Mark. Nature 2013, 499, 50–54. [Google Scholar] [CrossRef]
- Ochs, F.; Karemore, G.; Miron, E.; Brown, J.; Sedlackova, H.; Rask, M.-B.; Lampe, M.; Buckle, V.; Schermelleh, L.; Lukas, J.; et al. Stabilization of Chromatin Topology Safeguards Genome Integrity. Nature 2019, 574, 571–574. [Google Scholar] [CrossRef]
- Zgheib, O.; Pataky, K.; Brugger, J.; Halazonetis, T.D. An Oligomerized 53BP1 Tudor Domain Suffices for Recognition of DNA Double-Strand Breaks. Mol. Cell Biol. 2009, 29, 1050–1058. [Google Scholar] [CrossRef]
- Pommier, Y.; Leo, E.; Zhang, H.; Marchand, C. DNA Topoisomerases and Their Poisoning by Anticancer and Antibacterial Drugs. Chem. Biol. 2010, 17, 421–433. [Google Scholar] [CrossRef]
- Wu, C.-C.; Li, T.-K.; Farh, L.; Lin, L.-Y.; Lin, T.-S.; Yu, Y.-J.; Yen, T.-J.; Chiang, C.-W.; Chan, N.-L. Structural Basis of Type II Topoisomerase Inhibition by the Anticancer Drug Etoposide. Science 2011, 333, 459–462. [Google Scholar] [CrossRef] [PubMed]
- Nagasaka, K.; Davidson, I.F.; Stocsits, R.R.; Tang, W.; Wutz, G.; Batty, P.; Panarotto, M.; Litos, G.; Schleiffer, A.; Gerlich, D.W.; et al. Cohesin Mediates DNA Loop Extrusion and Sister Chromatid Cohesion by Distinct Mechanisms. Mol. Cell 2023, 83, 3049–3063.e6. [Google Scholar] [CrossRef]
- Ström, L.; Lindroos, H.B.; Shirahige, K.; Sjögren, C. Postreplicative Recruitment of Cohesin to Double-Strand Breaks Is Required for DNA Repair. Mol. Cell 2004, 16, 1003–1015. [Google Scholar] [CrossRef]
- Ünal, E.; Arbel-Eden, A.; Sattler, U.; Shroff, R.; Lichten, M.; Haber, J.E.; Koshland, D. DNA Damage Response Pathway Uses Histone Modification to Assemble a Double-Strand Break-Specific Cohesin Domain. Mol. Cell 2004, 16, 991–1002. [Google Scholar] [CrossRef]
- Ünal, E.; Heidinger-Pauli, J.M.; Koshland, D. DNA Double-Strand Breaks Trigger Genome-Wide Sister-Chromatid Cohesion Through Eco1 (Ctf7). Science 2007, 317, 245–248. [Google Scholar] [CrossRef] [PubMed]
- Ström, L.; Karlsson, C.; Lindroos, H.B.; Wedahl, S.; Katou, Y.; Shirahige, K.; Sjögren, C. Postreplicative Formation of Cohesion Is Required for Repair and Induced by a Single DNA Break. Science 2007, 317, 242–245. [Google Scholar] [CrossRef]
- Schmitz, J.; Watrin, E.; Lénárt, P.; Mechtler, K.; Peters, J.-M. Sororin Is Required for Stable Binding of Cohesin to Chromatin and for Sister Chromatid Cohesion in Interphase. Curr. Biol. 2007, 17, 630–636. [Google Scholar] [CrossRef] [PubMed]
- Kilic, S.; Lezaja, A.; Gatti, M.; Bianco, E.; Michelena, J.; Imhof, R.; Altmeyer, M. Phase Separation of 53BP1 Determines Liquid-like Behavior of DNA Repair Compartments. EMBO J. 2019, 38, e101379. [Google Scholar] [CrossRef]
- Shibata, A.; Jeggo, P.A. Roles for 53BP1 in the Repair of Radiation-Induced DNA Double Strand Breaks. DNA Repair. 2020, 93, 102915. [Google Scholar] [CrossRef]
- Zimmermann, M.; De Lange, T. 53BP1: Pro Choice in DNA Repair. Trends Cell Biol. 2014, 24, 108–117. [Google Scholar] [CrossRef]
- Panier, S.; Boulton, S.J. Double-Strand Break Repair: 53BP1 Comes into Focus. Nat. Rev. Mol. Cell Biol. 2014, 15, 7–18. [Google Scholar] [CrossRef]
- Dion, V.; Kalck, V.; Seeber, A.; Schleker, T.; Gasser, S.M. Cohesin and the Nucleolus Constrain the Mobility of Spontaneous Repair Foci. EMBO Rep. 2013, 14, 984–991. [Google Scholar] [CrossRef]
- Cheblal, A.; Challa, K.; Seeber, A.; Shimada, K.; Yoshida, H.; Ferreira, H.C.; Amitai, A.; Gasser, S.M. DNA Damage-Induced Nucleosome Depletion Enhances Homology Search Independently of Local Break Movement. Mol. Cell 2020, 80, 311–326.e4. [Google Scholar] [CrossRef]
- Dion, V.; Kalck, V.; Horigome, C.; Towbin, B.D.; Gasser, S.M. Increased Mobility of Double-Strand Breaks Requires Mec1, Rad9 and the Homologous Recombination Machinery. Nat. Cell Biol. 2012, 14, 502–509. [Google Scholar] [CrossRef]
- Miné-Hattab, J.; Rothstein, R. Increased Chromosome Mobility Facilitates Homology Search during Recombination. Nat. Cell Biol. 2012, 14, 510–517. [Google Scholar] [CrossRef]
- Soutoglou, E.; Dorn, J.F.; Sengupta, K.; Jasin, M.; Nussenzweig, A.; Ried, T.; Danuser, G.; Misteli, T. Positional Stability of Single Double-Strand Breaks in Mammalian Cells. Nat. Cell Biol. 2007, 9, 675–682. [Google Scholar] [CrossRef]
- Jakob, B.; Splinter, J.; Durante, M.; Taucher-Scholz, G. Live Cell Microscopy Analysis of Radiation-Induced DNA Double-Strand Break Motion. Proc. Natl. Acad. Sci. USA 2009, 106, 3172–3177. [Google Scholar] [CrossRef]
- Roukos, V.; Voss, T.C.; Schmidt, C.K.; Lee, S.; Wangsa, D.; Misteli, T. Spatial Dynamics of Chromosome Translocations in Living Cells. Science 2013, 341, 660–664. [Google Scholar] [CrossRef]
- Karanam, K.; Kafri, R.; Loewer, A.; Lahav, G. Quantitative Live Cell Imaging Reveals a Gradual Shift between DNA Repair Mechanisms and a Maximal Use of HR in Mid S Phase. Mol. Cell 2012, 47, 320–329. [Google Scholar] [CrossRef]
- Shahar, O.D.; Ram, E.V.S.R.; Shimshoni, E.; Hareli, S.; Meshorer, E.; Goldberg, M. Live Imaging of Induced and Controlled DNA Double-Strand Break Formation Reveals Extremely Low Repair by Homologous Recombination in Human Cells. Oncogene 2012, 31, 3495–3504. [Google Scholar] [CrossRef]
- Schrank, B.R.; Aparicio, T.; Li, Y.; Chang, W.; Chait, B.T.; Gundersen, G.G.; Gottesman, M.E.; Gautier, J. Nuclear ARP2/3 Drives DNA Break Clustering for Homology-Directed Repair. Nature 2018, 559, 61–66. [Google Scholar] [CrossRef] [PubMed]
- Isono, M.; Niimi, A.; Oike, T.; Hagiwara, Y.; Sato, H.; Sekine, R.; Yoshida, Y.; Isobe, S.-Y.; Obuse, C.; Nishi, R.; et al. BRCA1 Directs the Repair Pathway to Homologous Recombination by Promoting 53BP1 Dephosphorylation. Cell Rep. 2017, 18, 520–532. [Google Scholar] [CrossRef] [PubMed]
- Shibata, A. Regulation of Repair Pathway Choice at Two-Ended DNA Double-Strand Breaks. Mutat. Res./Fundam. Mol. Mech. Mutagen. 2017, 803–805, 51–55. [Google Scholar] [CrossRef] [PubMed]
- Babokhov, M.; Hibino, K.; Itoh, Y.; Maeshima, K. Local Chromatin Motion and Transcription. J. Mol. Biol. 2020, 432, 694–700. [Google Scholar] [CrossRef]
- Gu, B.; Swigut, T.; Spencley, A.; Bauer, M.R.; Chung, M.; Meyer, T.; Wysocka, J. Transcription-Coupled Changes in Nuclear Mobility of Mammalian Cis-Regulatory Elements. Science 2018, 359, 1050–1055. [Google Scholar] [CrossRef]
- Germier, T.; Kocanova, S.; Walther, N.; Bancaud, A.; Shaban, H.A.; Sellou, H.; Politi, A.Z.; Ellenberg, J.; Gallardo, F.; Bystricky, K. Real-Time Imaging of a Single Gene Reveals Transcription-Initiated Local Confinement. Biophys. J. 2017, 113, 1383–1394. [Google Scholar] [CrossRef]
- Shinkai, S.; Nozaki, T.; Maeshima, K.; Togashi, Y. Dynamic Nucleosome Movement Provides Structural Information of Topological Chromatin Domains in Living Human Cells. PLoS Comput. Biol. 2016, 12, e1005136. [Google Scholar] [CrossRef] [PubMed]
- Nozaki, T.; Imai, R.; Tanbo, M.; Nagashima, R.; Tamura, S.; Tani, T.; Joti, Y.; Tomita, M.; Hibino, K.; Kanemaki, M.T.; et al. Dynamic Organization of Chromatin Domains Revealed by Super-Resolution Live-Cell Imaging. Mol. Cell 2017, 67, 282–293.e7. [Google Scholar] [CrossRef]
- Nagashima, R.; Hibino, K.; Ashwin, S.S.; Babokhov, M.; Fujishiro, S.; Imai, R.; Nozaki, T.; Tamura, S.; Tani, T.; Kimura, H.; et al. Single Nucleosome Imaging Reveals Loose Genome Chromatin Networks via Active RNA Polymerase II. J. Cell Biol. 2019, 218, 1511–1530. [Google Scholar] [CrossRef]
- Daugird, T.A.; Shi, Y.; Holland, K.L.; Rostamian, H.; Liu, Z.; Lavis, L.D.; Rodriguez, J.; Strahl, B.D.; Legant, W.R. Correlative Single Molecule Lattice Light Sheet Imaging Reveals the Dynamic Relationship between Nucleosomes and the Local Chromatin Environment. Nat. Commun. 2024, 15, 4178. [Google Scholar] [CrossRef]
- Shaban, H.A.; Barth, R.; Recoules, L.; Bystricky, K. Hi-D: Nanoscale Mapping of Nuclear Dynamics in Single Living Cells. Genome Biol. 2020, 21, 95. [Google Scholar] [CrossRef]
- Nozaki, T.; Shinkai, S.; Ide, S.; Higashi, K.; Tamura, S.; Shimazoe, M.A.; Nakagawa, M.; Suzuki, Y.; Okada, Y.; Sasai, M.; et al. Condensed but Liquid-like Domain Organization of Active Chromatin Regions in Living Human Cells. Sci. Adv. 2023, 9, eadf1488. [Google Scholar] [CrossRef]
- Saxton, M.N.; Morisaki, T.; Krapf, D.; Kimura, H.; Stasevich, T.J. Live-Cell Imaging Uncovers the Relationship between Histone Acetylation, Transcription Initiation, and Nucleosome Mobility. Sci. Adv. 2023, 9, eadh4819. [Google Scholar] [CrossRef]
- Yunusova, A.; Smirnov, A.; Shnaider, T.; Lukyanchikova, V.; Afonnikova, S.; Battulin, N. Evaluation of the OsTIR1 and AtAFB2 AID Systems for Genome Architectural Protein Degradation in Mammalian Cells. Front. Mol. Biosci. 2021, 8, 757394. [Google Scholar] [CrossRef]
- Mali, P.; Yang, L.; Esvelt, K.M.; Aach, J.; Guell, M.; DiCarlo, J.E.; Norville, J.E.; Church, G.M. RNA-Guided Human Genome Engineering via Cas9. Science 2013, 339, 823–826. [Google Scholar] [CrossRef]
- Cong, L.; Ran, F.A.; Cox, D.; Lin, S.; Barretto, R.; Habib, N.; Hsu, P.D.; Wu, X.; Jiang, W.; Marraffini, L.A.; et al. Multiplex Genome Engineering Using CRISPR/Cas Systems. Science 2013, 339, 819–823. [Google Scholar] [CrossRef]
- Norrander, J.; Kempe, T.; Messing, J. Construction of Improved M13 Vectors Using Oligodeoxynucleotide-Directed Mutagenesis. Gene 1983, 26, 101–106, Erratum in Gene 1984, 27, 130. https://doi.org/10.1016/0378-1119(84)90249-X. [Google Scholar] [CrossRef] [PubMed]
- DeKelver, R.C.; Choi, V.M.; Moehle, E.A.; Paschon, D.E.; Hockemeyer, D.; Meijsing, S.H.; Sancak, Y.; Cui, X.; Steine, E.J.; Miller, J.C.; et al. Functional Genomics, Proteomics, and Regulatory DNA Analysis in Isogenic Settings Using Zinc Finger Nuclease-Driven Transgenesis into a Safe Harbor Locus in the Human Genome. Genome Res. 2010, 20, 1133–1142. [Google Scholar] [CrossRef]
- Stewart, S.A.; Dykxhoorn, D.M.; Palliser, D.; Mizuno, H.; Yu, E.Y.; An, D.S.; Sabatini, D.M.; Chen, I.S.Y.; Hahn, W.C.; Sharp, P.A.; et al. Lentivirus-Delivered Stable Gene Silencing by RNAi in Primary Cells. RNA 2003, 9, 493–501. [Google Scholar] [CrossRef]
- Lomov, N.A.; Viushkov, V.S.; Zamalutdinov, A.V.; Sboeva, M.D.; Rubtsov, M.A. Direct ENIT: An Easy and Reliable Tool for gRNA Efficacy Verification by Tracking Induced Chromosomal Translocation. MethodsX 2020, 7, 101104. [Google Scholar] [CrossRef]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An Open-Source Platform for Biological-Image Analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef]
- The Galaxy Community. The Galaxy Platform for Accessible, Reproducible, and Collaborative Data Analyses: 2024 Update. Nucleic Acids Res. 2024, 52, W83–W94. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast Gapped-Read Alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Ramirez, F.; Ryan, D.P.; Gruning, B.; Bhardwaj, V.; Kilpert, F.; Richter, A.S.; Heyne, S.; Dundar, F.; Manke, T. deepTools2: A next Generation Web Server for Deep-Sequencing Data Analysis. Nucleic Acids Res. 2016, 44, W160–W165. [Google Scholar] [CrossRef]
- Robinson, J.T.; Thorvaldsdóttir, H.; Winckler, W.; Guttman, M.; Lander, E.S.; Getz, G.; Mesirov, J.P. Integrative Genomics Viewer. Nat. Biotechnol. 2011, 29, 24–26. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, T.; Meyer, C.A.; Eeckhoute, J.; Johnson, D.S.; Bernstein, B.E.; Nusbaum, C.; Myers, R.M.; Brown, M.; Li, W.; et al. Model-Based Analysis of ChIP-Seq (MACS). Genome Biol. 2008, 9, R137. [Google Scholar] [CrossRef] [PubMed]
- Tinevez, J.-Y.; Perry, N.; Schindelin, J.; Hoopes, G.M.; Reynolds, G.D.; Laplantine, E.; Bednarek, S.Y.; Shorte, S.L.; Eliceiri, K.W. TrackMate: An Open and Extensible Platform for Single-Particle Tracking. Methods 2017, 115, 80–90. [Google Scholar] [CrossRef] [PubMed]
- Ershov, D.; Phan, M.-S.; Pylvänäinen, J.W.; Rigaud, S.U.; Le Blanc, L.; Charles-Orszag, A.; Conway, J.R.W.; Laine, R.F.; Roy, N.H.; Bonazzi, D.; et al. TrackMate 7: Integrating State-of-the-Art Segmentation Algorithms into Tracking Pipelines. Nat. Methods 2022, 19, 829–832. [Google Scholar] [CrossRef]
- Bronstein, I.; Israel, Y.; Kepten, E.; Mai, S.; Shav-Tal, Y.; Barkai, E.; Garini, Y. Transient Anomalous Diffusion of Telomeres in the Nucleus of Mammalian Cells. Phys. Rev. Lett. 2009, 103, 018102. [Google Scholar] [CrossRef] [PubMed]


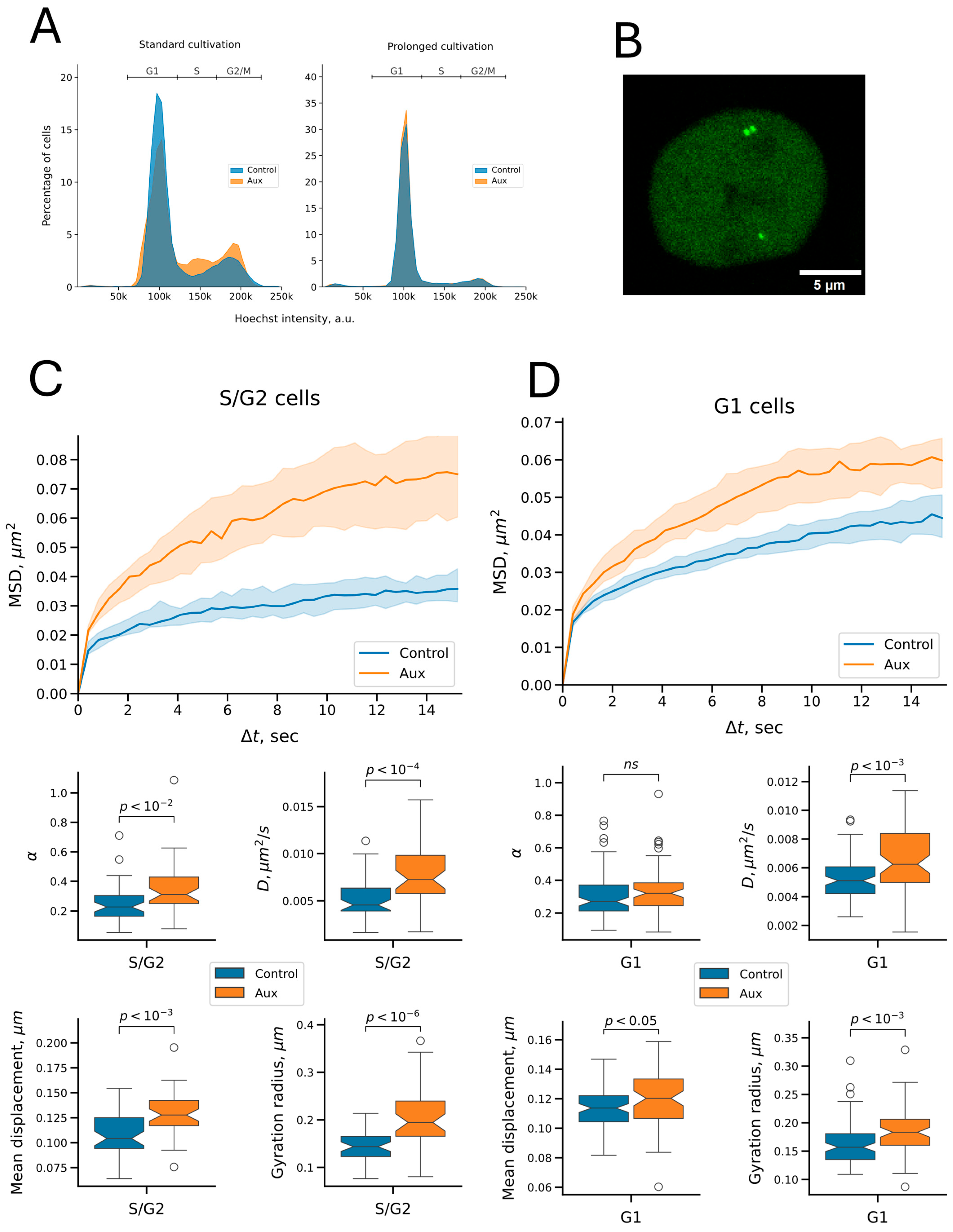
| Parameter | Chromatin State | Untreated Cells | Auxin Treatment | Median Fold Change (Aux/Control) | p Adjusted 1 |
|---|---|---|---|---|---|
| Anomalous exponent | unreplicated | 0.27 | 0.32 | 1.19 | 1.443 × 10−1 |
| replicated | 0.23 | 0.31 | 1.37 | 1.616 × 10−3 | |
| Diffusion coefficient, μm2/s | unreplicated | 0.0051 | 0.0062 | 1.23 | 6.470 × 10−4 |
| replicated | 0.0046 | 0.0072 | 1.59 | 5.203 × 10−5 | |
| Mean displacement, μm | unreplicated | 0.11 | 0.12 | 1.06 | 1.470 × 10−2 |
| replicated | 0.10 | 0.13 | 1.22 | 6.470 × 10−4 | |
| Gyration radius, μm | unreplicated | 0.16 | 0.18 | 1.17 | 6.470 × 10−4 |
| replicated | 0.14 | 0.19 | 1.35 | 8.067 × 10−7 |
| Parameter | Chromatin State | Treatment | C6 Locus | BP1-2 Foci | p-Value 1 | p Adjusted 2 |
|---|---|---|---|---|---|---|
| Anomalous exponent | unreplicated | control | 0.270573 | 0.178984 | 3.551 × 10−7 | 1.065 × 10−6 |
| unreplicated | auxins | 0.321870 | 0.184729 | 1.863 × 10−8 | 7.453 × 10−8 | |
| replicated | control | 0.227226 | 0.170984 | 4.427 × 10−3 | 4.427 × 10−3 | |
| replicated | auxins | 0.311817 | 0.187843 | 1.321 × 10−6 | 2.641 × 10−6 | |
| Diffusion coefficient, μm2/s | unreplicated | control | 0.005100 | 0.001881 | 7.646 × 10−19 | 1.147 × 10−17 |
| unreplicated | auxins | 0.006248 | 0.001900 | 4.930 × 10−15 | 5.916 × 10−14 | |
| replicated | control | 0.004559 | 0.001874 | 7.969 × 10−11 | 4.781 × 10−10 | |
| replicated | auxins | 0.007236 | 0.001970 | 1.104 × 10−11 | 7.729 × 10−11 | |
| Mean displacement, μm | unreplicated | control | 0.113697 | 0.075425 | 1.342 × 10−18 | 1.879 × 10−17 |
| unreplicated | auxins | 0.120304 | 0.075046 | 1.171 × 10−14 | 1.288 × 10−13 | |
| replicated | control | 0.104229 | 0.072040 | 6.333 × 10−10 | 3.167 × 10−9 | |
| replicated | auxins | 0.127673 | 0.077045 | 8.661 × 10−12 | 6.929 × 10−11 | |
| Gyration radius 3, μm | unreplicated | control | 0.141783 | 0.084200 | 2.831 × 10−19 | 4.529 × 10−18 |
| unreplicated | auxins | 0.172551 | 0.081000 | 3.094 × 10−15 | 4.022 × 10−14 | |
| replicated | control | 0.129927 | 0.081016 | 4.896 × 10−12 | 4.896 × 10−11 | |
| replicated | auxins | 0.183876 | 0.085393 | 6.255 × 10−12 | 5.629 × 10−11 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Viushkov, V.S.; Lomov, N.A.; Kalitina, P.O.; Potashnikova, D.M.; Shtompel, A.S.; Ulianov, S.V.; Razin, S.V.; Rubtsov, M.A. The Influence of Cohesin on the Short-Scale Dynamics of Intact and Damaged Chromatin in Different Phases of the Cell Cycle. Int. J. Mol. Sci. 2025, 26, 8837. https://doi.org/10.3390/ijms26188837
Viushkov VS, Lomov NA, Kalitina PO, Potashnikova DM, Shtompel AS, Ulianov SV, Razin SV, Rubtsov MA. The Influence of Cohesin on the Short-Scale Dynamics of Intact and Damaged Chromatin in Different Phases of the Cell Cycle. International Journal of Molecular Sciences. 2025; 26(18):8837. https://doi.org/10.3390/ijms26188837
Chicago/Turabian StyleViushkov, Vladimir S., Nikolai A. Lomov, Polina O. Kalitina, Daria M. Potashnikova, Anastasia S. Shtompel, Sergey V. Ulianov, Sergey V. Razin, and Mikhail A. Rubtsov. 2025. "The Influence of Cohesin on the Short-Scale Dynamics of Intact and Damaged Chromatin in Different Phases of the Cell Cycle" International Journal of Molecular Sciences 26, no. 18: 8837. https://doi.org/10.3390/ijms26188837
APA StyleViushkov, V. S., Lomov, N. A., Kalitina, P. O., Potashnikova, D. M., Shtompel, A. S., Ulianov, S. V., Razin, S. V., & Rubtsov, M. A. (2025). The Influence of Cohesin on the Short-Scale Dynamics of Intact and Damaged Chromatin in Different Phases of the Cell Cycle. International Journal of Molecular Sciences, 26(18), 8837. https://doi.org/10.3390/ijms26188837






