Claspin and Cancer: Where Are We Now?
Abstract
1. Introduction
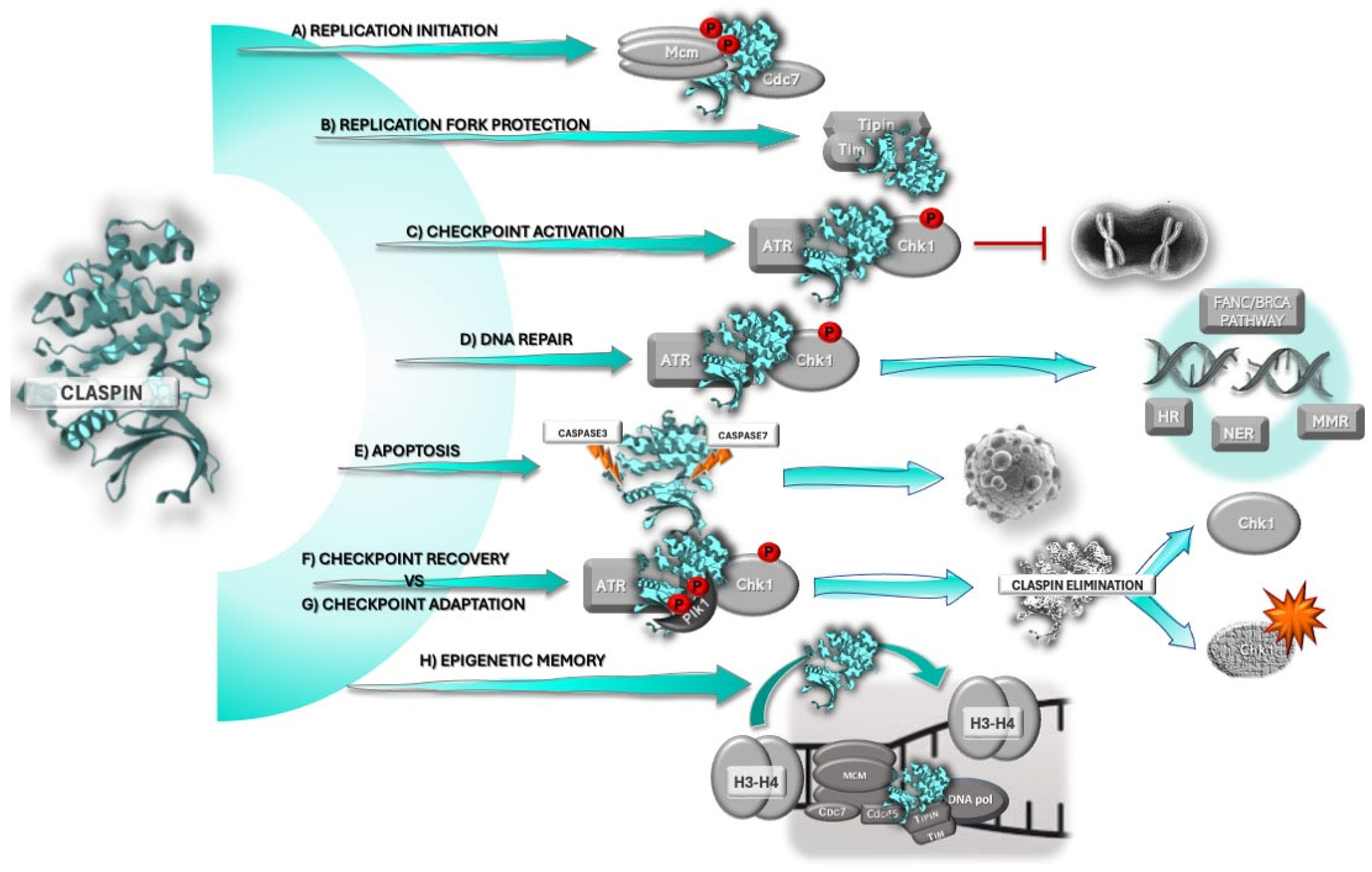
2. Claspin
2.1. Claspin and the DNA Damage Response
2.2. Claspin’s Role in DNA Replication and Replication Stress
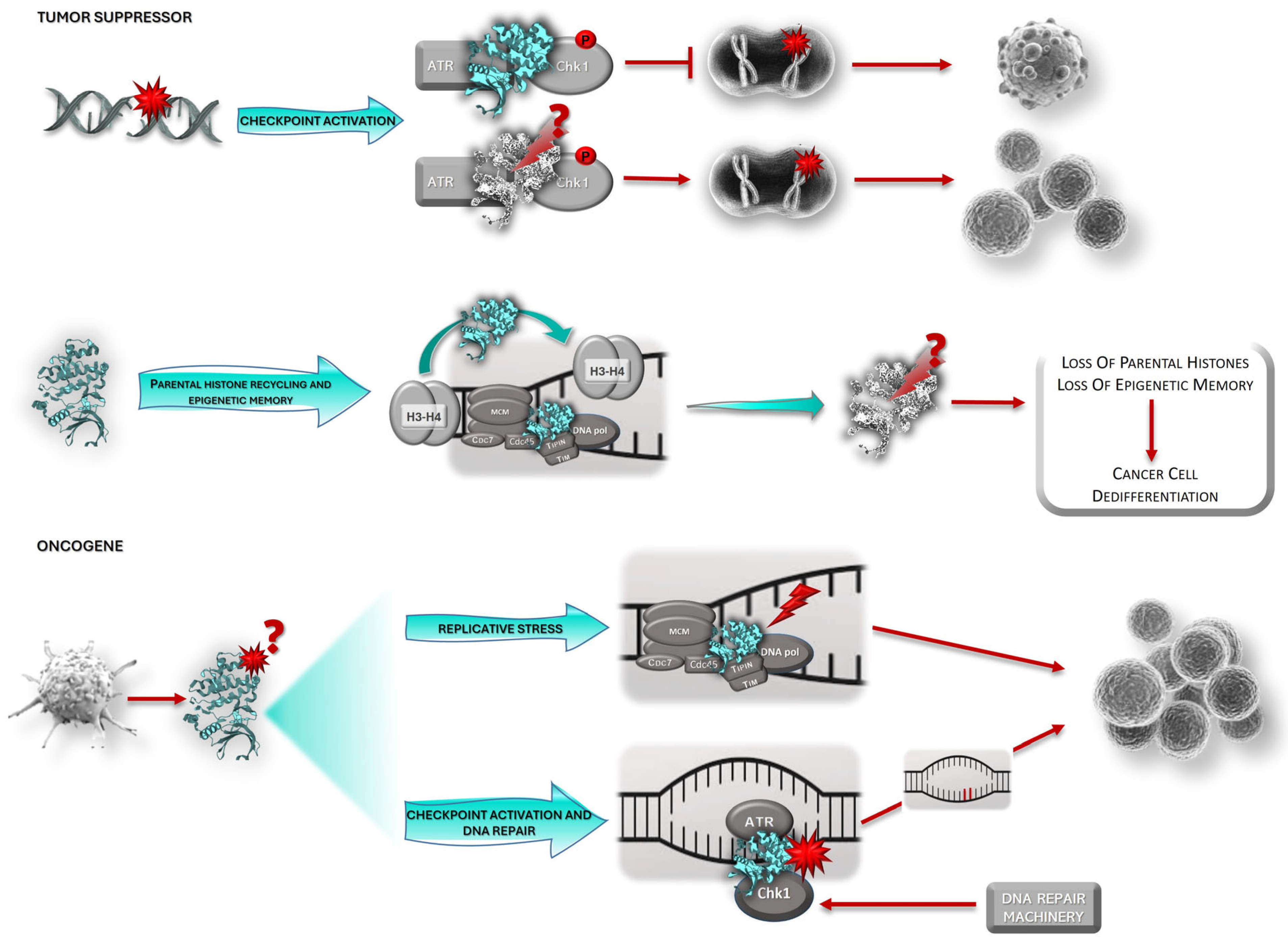
3. Claspin and Cancer
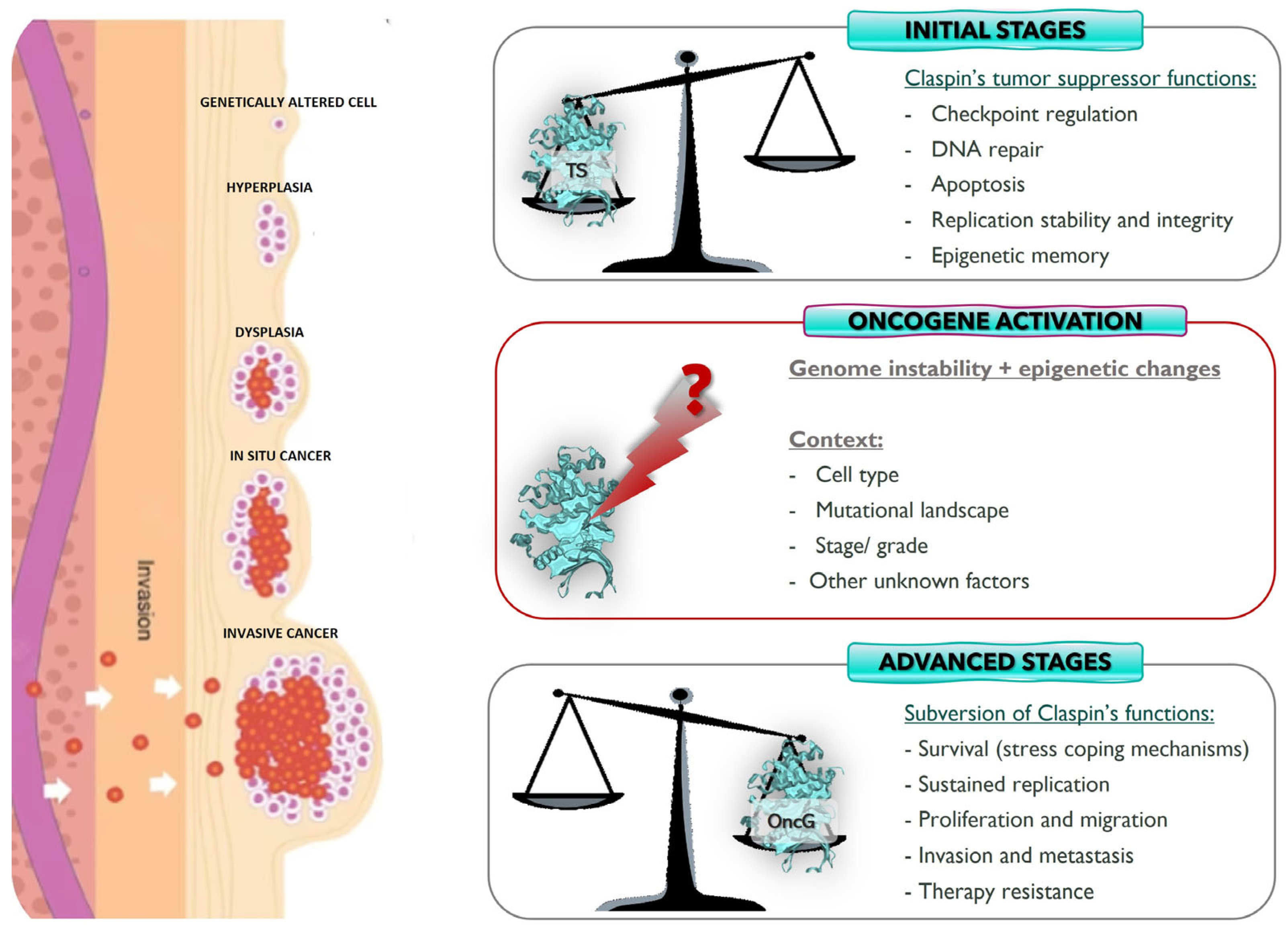
3.1. Claspin: Tumor Suppressor or Oncogene?
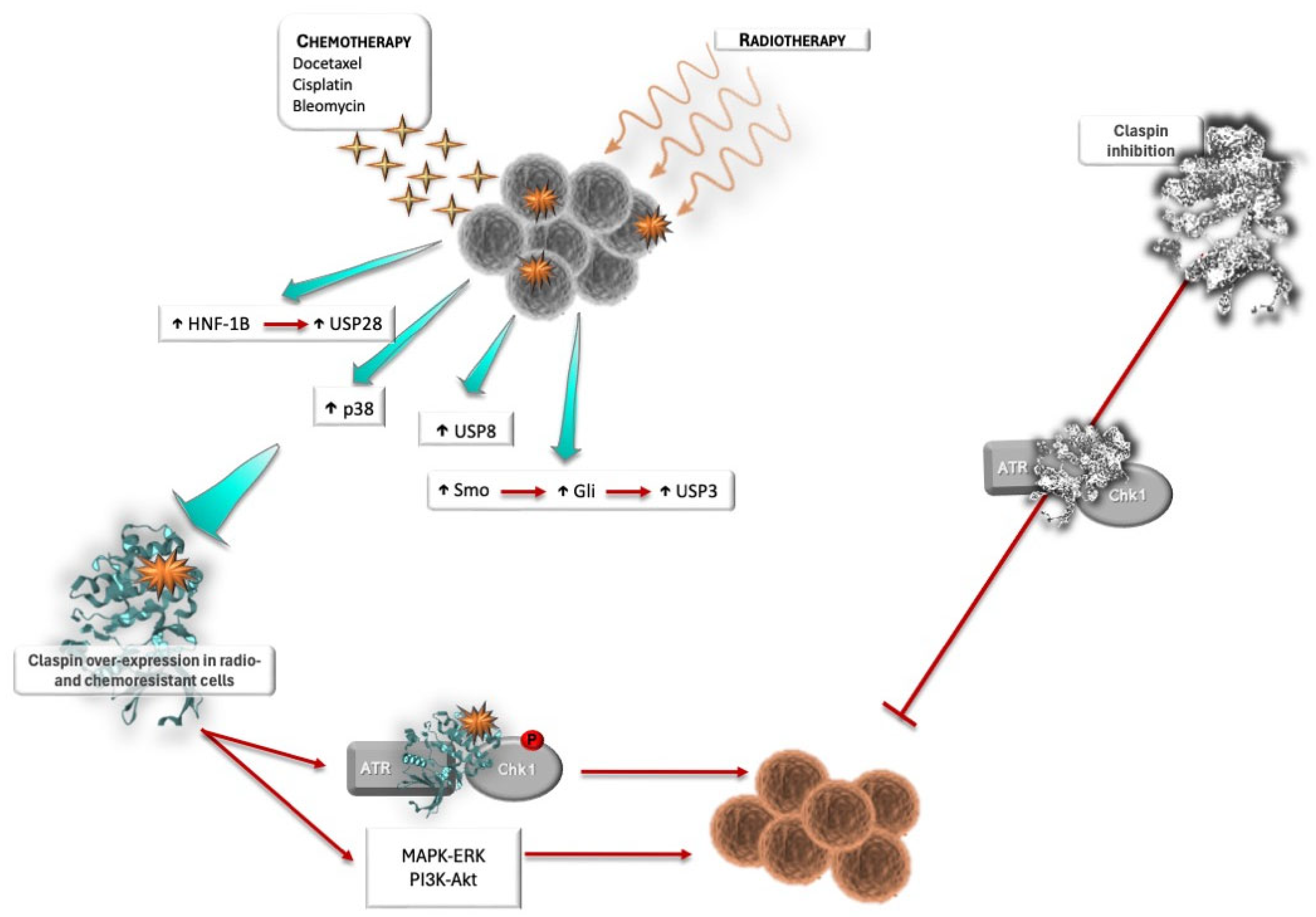
3.2. Claspin and Response to Therapy
4. Final Thoughts and Perspectives
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| AKT | AK strain transforming |
| ALDH | Aldehyde dehydrogenase |
| APC | Anaphase promoting complex |
| ATM | Ataxia telangiectasia mutated |
| ATR | Ataxia telangiectasia and Rad3-related protein |
| ATRIP | ATR interacting protein |
| BARD1 | BRCA1-Associated RING Domain 1 |
| BRCA1 | Breast Cancer gene 1 |
| Bub1 | Budding uninhibited by benzimidazoles 1 |
| Cdc | Cell division cycle |
| Cdh1 | Cdc20 homolog 1 |
| CDKs | Cyclin-dependent kinases |
| Chk1/2 | Checkpoint kinase 1/2 |
| CK1γ1 | Casein kinase 1 gamma 1 |
| DDB | Damaged DNA binding protein |
| EGFR | Epidermal Growth Factor Receptor |
| ERK | Extracellular signal-regulated kinase |
| ETAA1 | Ewing’s tumor-associated antigen 1 |
| FANC | Fanconi |
| FANCD2 | Fanconi Anemia Complementation Group D2 |
| GSK3-β | Glycogen Synthase Kinase 3-beta |
| HER2 | Human Epidermal Growth Factor Receptor 2 |
| HNF-1β | hepatocyte nuclear factor 1-beta |
| Ki67 | Antigen Kiel 67 |
| MAPK | Mitogen-activated protein kinases |
| MCM | Mini-chromosome maintenance |
| MSH6 | MutS homolog 6 |
| mTOR | Mammalian target of Rapamycin |
| OZF | Only Zinc-Finger |
| PCNA | Proliferating cell nuclear antigen |
| PDK1 | Phosphoinositide-dependent kinase 1 |
| PD-L1 | Programmed death ligand 1 |
| PI3K | Phosphoinositide 3-kinase |
| PMS2 | Postmeiotic segregation increased 2 |
| Pol/POL | Polymerases |
| pRb | Retinoblastoma protein |
| RAD51 | DNA repair protein |
| Ras | Rat sarcoma oncogene |
| RFC | Replication Factor C |
| RPA | Replication protein A |
| SCFβTrCP | Skp1-Cullin-F-box-protein ubiquitin ligase complex |
| SLF2 | SMC5-SMC6 Complex Localization Factor 2 |
| STAT3 | Signal transducer and activator of transcription 3 |
| Tim | Timeless |
| TOPBP1 | Topoisomerase II binding protein 1 |
| TRIM21 | Tripartite motif protein 21 |
| USP | Ubiquitin-specific proteases |
| 3′-UTR | 3′-untranslated region |
| XPC | Xeroderma pigmentosum protein C |
References
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Hanahan, D. Hallmarks of cancer: New dimensions. Cancer Discov. 2022, 12, 31–46. [Google Scholar] [CrossRef] [PubMed]
- Jones, R.M.; Petermann, E. Replication fork dynamics and the DNA damage response. Biochem. J. 2012, 443, 13–26. [Google Scholar] [CrossRef] [PubMed]
- Atemin, A.; Ivanona, A.; Kanev, P.-B.; Uzunova, S.; Nedelcheva-Veleva, M.; Stoynov, S. Dynamics of replication-associated protein levels through the cell cycle. Int. J. Mol. Sci. 2024, 25, 8230. [Google Scholar] [CrossRef] [PubMed]
- Kumagai, A.; Dunphy, W.G. Claspin, a novel protein required for the activation of Chk1 during a DNA replication checkpoint response in Xenopus egg extracts. Mol. Cell 2000, 6, 839–849. [Google Scholar] [CrossRef]
- Azenha, D.; Lopes, M.C.; Martins, T.C. Claspin functions in cell homeostasis—A link to cancer? DNA Repair 2017, 59, 27–33. [Google Scholar] [CrossRef]
- Clarke, C.A.; Bennett, L.N.; Clarke, P.R. Cleavage of claspin by caspase-7 during apoptosis inhibits the Chk1 pathway. J. Biol. Chem. 2005, 280, 35337–35345. [Google Scholar] [CrossRef]
- Semple, J.I.; Smits, V.A.; Fernaud, J.R.; Mamely, I.; Freire, R. Cleavage and degradation of Claspin during apoptosis by caspases and the proteasome. Cell Death Differ. 2007, 14, 1433–1442. [Google Scholar] [CrossRef]
- Masai, H.; Yang, C.C.; Matsumoto, S. Mrc1/Claspin: A new role for regulation of origin firing. Curr. Genet. 2017, 63, 813–818. [Google Scholar] [CrossRef]
- Smith-Roe, S.L.; Patel, S.S.; Zhou, Y.; Simpson, D.A.; Rao, S.; Ibrahim Cordeiro-Stone, M.; Kaufmann, W.K. Separation of intra-S checkpoint protein contributions to DNA replication fork protection and genomic stability in normal human fibroblasts. Cell Cycle 2013, 12, 332–345. [Google Scholar] [CrossRef]
- Charlton, S.J.; Flury, V.; Kanoh, Y.; Genzor, A.V.; Kollenstart, L.; Ao, W.; Brogger, P.; Weisser, M.B.; Adamus, M.; Alcaraz, N.; et al. The fork protection complex promotes parental histone recycling and epigenetic memory. Cell 2024, 187, 5029–5047. [Google Scholar] [CrossRef] [PubMed]
- Smits, V.A.; Gillespie, D.A. DNA damage control: Regulation and functions of checkpoint kinase 1. FEBS J. 2015, 282, 3681–3692. [Google Scholar] [CrossRef] [PubMed]
- Chini, C.C.; Chen, J. Human claspin is required for replication checkpoint control. J. Biol. Chem. 2003, 278, 30057–30062. [Google Scholar] [CrossRef] [PubMed]
- Madgwick, S.; Luli, S.; Sellier, H.; Butterworth, J.A.; Leslie, J.; Moore, A.J.; Corbin, E.K.; Yemm, A.I.; Chiremba, R.T.; Tiniakos, D.; et al. Claspin haploinsufficiency leads to defects in fertility, hyperplasia and an increased oncogenic potential. Biochem. J. 2022, 479, 2115–2130. [Google Scholar] [CrossRef]
- Deng, C.-X.; Xu, X. Generation and analysis of Brca1 conditional knockout mice. Methods Mol. Biol. 2004, 280, 185–200. [Google Scholar] [CrossRef]
- Dine, J.; Deng, C.-X. Mouse models of BRCA1 and their application to breast cancer research. Cancer Metastasis Rev. 2013, 32, 25–37. [Google Scholar] [CrossRef]
- Liu, Q.; Guntuku, S.; Cui, X.S.; Matsuoka, S.; Cortez, D.; Tamai, K.; Luo, G.; Carattini-Rivera, S.; DeMayo, F.; Bradley, A.; et al. Chk1 is an essential kinase that is regulated by Atr and required for the G(2)/M DNA damage checkpoint. Genes Dev. 2000, 14, 1448–1459. [Google Scholar] [CrossRef]
- Takai, H.; Tominaga, K.; Motoyama, N.; Minamishima, Y.A.; Nagahama, H.; Tsukiyama, T.; Ikeda, K.; Nakayama, K.; Nakanishi, M.; Nakayama, K. Aberrant cell cycle checkpoint function and early embryonic death in Chk1(−/−) mice. Genes Dev. 2000, 14, 1439–1447. [Google Scholar] [CrossRef]
- Aze, A.; Zhou, J.C.; Costa, A.; Costanzo, V. DNA replication and homologous recombination factors: Acting together to maintain genome stability. Chromosoma 2013, 122, 401–413. [Google Scholar] [CrossRef]
- Jones, M.L.; Baris, Y.; Taylor, M.R.G.; Yeeles, J.T.P. Structure of a human replisome shows the organization and interactions of a DNA replication machine. Embo J. 2021, 40, e108819. [Google Scholar] [CrossRef]
- Pellegrini, L. The CMG DNA helicase and the core replisome. Curr. Opin. Struct. Biol. 2023, 81, 102612. [Google Scholar] [CrossRef] [PubMed]
- Petermann, E.; Helleday, T.; Caldecott, K.W. Claspin promotes normal replication fork rates in human cells. Mol. Biol. Cell 2008, 19, 2373–2378. [Google Scholar] [CrossRef]
- Guervilly, J.H.; Mace-Aime, G.; Rosselli, F. Loss of CHK1 function impedes DNA damage-induced FANCD2 monoubiquitination but normalizes the abnormal G2 arrest in Fanconi anemia. Hum. Mol. Genet. 2008, 17, 679–689. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Fang, Y.; Shao, H.; Lindsey-Boltz, L.; Sancar, A.; Modrich, P. Interactions of human mismatch repair proteins MutSα and MutLα with proteins of the ATR-Chk1 pathway. J. Biol. Chem. 2010, 285, 5974–5982. [Google Scholar] [CrossRef] [PubMed]
- Prætorius-Ibba, M.; Wang, Q.-E.; Wani, G.; El-Mahdy, M.A.; Zhu, Q.; Qin, S.; Wani, A.A. Role of Claspin in regulation of nucleotide excision repair factor DDB2. DNA Repair 2007, 6, 578–587. [Google Scholar] [CrossRef]
- Mailand, N.; Bekker-Jensen, S.; Bartek, J.; Lukas, J. Destruction of claspin by SCFβTrCP restrains Chk1 activation and facilitates recovery from genotoxic stress. Mol. Cell 2006, 23, 307–318. [Google Scholar] [CrossRef]
- Mamely, I.; van Vugt, M.A.; Smits, V.A.; Semple, J.I.; Lemmens, B.; Perrakis, A.; Medema, R.H.; Freire, R. Polo-like kinase-1 controls proteasome-dependent degradation of claspin during checkpoint recovery. Curr. Biol. 2006, 16, 1950–1955. [Google Scholar] [CrossRef]
- Peschiaroli, A.; Dorrello, N.V.; Guardavaccaro, D.; Venere, M.; Halazonetis, T.; Sherman, N.E.; Pagano, M. SCFβTrCP-mediated degradation of claspin regulates recovery from the DNA replication checkpoint response. Mol. Cell 2006, 23, 319–329. [Google Scholar] [CrossRef]
- Bennett, L.N.; Clarke, P.R. Regulation of Claspin degradation by the ubiquitin-proteasome pathway during the cell cycle and in response to ATR-dependent checkpoint activation. FEBS Lett. 2006, 580, 4176–4181. [Google Scholar] [CrossRef]
- Faustrup, H.; Bekker-Jensen, S.; Bartek, J.; Lukas, J.; Mailand, N. USP7 counteracts SCFβTrCP- but not APC/Cdh1-mediated proteolysis of Claspin. J. Cell Biol. 2009, 184, 13–19. [Google Scholar] [CrossRef]
- Martín, Y.; Cabrera, E.; Amoedo, H.; Hernández-Pérez, S.; Domínguez-Kelly, R.; Freire, R. USP29 controls the stability of checkpoint adaptor Claspin by deubiquitination. Oncogene 2015, 34, 1058–1063. [Google Scholar] [CrossRef]
- McGarry, E.; Gaboriau, D.; Rainey, M.D.; Restuccia, U.; Bachi, A.; Santocanale, C. The deubiquitinase USP9X maintains DNA replication fork stability and DNA damage checkpoint responses by regulating CLASPIN during S-phase. Cancer Res. 2016, 76, 2384–2393. [Google Scholar] [CrossRef]
- Zhang, D.; Zaugg, K.; Mak, T.W.; Elledge, S.J. A role for the deubiquitinating enzyme USP28 in control of the DNA-damage response. Cell 2006, 126, 529–542. [Google Scholar] [CrossRef] [PubMed]
- Lee, T.H.; Choi, J.Y.; Park, J.M.; Kang, T.H. Posttranscriptional control of the RS response via TTP-mediated Claspin mRNA stabilization. Oncogene 2020, 39, 3245–3257. [Google Scholar] [CrossRef] [PubMed]
- Awasthi, P.; Foiani, M.; Kumar, A. ATM and ATR signaling at a glance. J. Cell Sci. 2015, 128, 4255–4262. [Google Scholar] [CrossRef] [PubMed]
- Marechal, A.; Zou, L. RPA-coated single-stranded DNA as a platform for post-translational modifications in the DNA damage response. Cell Res. 2015, 25, 9–23. [Google Scholar] [CrossRef]
- Smith, H.L.; Southgate, H.; Tweddle, D.A.; Curtin, N.J. DNA damage checkpoint kinases in cancer. Expert Rev. Mol. Med. 2020, 22, e2. [Google Scholar] [CrossRef]
- Marechal, A.; Zou, L. DNA damage sensing by the ATM and ATR kinases. Cold Spring Harb. Perspect. Biol. 2013, 5, a012716. [Google Scholar] [CrossRef]
- Gonzalez Besteiro, M.A.; Gottifredi, V. The fork and the kinase: A DNA replication tale from a CHK1 perspective. Mutat. Res./Rev. Mutat. Res. 2015, 763, 168–180. [Google Scholar] [CrossRef]
- Clarke, C.A.L.; Clarke, P.R. DNA-dependent phosphorylation of Chk1 and Claspin in a human cell-free system. Biochem. J. 2005, 388, 705–712. [Google Scholar] [CrossRef]
- Lin, S.Y.; Li, K.; Stewart, G.S.; Elledge, S.J. Human Claspin works with BRCA1 to both positively and negatively regulate cell proliferation. Proc. Natl. Acad. Sci. USA 2004, 101, 6484–6489. [Google Scholar] [CrossRef] [PubMed]
- Yoo, H.Y.; Jeong, S.-Y.; Dunphy, W.G. Site-specific phosphorylation of a checkpoint mediator protein controls its responses to different DNA structures. Genes Dev. 2006, 20, 772–783. [Google Scholar] [CrossRef] [PubMed]
- Azenha, D.; Lopes, M.C.; Martins, T.C. Claspin: From replication stress and DNA damage responses to cancer therapy. Adv. Protein Chem. Struct. Biol. 2019, 115, 203–246. [Google Scholar] [CrossRef] [PubMed]
- Bahassi, E.M.; Ovesen, J.L.; Riesenberg, A.L.; Bernstein, W.Z.; Hasty, P.E.; Stambrook, P.J. The checkpoint kinases Chk1 and Chk2 regulate the functional associations between hBRCA2 and Rad51 in response to DNA damage. Oncogene 2008, 27, 3977–3985. [Google Scholar] [CrossRef]
- Sørensen, C.S.; Hansen, L.T.; Dziegielewski, J.; Syljuåsen, R.G.; Lundin, C.; Bartek, J.; Helleday, T. The cell-cycle checkpoint kinase Chk1 is required for mammalian homologous recombination repair. Nat. Cell Biol. 2005, 7, 195–201. [Google Scholar] [CrossRef]
- Sato, K.; Sundaramoorthy, E.; Rajendra, E.; Hattori, H.; Jeyasekharan, A.D.; Ayoub, N.; Shiess, R.; Aebersold, R.; Nishikawa, H.; Sedukhina, A.S.; et al. A DNA-damage selective role for BRCA1 E3 ligase in Claspin ubiquitylation, CHK1 activation, and DNA repair. Curr. Biol. 2012, 22, 1659–1666. [Google Scholar] [CrossRef]
- Yoo, H.Y.; Kumagai, A.; Shevchenko, A.; Shevchenko, A.; Dunphy, W.G. Adaptation of a DNA replication checkpoint response depends upon inactivation of Claspin by the Polo-like kinase. Cell 2004, 117, 575–588. [Google Scholar] [CrossRef]
- Syljuåsen, R.G.; Jensen, S.; Bartek, J.; Lukas, J. Adaptation to the ionizing radiation-induced G2 checkpoint occurs in human cells and depends on checkpoint kinase 1 and Polo-like kinase 1 kinases. Cancer Res. 2006, 66, 10253–10257. [Google Scholar] [CrossRef]
- Scorah, J.; McGowan, C.H. Claspin and Chk1 regulate replication fork stability by different mechanisms. Cell Cycle 2009, 8, 1036–1043. [Google Scholar] [CrossRef]
- Lee, J.; Gold, D.A.; Shevchenko, A.; Shevchenko, A.; Dunphy, W.G. Roles of replication fork-interacting and Chk1-activating domains from Claspin in a DNA replication checkpoint response. Mol. Biol. Cell 2005, 16, 5269–5282. [Google Scholar] [CrossRef]
- Uno, S.; Masai, H. Efficient expression and purification of human replication fork-stabilizing factor, Claspin, from mammalian cells: DNA-binding activity and novel protein interactions. Genes Cells 2011, 16, 842–856. [Google Scholar] [CrossRef]
- Sar, F.; Lindsey-Boltz, L.A.; Subramanian, D.; Croteau, D.L.; Hutsell, S.Q.; Griffith, J.D.; Sancar, A. Human Claspin is a ring-shaped DNA-binding protein with high affinity to branched DNA structures. J. Biol. Chem. 2004, 279, 39289–39295. [Google Scholar] [CrossRef] [PubMed]
- Broderick, R.; Rainey, M.D.; Santocanale, C.; Nasheuer, H.P. Cell cycle-dependent formation of Cdc45-Claspin complexes in human cells is compromised by UV-mediated DNA damage. FEBS J. 2013, 280, 4888–4902. [Google Scholar] [CrossRef] [PubMed]
- Feu, S.; Unzueta, F.; Llopis, A.; Semple, J.I.; Ercilla, A.; Guaita-Esteruelas, S.; Jaumont, M.; Freire, R.; Agell, N. OZF is a Claspin-interacting protein essential to maintain the replication fork progression rate under RS. FASEB J. 2020, 34, 6907–6919. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.C.; Suzuki, M.; Yamakawa, S.; Uno, S.; Ishii, A.; Yamazaki, S.; Fukatsu, R.; Fujisawa, R.; Sakimura, K.; Tsurimoto, T.; et al. Claspin recruits Cdc7 kinase for initiation of DNA replication in human cells. Nat. Commun. 2016, 7, 12135. [Google Scholar] [CrossRef]
- Smith-Roe, S.L.; Patel, S.S.; Simpson, D.A.; Zhou, Y.C.; Rao, S.; Ibrahim, J.G.; Kaiser-Rogers, K.A.; Cordeiro-Stone, M.; Kaufmann, W.K. Timeless functions independently of the Tim-Tipin complex to promote sister chromatid cohesion in normal human fibroblasts. Cell Cycle 2011, 10, 1618–1624. [Google Scholar] [CrossRef]
- Liu, G.; Chen, X.; Gao, Y.; Lewis, T.; Barthelemy, J.; Leffak, M. Altered replication in human cells promotes DMPK (CTG)(n) (CAG)(n) repeat instability. Mol. Cell. Biol. 2012, 32, 1618–1632. [Google Scholar] [CrossRef]
- Yang, X.H.; Shiotani, B.; Classon, M.; Zou, L. Chk1 and Claspin potentiate PCNA ubiquitination. Genes Dev. 2008, 22, 1147–1152. [Google Scholar] [CrossRef]
- Focarelli, M.L.; Soza, S.; Mannini, L.; Paulis, M.; Montecucco, A.; Musio, A. Claspin inhibition leads to fragile site expression. Genes Chrom. Canc. 2009, 48, 1083–1090. [Google Scholar] [CrossRef]
- Dillon, L.W.; Burrow, A.A.; Wang, Y.H. DNA instability at chromosomal fragile sites in cancer. Curr. Genom. 2010, 11, 326–337. [Google Scholar] [CrossRef]
- Casper, A.M.; Nghiem, P.; Arlt, M.F.; Glover, T.W. ATR regulates fragile site stability. Cell 2002, 111, 779–789. [Google Scholar] [CrossRef] [PubMed]
- Durkin, S.G.; Arlt, M.F.; Howlett, N.G.; Glover, T.W. Depletion of CHK1, but not CHK2, induces chromosomal instability and breaks at common fragile sites. Oncogene 2006, 25, 4381–4388. [Google Scholar] [CrossRef] [PubMed]
- Koundrioukoff, S.; Carignon, S.; Techer, H.; Letessier, A.; Brison, O.; Debatisse, M. Stepwise activation of the ATR signaling pathway upon increasing RS impacts fragile site integrity. PLoS Genet. 2013, 9, e1003643. [Google Scholar] [CrossRef] [PubMed]
- Franchitto, A.; Pichierri, P. Replication fork recovery and regulation of common fragile sites stability. Cell. Mol. Life Sci. 2014, 71, 4507–4517. [Google Scholar] [CrossRef]
- Khamidullina, A.I.; Abramenko, Y.E.; Bruter, A.V.; Tatarskiy, V.V. Key proteins of replication stress response and cell cycle control as cancer therapy targets. Int. J. Mol. Sci. 2024, 25, 1263. [Google Scholar] [CrossRef]
- Kim, J.M.; Kakusho, N.; Yamada, M.; Kanoh, Y.; Takemoto, N.; Masai, H. Cdc7 kinase mediates Claspin phosphorylation in DNA replication checkpoint. Oncogene 2008, 27, 3475–3482. [Google Scholar] [CrossRef]
- Rainey, M.D.; Harhen, B.; Wang, G.N.; Murphy, P.V.; Santocanale, C. Cdc7-dependent and -independent phosphorylation of Claspin in the induction of the DNA replication checkpoint. Cell Cycle 2013, 12, 1560–1568. [Google Scholar] [CrossRef]
- Yang, C.C.; Kato, H.; Shindo, M.; Masai, H. Cdc7 activates replication checkpoint by phosphorylating the Chk1 binding domain of Claspin in human cells. eLife 2019, 8, e50796. [Google Scholar] [CrossRef]
- Ulsamer, A.; Martínez-Limón, A.; Bader, S.; Rodríguez-Acebes, S.; Freire, R.; Méndez, J.; Nadal, E.; Posas, F. Regulation of Claspin by the p38 stress-activated protein kinase protects cells from DNA damage. Cell Rep. 2022, 40, 111375. [Google Scholar] [CrossRef]
- Koganti, S.; Hui-Yuen, J.; McAllister, S.; Gardner, B.; Grasser, F.; Palendira, U.; Tangye, S.G.; Freeman, A.F.; Bhaduri-McIntosh, S. STAT3 interrupts ATR-Chk1 signaling to allow oncovirus-mediated cell proliferation. Proc. Natl. Acad. Sci. USA 2014, 111, 4946–4951. [Google Scholar] [CrossRef]
- Spardy, N.; Covella, K.; Cha, E.; Hoskins, E.E.; Wells, S.I.; Duensing, A.; Duensing, S. Human papillomavirus 16 E7 oncoprotein attenuates DNA damage checkpoint control by increasing the proteolytic turnover of Claspin. Cancer Res. 2009, 69, 7022–7029. [Google Scholar] [CrossRef] [PubMed]
- Studach, L.; Wang, W.H.; Weber, G.; Tang, J.; Hullinger, R.L.; Malbrue, R.; Liu, X.; Andrisani, O. Polo-like kinase 1 activated by the hepatitis B virus X protein attenuates both the DNA damage checkpoint and DNA repair resulting in partial polyploidy. J. Biol. Chem. 2010, 285, 30282–30293. [Google Scholar] [CrossRef] [PubMed]
- Sun, A.; Bagella, L.; Tutton, S.; Romano, G.; Giordano, A. From G0 to S phase: A view of the roles played by the retinoblastoma (Rb) family members in the Rb-E2F pathway. J. Cell Biochem. 2007, 102, 1400–1404. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Clements, A.; Zhao, K.; Marmorstein, R. Structure of the human papillomavirus E7 oncoprotein and its mechanism for inactivation of the retinoblastoma tumor suppressor. J. Biol. Chem. 2006, 281, 578–586. [Google Scholar] [CrossRef]
- Scheffner, M.; Huibregtse, J.M.; Vierstra, R.D.; Howley, P.M. The HPV-16 E6 and E6-AP complex functions as a ubiquitin-protein ligase in the ubiquitination of p53. Cell 1993, 75, 495–505. [Google Scholar] [CrossRef]
- Rodriguez-Bravo, V.; Guaita-Esteruelas, S.; Florensa, R.; Bachs, O.; Agell, N. Chk1- and Claspin-dependent but ATR/ATM- and Rad17-independent DNA replication checkpoint response in HeLa cells. Cancer Res. 2006, 66, 8672–8679. [Google Scholar] [CrossRef]
- Zhang, L.; Wirth, M.; Patra, U.; Stroh, J.; Isaakidis, K.; Rieger, L.; Kossatz, S.; Milanovic, M.; Zang, C.; Demel, U.; et al. Actionable loss of SLF2 drives B-cell lymphomagenesis and impairs the DNA damage response. EMBO Mol. Med. 2023, 15, e16431. [Google Scholar] [CrossRef]
- Wang, C.; Yang, C.; Ji, J.; Jiang, J.; Shi, M.; Cai, Q.; Yu, Y.; Zhu, Z.; Zhang, J. Deubiquitinating enzyme USP20 is a positive regulator of claspin and suppresses the malignant characteristics of gastric cancer cells. Int. J. Oncol. 2017, 50, 1136–1146. [Google Scholar] [CrossRef]
- Bold, I.T.; Specht, A.-K.; Droste, C.F.; Zielinski, A.; Meyer, F.; Clauditz, T.S.; Munsher, A.; Werner, S.; Rothkamm, K.; Petersen, C.; et al. DNA damage response during replication correlates with CIN70 score and determines survival in HNSCC patients. Cancers 2021, 13, 1194. [Google Scholar] [CrossRef]
- Hunter, J.E.; Butterworth, J.A.; Sellier, H.; Luli, S.; Floudas, A.; Moore, A.J.; Thomas, H.D.; Campbell, K.; Kenneth, N.; Chiremba, T.; et al. Regulation of checkpoint kinase signalling and tumorigenesis by the NF-κB regulated gene, CLSPN. BioRxiv 2018, arXiv:10.1101/358291, 358291. [Google Scholar]
- Azenha, D.; Hernandez-Perez, S.; Martin, Y.; Viegas, M.S.; Martins, A.; Lopes, M.C.; Lam, E.W.; Freire, R.; Martins, T.C. Implications of CLSPN variants in cellular function and susceptibility to cancer. Cancers 2020, 12, 2396. [Google Scholar] [CrossRef]
- Erkko, H.; Pylkas, K.; Karppinen, S.M.; Winqvist, R. Germline alterations in the CLSPN gene in breast cancer families. Cancer Lett. 2008, 261, 93–97. [Google Scholar] [CrossRef] [PubMed]
- Madeira, A.; Azenha, D.; Correia, L.; Goncalves, V.; Ferreira, M.; Lacerda, M.; Lopes, C.; Martins, T.C. Claspin mutations and loss of function may contribute to breast carcinogenesis and gliomagenesis. Eur. J. Cancer 2012, 48, S172. [Google Scholar] [CrossRef]
- Madeira, A.; Gonçalves, V.; Ferreira, M.; Lacerda, M.; Martins, T.C. Loss of expression of Claspin in tumour cells may be involved in breast carcinogenesis. Eur. J. Cancer 2008, 6, S43. [Google Scholar] [CrossRef]
- Wang, X.; Szabo, C.; Qian, C.; Amadio, P.G.; Thibodeau, S.N.; Cerhan, J.R.; Petersen, G.M.; Liu, W.; Couch, F.J. Mutational analysis of thirty-two double-strand DNA break repair genes in breast and pancreatic cancers. Cancer Res. 2008, 68, 971–975. [Google Scholar] [CrossRef]
- Zhang, J.; Song, Y.-H.; Brannigan, B.W.; Wahrer, D.C.R.; Schiripo, T.A.; Harris, P.L.; Haserlat, S.M.; Ulkus, L.E.; Shannon, K.M.; Garber, J.E.; et al. Prevalence and functional analysis of sequence variants in the ATR checkpoint mediator Claspin. Mol. Cancer Res. 2009, 7, 1510–1516. [Google Scholar] [CrossRef]
- Serçin, O.; Kemp, M.G. Characterization of functional domains in human Claspin. Cell Cycle 2011, 10, 1599–1606. [Google Scholar] [CrossRef]
- Thutkawkorapin, J.; Lindblom, A.; Tham, E. Exome sequencing in 51 early onset non-familial CRC cases. Mol. Genet. Genom. Med. 2019, 7, e605. [Google Scholar] [CrossRef]
- Smits, V.A.J.; Cabrera, E.; Freire, R.; Gillespie, D.A. Claspin—Checkpoint adaptor and DNA replication factor. FEBS J. 2019, 286, 441–455. [Google Scholar] [CrossRef]
- Hsieh, M.-J.; Lo, Y.-S.; Ho, H.-Y.; Lin, C.-C.; Chuang, Y.-C. The interaction between CLSPN gene polymorphisms and alcohol consumption contributes to oral cancer progression. Int. J. Mol. Sci. 2024, 25, 1098. [Google Scholar] [CrossRef]
- Johmura, Y.; Yamashita, E.; Shimada, M.; Nakanishi, K.; Nakanishi, M. Defective DNA repair increases susceptibility to senescence through extension of Chk1-mediated G2 checkpoint activation. Sci. Rep. 2016, 6, 31194. [Google Scholar] [CrossRef]
- Zhu, X.; Xue, J.; Jiang, X.; Gong, Y.; Gao, C.; Cao, T.; Li, Q.; Bai, L.; Li, Y.; Xu, G.; et al. TRIM21 suppresses CHK1 activation by preferentially targeting CLASPIN for K63-linked ubiquitination. Nucleic Acids Res. 2022, 50, 1517–1530. [Google Scholar] [CrossRef]
- Benevolo, M.; Musio, A.; Vocaturo, A.; Donà, M.; Rollo, F.; Terrenato, I.; Carosi, M.; Pescarmona, E.; Vocaturo, G.; Mottolese, M. Claspin as a biomarker of human papillomavirus-related high grade lesions of uterine cervix. J. Transl. Med. 2012, 10, 132. [Google Scholar] [CrossRef] [PubMed]
- Bianco, J.N.; Bergoglio, V.; Lin, Y.L.; Pillaire, M.J.; Schmitz, A.L.; Gilhodes, J.; Lusque, A.; Mazières, J.; Lacroix-Triki, M.; Roumeliotis, T.I.; et al. Overexpression of Claspin and Timeless protects cancer cells from RS in a checkpoint-independent manner. Nat. Commun. 2019, 10, 910. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, G.; Sentani, K.; Babasaki, T.; Sekino, Y.; Shigematsu, Y.; Hayashi, T.; Oue, N.; Teishima, J.; Matsubara, A.; Sasaki, N.; et al. Claspin overexpression is associated with high-grade histology and poor prognosis in renal cell carcinoma. Cancer Sci. 2020, 111, 1020–1027. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, G.; Hayashi, T.; Sentani, K.; Babasaki, T.; Sekino, Y.; Inoue, S.; Uraoka, N.; Hanamoto, M.; Nose, H.; Teishima, J.; et al. Clinicopathological significance of claspin overexpression and its efficacy as a novel biomarker for the diagnosis of urothelial carcinoma. Virchows Arch. 2022, 480, 621–633. [Google Scholar] [CrossRef]
- Kobayashi, G.; Sentani, K.; Hattori, T.; Yamamoto, Y.; Imai, T.; Sakamoto, N.; Kuraoka, K.; Oue, N.; Sasaki, N.; Taniyama, K.; et al. Clinicopathological significance of claspin overexpression and its association with spheroid formation in gastric cancer. Hum. Pathol. 2019, 84, 8–17. [Google Scholar] [CrossRef]
- Jia, Y.; Cheng, X.; Liang, W.; Lin, S.; Li, P.; Yan, Z.; Zhang, M.; Ma, W.; Hu, C.; Wang, B.; et al. CLSPN is a potential biomarker associated with poor prognosis in low-grade gliomas based on a multi-database analysis. Curr. Res. Translat. Med. 2022, 70, 103345. [Google Scholar] [CrossRef]
- Babasaki, T.; Sentani, K.; Sekino, Y.; Kobayashi, G.; Pham, Q.T.; Katsuya, N.; Akabane, S.; Taniyama, D.; Hayashi, T.; Shiota, M.; et al. Overexpression of Claspin promotes docetaxel resistance and is associated with prostate-specific antigen recurrence in prostate cancer. Cancer Med. 2021, 10, 5574–5588. [Google Scholar] [CrossRef]
- Cai, C.; Luo, J.; Liu, Q.; Liu, Z.; Zhao, Y.; Wu, X.; Yuegao, Y.; Lei, Y.; Lu, J.; Wang, Y.; et al. Claspin overexpression promotes tumor progression and predicts poor clinical outcome in prostate cancer. Gene Ther. Mol. Biol. 2021, 25, 131–139, Correction in Gene Ther. Mol. Biol. 2021, 25, 382. https://doi.org/10.1089/gtmb.2020.0226.correx. [Google Scholar] [CrossRef]
- Elbasateeny, S.; Abdelwahab, M.; Mahmoud, A.; Malek, M.; Yassin, M.; Ismail, A.; Ibrahim, H. Immunohistochemical expression of Claspin and TopBP1 in prostatic adenocarcinoma: Correlation with clinicopathological parameters and prognostic significance. Egypt. J. Hosp. Med. 2022, 89, 6422–6431. [Google Scholar] [CrossRef]
- Lin, C.; Yuan, G.; Hu, Z.; Zeng, Y.; Qiu, X.; Yu, H.; He, S. Bioinformatics analysis of the interactions among lncRNA, miRNA, and mRNA expression, genetic mutations, and epigenetic modifications in hepatocellular carcinoma. Mol. Med. Rep. 2019, 19, 1356–1364. [Google Scholar] [CrossRef] [PubMed]
- Liao, X.; Wang, X.; Huang, K.; Han, C.; Deng, J.; Yu, T.; Yang, C.; Huang, R.; Liu, X.; Yu, L.; et al. Integrated analysis of competing endogenous RNA network revealing potential prognostic biomarkers of hepatocellular carcinoma. J. Cancer 2019, 10, 3267–3283. [Google Scholar] [CrossRef] [PubMed]
- Hein, J.; Boichuk, S.; Wu, J.; Cheng, Y.; Freire, R.; Jat, P.S.; Roberts, T.M.; Gjoerup, O.V. Simian virus 40 large T antigen disrupts genome integrity and activates a DNA damage response via Bub1 binding. J. Virol. 2009, 83, 117–127. [Google Scholar] [CrossRef] [PubMed]
- Rohaly, G.; Korf, K.; Dehde, S.; Dornreiter, I. Simian virus 40 activates ATR-Delta p53 signaling to override cell cycle and DNA replication control. J. Virol. 2010, 84, 10727–10747. [Google Scholar] [CrossRef]
- Senturk, E.; Manfredi, J.J. p53 and cell cycle effects after DNA damage. Methods Mol. Biol. 2013, 962, 49–61. [Google Scholar] [CrossRef]
- Marei, H.E.; Althani, A.; Afifi, N.; Hasan, A.; Caceci, T.; Pozzoli, G.; Morrione, A.; Giordano, A.; Cenciarelli, C.; Cenciarelli, C. p53 signaling in cancer progression and therapy. Cancer Cell Int. 2021, 21, 703. [Google Scholar] [CrossRef]
- Allera-Moreau, C.; Rouquette, I.; Lepage, B.; Oumouhou, N.; Walschaerts, M.; Leconte, E.; Schilling, V.; Gordien, K.; Brouchet, L.; Delisle, M.B.; et al. DNA replication stress response involving PLK1, CDC6, POLQ, RAD51 and CLASPIN upregulation prognoses the outcome of early/mid-stage non-small cell lung cancer patients. Oncogenesis 2012, 1, e30. [Google Scholar] [CrossRef]
- Briu, L.M.; Maric, C.; Cadoret, J.C. Replication stress, genomic instability, and replication timing: A complex relationship. Int. J. Mol. Sci. 2021, 22, 4764. [Google Scholar] [CrossRef]
- Pasero, P.; Tourrière, H. Overexpression of the Fork Protection Complex: A strategy to tolerate oncogene-induced RS in cancer cells. Cell Cycle 2019, 6, 1607455. [Google Scholar] [CrossRef]
- Bartek, J.; Mistrik, M.; Bartkova, J. Thresholds of replication stress signaling in cancer development and treatment. Nat. Struct. Mol. Biol. 2012, 19, 5. [Google Scholar] [CrossRef]
- Lopez-Contreras, A.J.; Fernadez-Capetillo, O. The ATR barrier to replication-born DNA damage. DNA Repair 2010, 9, 1249–1255. [Google Scholar] [CrossRef]
- Gilad, O.; Nabet, B.Y.; Ragland, R.L.; Schoppy, D.W.; Smith, K.D.; Durham, A.C.; Brown, E.J. Combining ATR suppression with oncogenic Ras synergistically increases genomic instability, causing synthetic lethality or tumorigenesis in a dosage-dependent manner. Cancer Res. 2010, 70, 9693–9702. [Google Scholar] [CrossRef]
- Fang, Y.; Tsao, C.C.; Goodman, B.K.; Furumai, R.; Tirado, C.A.; Abraham, R.T.; Wang, X.F. ATR functions as a gene dosage-dependent tumor suppressor on a mismatch repair-deficient background. EMBO J. 2004, 23, 3164–3174. [Google Scholar] [CrossRef]
- Lopez-Contreras, A.J.; Gutierrez-Martinez, P.; Specks, J.; Rodrigo-Perez, S.; Fernadez-Capetillo, O. An extra allele of Chk1 limits oncogene-induced replicative stress and promotes transformation. J. Exp. Med. 2012, 209, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Black, E.M.; Joo, Y.K.; Kabeche, L. Keeping relApse in Chk: Molecular mechanisms of Chk1 inhibitor resistance in lymphoma. Biochem. J. 2022, 479, 2345–2349. [Google Scholar] [CrossRef]
- Wee, P.; Wang, Z. Epidermal growth factor receptor cell proliferation signaling pathways. Cancers 2017, 9, 52. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.C.; Masai, H. Claspin is Required for Growth Recovery from Serum Starvation through Regulating the PI3K-PDK1-mTOR Pathway in Mammalian Cells. Mol. Cell. Biol. 2023, 43, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Wang, Y.; Zhou, C.; Mei, W.; Zeng, C. PI3K/Akt/mTOR pathway and its role in cancer therapeutics: Are we making headway? Front. Oncol. 2022, 12, 819128. [Google Scholar] [CrossRef]
- Salmena, L.; Poliseno, L.; Tay, Y.; Kats, L.; Pandolfi, P.P. A ceRNA hypothesis: The Rosetta Stone of a hidden RNA language? Cell 2011, 146, 353–358. [Google Scholar] [CrossRef]
- Hu, T.; Lei, D.; Zhou, J.; Zhang, B. circRNA derived from CLSPN (circCLSPN) is an oncogene in human glioblastoma multiforme by regulating cell growth, migration, and invasion via ceRNA pathway. J. Biosci. 2021, 46, 66. [Google Scholar] [CrossRef]
- Tsimaratou, K.; Kletsas, D.; Kastrinakis, N.; Tsantoulis, P.; Evangelou, K.; Sideridou, M.; Liontos, M.; Poulias, I.; Venere, M.; Salmas, M.; et al. Evaluation of claspin as a proliferation marker in human cancer and normal tissues. J. Pathol. 2007, 211, 331–339. [Google Scholar] [CrossRef]
- Hsiao, H.W.; Yang, C.C.; Masai, H. Roles of Claspin in regulation of DNA replication, replication stress responses, and oncogenesis in human cells. Genome Instab. Dis. 2021, 2, 263–280. [Google Scholar] [CrossRef]
- Yamada, S.; Miyata, H.; Isono, M.; Hori, K.; Yanagawa, J.; Murai, A.; Minowa, T.; Mizue, Y.; Sasaki, K.; Murata, K.; et al. Cisplatin resistance driver Claspin is a target for immunotherapy in urothelial carcinoma. Cancer Immunol. Immunother. 2023, 72, 2057–2065. [Google Scholar] [CrossRef] [PubMed]
- Shen, L.; Shi, Q.; Wang, W. Double agents: Genes with both oncogenic and tumor-suppressor functions. Oncogenesis 2018, 7, 25. [Google Scholar] [CrossRef] [PubMed]
- Datta, N.; Chakraborty, S.; Basu, M.; Ghosh, M.K. Tumor suppressors having oncogenic functions: The double agents. Cells 2020, 10, 46. [Google Scholar] [CrossRef]
- Pitolli, C.; Wang, Y.; Candi, E.; Shi, Y.; Melino, G.; Amelio, I. p53-mediated tumor suppression: DNA-damage response and alternative mechanisms. Cancers 2019, 11, 1983. [Google Scholar] [CrossRef]
- Soussi, T.; Wiman, K.G. TP53: An oncogene in disguise. Cell Death Diff. 2015, 22, 1239–1249. [Google Scholar] [CrossRef]
- Zhang, C.; Liu, J.; Xu, D.; Zhang, T.; Hu, W.; Feng, Z. Gain-of-function mutant p53 in cancer progression and therapy. J. Mol. Cell Biol. 2020, 12, 674–687. [Google Scholar] [CrossRef]
- Choi, W.; Lee, E.S. Therapeutic targeting of DNA damage response in cancer. Int. J. Mol. Sci. 2022, 23, 1701. [Google Scholar] [CrossRef]
- Rose, M.; Burgess, J.T.; O’Byrne, K.; Richard, D.J.; Bolderson, E. PARP inhibitors: Clinical relevance, mechanisms of action and tumor resistance. Front. Cell Dev. Biol. 2020, 8, 564601. [Google Scholar] [CrossRef]
- Choi, S.; Yang, H.; Lee, S.; Ki, J.-H.; Nam, D.-H.; Yoo, H. TopBP1 and Claspin contribute to the radioresistance of lung cancer brain metastases. Mol. Cancer 2014, 13, 211. [Google Scholar] [CrossRef]
- Tu, Y.; Chen, Z.; Zhao, P.; Sun, G.; Bao, Z.; Chao, H.; Fan, L.; Li, C.; You, Y.; Qu, Y.; et al. Smoothened promotes glioblastoma radiation resistance via activating USP3-mediated claspin deubiquitination. Clin. Cancer Res. 2020, 26, 1749–1762. [Google Scholar] [CrossRef]
- Ito, F.; Yoshimoto, C.; Yamada, Y.; Sudo, T.; Kobayashi, H. The HNF-1β-USP28-Claspin pathway upregulates DNA damage-induced Chk1 activation in ovarian clear cell carcinoma. Oncotarget 2018, 9, 17512–17522. [Google Scholar] [CrossRef]
- Busato, F.; Khouzai, B.E.; Mognato, M. Biological mechanisms to reduce radioresistance and increase the efficacy of radiotherapy: State of the art. Int. J. Mol. Sci. 2022, 23, 10211. [Google Scholar] [CrossRef]
- Imran, M.; Saleem, S.; Chaudhuri, A.; Ali, J.; Baboota, S. Docetaxel: An update on its molecular mechanisms, therapeutic trajectory and nanotechnology in the treatment of breast, lung, and prostate cancer. J. Drug Deliv. Sci. Technol. 2020, 60, 101959. [Google Scholar] [CrossRef]
- Corno, C.; D’Arcy, P.; Bagnoli, M.; Paolini, B.; Constantino, M.; Carenini, N.; Corna, E.; Alberti, P.; Mezzanzanica, D.; Colombo, D.; et al. The deubiquitinase USP8 regulates ovarian cancer cell response to cisplatin by suppressing apoptosis. Front. Cell. Dev. Biol. 2022, 10, 1055067. [Google Scholar] [CrossRef]
- Aldossary, S.A. Review on pharmacology of cisplatin: Clinical use, toxicity, and mechanism of resistance of cisplatin. Biomed. Pharmacol. J. 2019, 12, 7–15. [Google Scholar] [CrossRef]
- Walz, A.; Ugolkov, A.; Chandra, S.; Kozikowski, A.; Carneiro, B.A.; O’Halloran, T.V.; Giles, F.J.; Billadeau, D.D.; Mazar, A.P. Molecular pathways: Revisiting glycogen synthase kinase-3β as a target for the treatment of cancer. Clin. Cancer Res. 2017, 23, 1891–1897. [Google Scholar] [CrossRef]
- Ugolkov, A.; Gaisina, I.; Zhang, J.-S.; Billadeau, D.D.; White, K.; Kozikowski, A.; Jain, S.; Cristofanilli, M.; Giles, F.; O’Halloran, T.; et al. GSK-3 inhibition overcomes chemoresistance in human breast cancer. Cancer Lett. 2016, 380, 384–392. [Google Scholar] [CrossRef]
- Cabrera, E.; Raninga, P.; Khanna, K.K.; Freire, R. GSK3-β Stimulates Claspin Degradation via β-TrCP Ubiquitin Ligase and Alters Cancer Cell Survival. Cancers 2019, 11, 1073. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Azenha, D.; Martins, T.C. Claspin and Cancer: Where Are We Now? Int. J. Mol. Sci. 2025, 26, 8828. https://doi.org/10.3390/ijms26188828
Azenha D, Martins TC. Claspin and Cancer: Where Are We Now? International Journal of Molecular Sciences. 2025; 26(18):8828. https://doi.org/10.3390/ijms26188828
Chicago/Turabian StyleAzenha, Diana, and Teresa C. Martins. 2025. "Claspin and Cancer: Where Are We Now?" International Journal of Molecular Sciences 26, no. 18: 8828. https://doi.org/10.3390/ijms26188828
APA StyleAzenha, D., & Martins, T. C. (2025). Claspin and Cancer: Where Are We Now? International Journal of Molecular Sciences, 26(18), 8828. https://doi.org/10.3390/ijms26188828





