The Mediterranean Habitat of the Nile Soft-Shelled Turtle (Trionyx triunguis): Genomic and Reproductive Insights into an Endangered Population
Abstract
1. Introduction
2. Results
2.1. mtDNA Analysis
2.2. RADseq Analysis
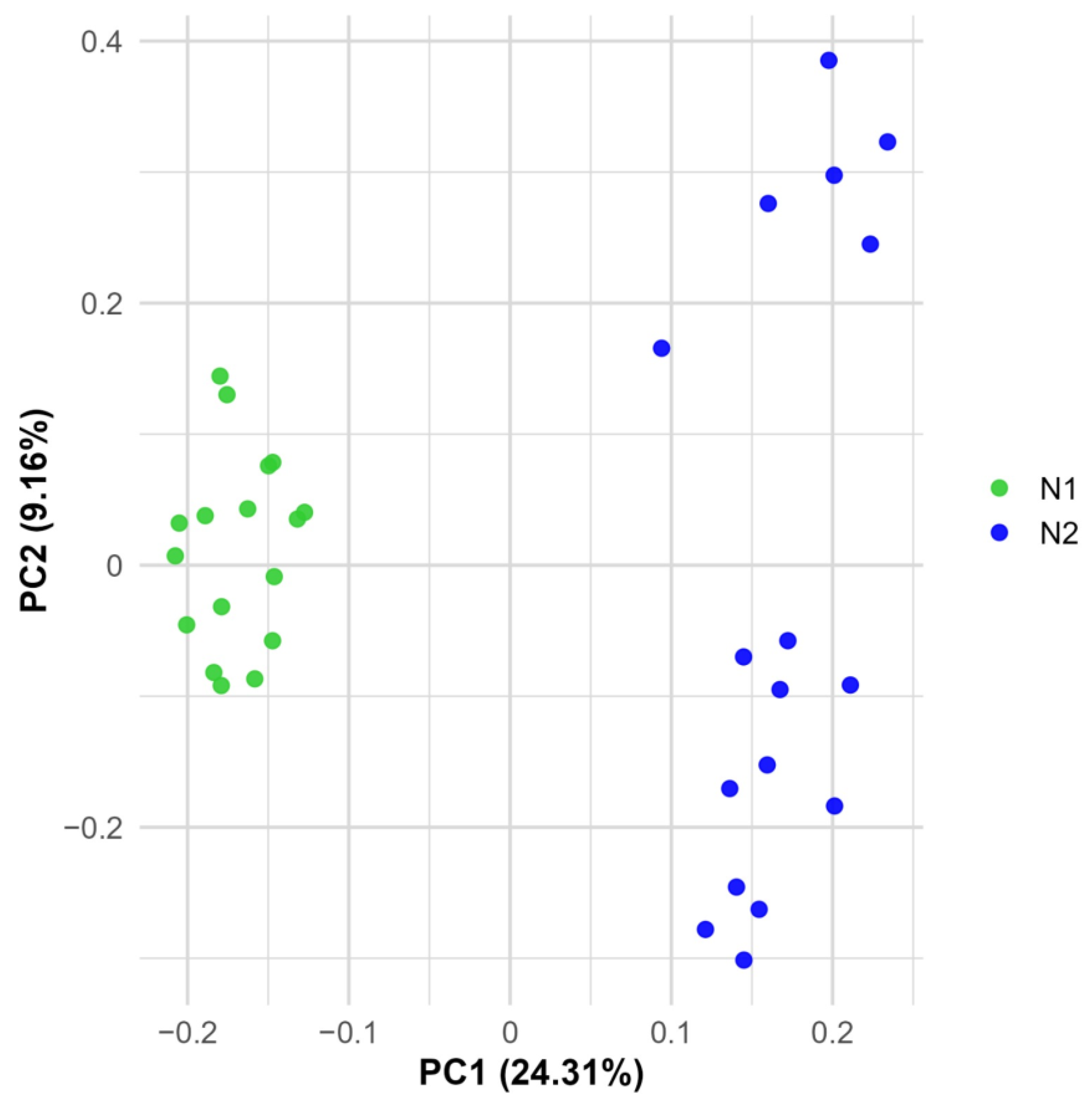
2.3. Population Structure
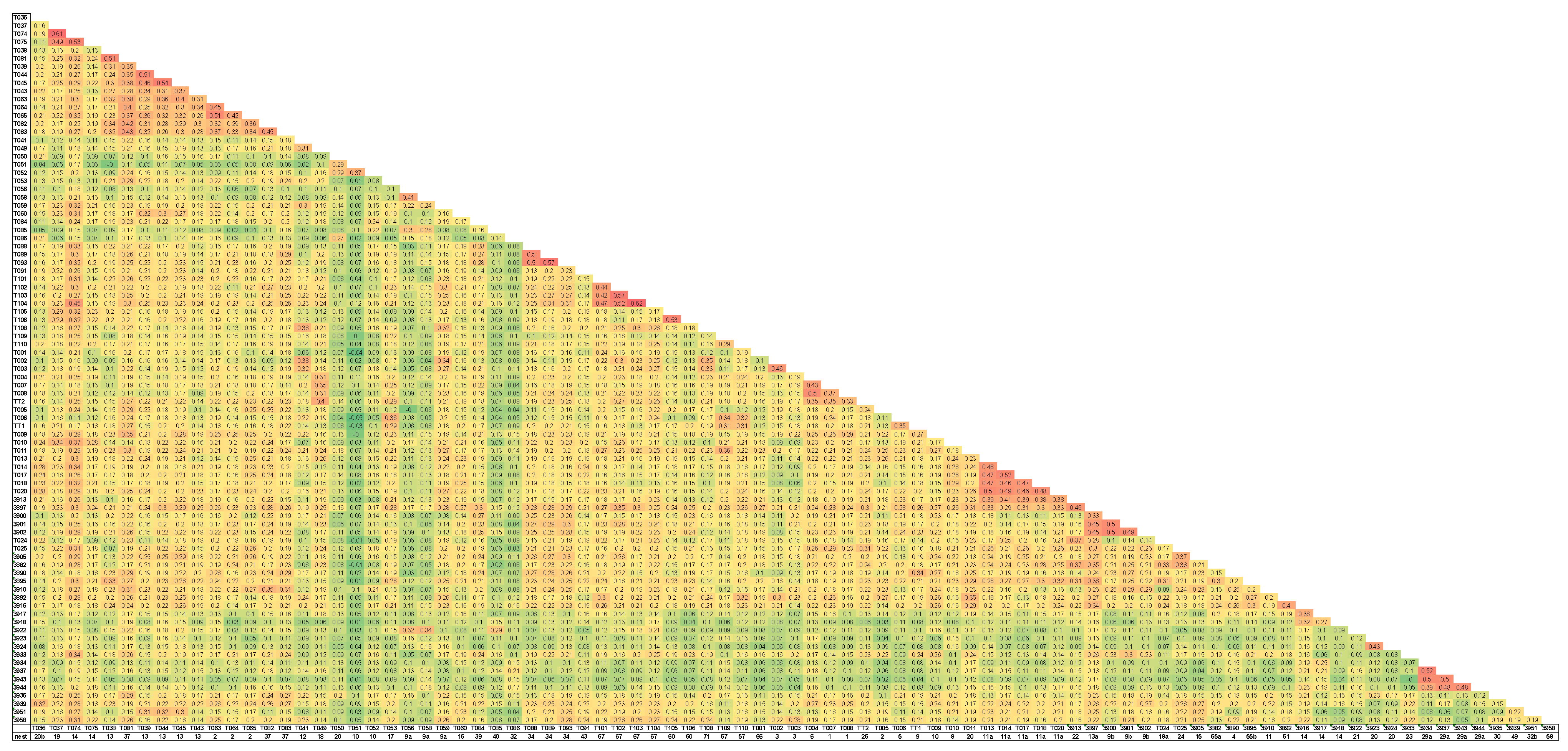
2.4. Normalized-IBS (Identity by State)
2.5. The Alexander Population
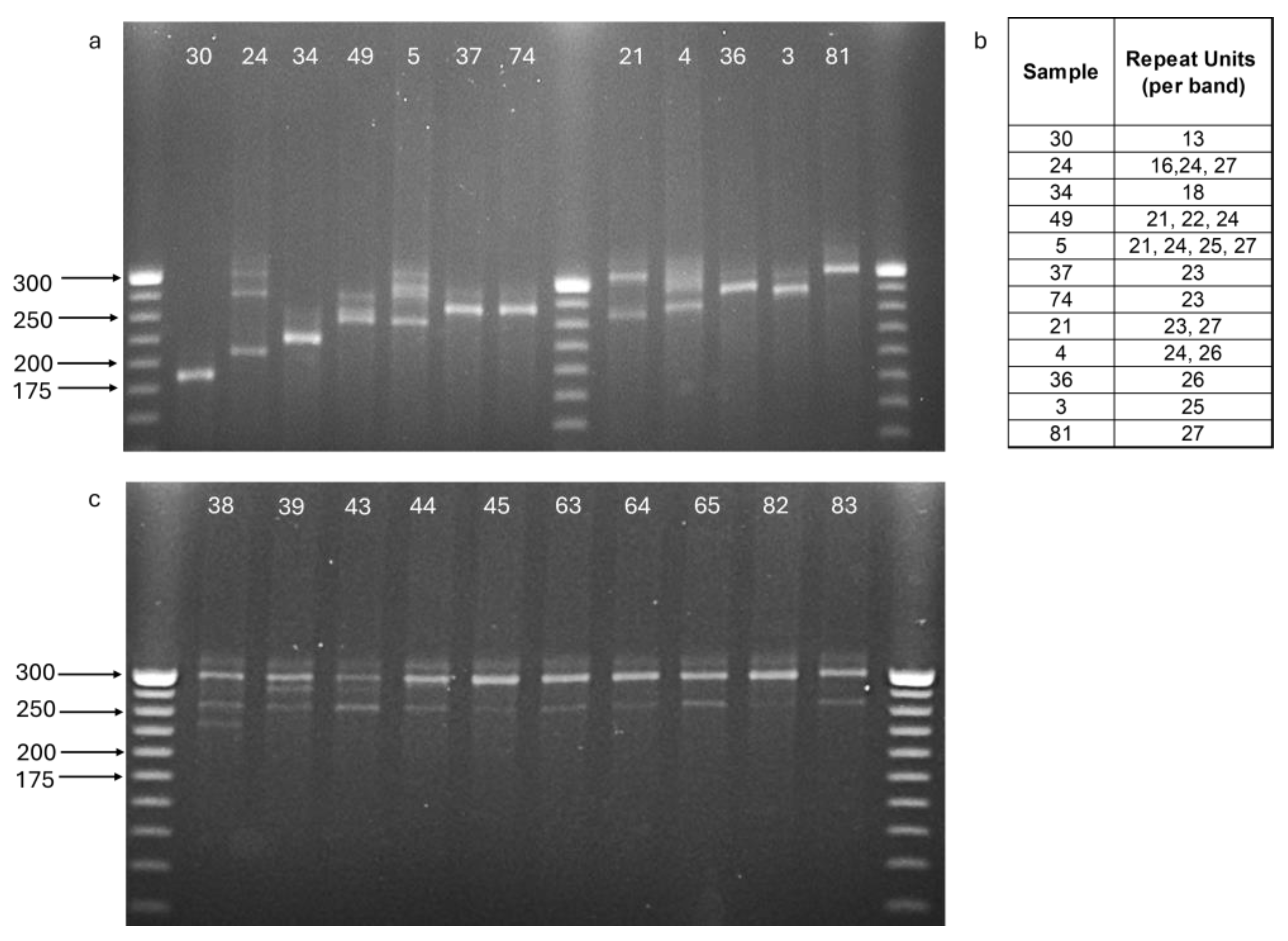
2.6. The Two Hula Nests
3. Discussion
4. Materials and Methods
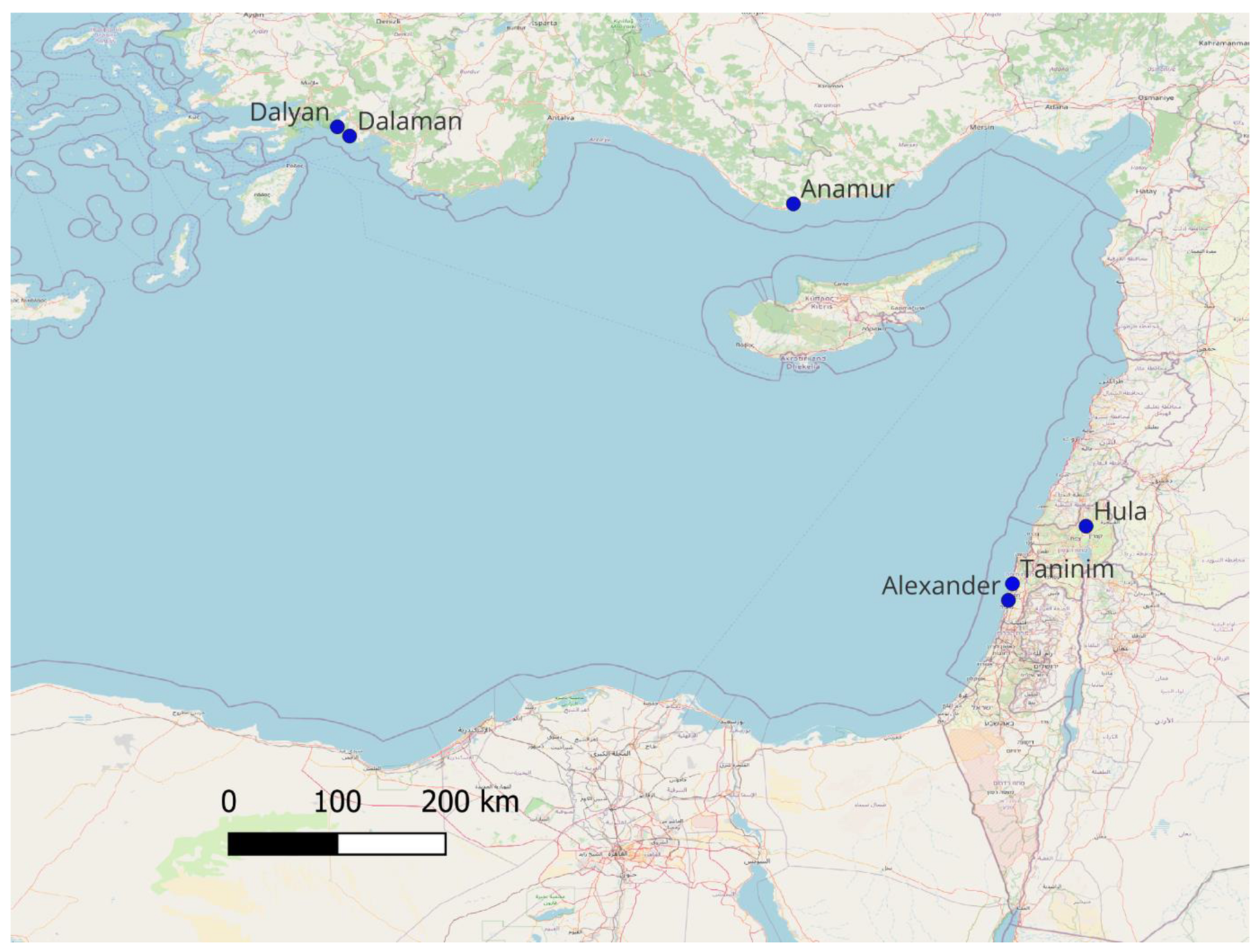
4.1. Sample Collection and DNA Extraction
4.2. mtDNA Haplotyping
4.3. RAD Laboratory
4.4. RAD Analysis
4.5. Reference Construction and SNP Calling
4.6. SNP Filtering and Population Genetic Analyses
4.7. Kinship Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| NPA | Nature and Parks Authority |
| IBS | Identity by State |
| mtDNA | Mitochondrial DNA |
References
- Meylan, P. The Phylogenetic Relationships of Soft-Shelled Turtles (Family Trionychidae). 1987, 186. Available online: https://iucn-tftsg.org/wp-content/uploads/file/Articles/Meylan_1987.pdf (accessed on 1 September 2025).
- Vitt, L.J.; Caldwell, J.P. Herpetology an Introductory Biology of Amphibians and Reptiles, 4th ed.; Academic Press: Cambridge, MA, USA, 2013; p. 531. [Google Scholar]
- Engstrom, T.N.; Shaffer, H.B.; McCord, W.P.; Baker, A. Multiple Data Sets, High Homoplasy, and the Phylogeny of Softshell Turtles (Testudines: Trionychidae). Syst. Biol. 2004, 53, 693–710. [Google Scholar] [CrossRef]
- Güçlü, Ö.; Ulger, C.; Türkozan, O. Genetic Variation of the Nile Soft-Shelled Turtle (Trionyx triunguis). Int. J. Mol. Sci. 2011, 12, 6418–6431. [Google Scholar] [CrossRef]
- Flower, M.S.S. Notes on the Recent Reptiles and Amphibians of Egypt, with a List of the Species recorded from that Kingdom. J. Zool. 1933, 103, 735–851. [Google Scholar] [CrossRef]
- Taskavak, E.; Akcinar, S.C. Marine records of the Nile soft-shelled turtle, Trionyx triunguis from Turkey. Mar. Biodivers. Rec. 2009, 2, e9. [Google Scholar] [CrossRef]
- Cattaneo, A.; Cattaneo, C.; Grano, M. Update on the herpetofauna of the DPlease, confirm all newly added/revised information. Highlighted.odecanese Archipelago (Greece). Biodivers. J. 2020, 10, 69–84. [Google Scholar] [CrossRef]
- Taşkavak, E. Cranial morphology of Rafetus euphraticus (Daudin, 1801) from Southeastern Anatolia. Amphib. Reptil. 1999, 20, 35–53. [Google Scholar] [CrossRef]
- Güçlü, Ö.; Ulger, C.; Türkozan, O.; Gemel, R.; Reimann, M.; Levy, Y.; Ergene, S.; Uçar, A.H.; Aymak, C. First Assessment of Mitochondrial DNA Diversity in the Endangered Nile Softshell Turtle, Trionyx triunguis, in the Mediterranean. Chelonian Conserv. Biol. 2009, 8, 222–226. [Google Scholar] [CrossRef]
- Kasparek, M. Priorities for the Conservation of the Nile Soft-Shelled Turtle, Trionyx triunguis, in the Mediterranean. Testudo 2001, 5, 49–59. [Google Scholar]
- Oruç, A. Trawl fisheries in the eastern Mediterranean and their impact on marine turtles. Zool. Middle East 2001, 24, 119–125. [Google Scholar] [CrossRef]
- Kasparek, M.; Kinzelbach, R. Distribution and Bionomics of the Nile Soft-Shelled Turtle, Trionyx Triunguis, in the Eastern Mediterranean. German Journal of Zoology Research 1991, 78, 138–159. [Google Scholar]
- Akçınar, S.C.; Taşkavak, E. Population size and structure of the African Softshell Turtle, Trionyx triunguis, in Dalaman, southwestern Turkey. Zool. Middle East 2017, 63, 202–209. [Google Scholar] [CrossRef]
- Rozner, O. Chapters in the Ecology of the Nile Soft-Shelled Turtle (Trionyx triunguis) in Nahal Alexander. Master’s Thesis, University of Haifa, Haifa, Israel, 2007. (In Hebrew). [Google Scholar]
- Gidis, M.; Spinks, P.Q.; Çevik, E.; Kaska, Y.; Shaffer, H.B. Shallow genetic divergence indicates a Congo–Nile riverine connection for the softshell turtle Trionyx triunguis. Conserv. Genet. 2010, 12, 589–594. [Google Scholar] [CrossRef]
- Shanas, U.; Gidiş, M.; Kaska, Y.; Kimalov, Y.; Rosner, O.; Ben-Shlomo, R. The Nile Soft-shell Turtle, Trionyx triunguis, of Israel and Turkey: Two genetically indistinguishable populations? Zool. Middle East 2012, 57, 61–68. [Google Scholar] [CrossRef][Green Version]
- Güçlü, Ö.; Durmuş, S.H.; Candan, K.; Beşer, N.; Türkyilmaz, S.; Yerli, S.; Bozdoğan, B. Development and characterization of new polymorphic microsatellite loci for Trionyx triunguis (Testudines: Trionychidae) in the Mediterranean Basin. Amphibia-Reptilia 2015, 36, 318–324. [Google Scholar] [CrossRef]
- Güçlü, Ö.; Bozdoğan, B. Genetic Diversity and Structure of the Nile Soft-Shelled Turtle (Trionyx triunguis) Along the Mediterranean coast of Turkey. Russ. J. Herpetol. 2020, 27, 307–315. [Google Scholar] [CrossRef]
- Tikochinski, Y.; Bendelac, R.; Barash, A.; Daya, A.; Levy, Y.; Friedmann, A. Mitochondrial DNA STR analysis as a tool for studying the green sea turtle (Chelonia mydas) populations: The Mediterranean Sea case study. Mar. Genom. 2012, 6, 17–24. [Google Scholar] [CrossRef]
- Ohana, T.; Arantes, L.S.; Tikochinski, G.; Cohen, R.; Rybak, O.; Gaspar, A.; Sparmann, S.; Mbedi, S.; Levy, Y.; Mazzoni, C.J.; et al. Green Turtle Conservation in the Genomic Era—Monitoring an Endangered Mediterranean Population and Its Breeding Habits. Ecol. Evol. 2025, 15, e71124. [Google Scholar] [CrossRef]
- Shamblin, B.M.; Bolten, A.B.; Abreu-Grobois, F.A.; Bjorndal, K.A.; Cardona, L.; Carreras, C.; Clusa, M.; Monzón-Argüello, C.; Nairn, C.J.; Nielsen, J.T.; et al. Geographic Patterns of Genetic Variation in a Broadly Distributed Marine Vertebrate: New Insights into Loggerhead Turtle Stock Structure from Expanded Mitochondrial DNA Sequences. PLoS ONE 2014, 9, e85956. [Google Scholar] [CrossRef]
- Tikochinski, Y.; Bradshaw, P.; Mastrogiacomo, A.; Broderick, A.; Daya, A.; Demetropoulos, A.; Demetropoulos, S.; Eliades, N.; Fuller, W.; Godley, B.; et al. Mitochondrial DNA short tandem repeats unveil hidden population structuring and migration routes of an endangered marine turtle. Aquat. Conserv. Mar. Freshw. Ecosyst. 2018, 28, 788–797. [Google Scholar] [CrossRef]
- Tikochinski, Y.; Carreras, C.; Tikochinski, G.; Vilaça, S.T. Author Correction: Population-Specific Signatures of Intra-Individual Mitochondrial DNA Heteroplasmy and Their Potential Evolutionary Advantages. Sci. Rep. 2020, 10, 13180. [Google Scholar] [CrossRef]
- Andrews, K.R.; Good, J.M.; Miller, M.R.; Luikart, G.; Hohenlohe, P.A. Harnessing the power of RADseq for ecological and evolutionary genomics. Nat. Rev. Genet. 2016, 17, 81–92. [Google Scholar] [CrossRef] [PubMed]
- Driller, M.; Arantes, L.S.; Torres Vilaça, S.; Carrasco-Valenzuela, T.; Heeger, F.; Mbedi, S.; Chevallier, D.; De Thoisy, B.; Mazzoni, C.J. Achieving High-Quality DdRAD-like Reference Catalogs for Non-Model Species: The Power of Over-lapping Paired-End Reads. bioRxiv 2021. [Google Scholar] [CrossRef]
- Pearse, D.E.; Avise, J.C. Turtle Mating Systems: Behavior, Sperm Storage, and Genetic Paternity. J. Hered. 2001, 92, 206–211. [Google Scholar] [CrossRef] [PubMed]
- Uller, T.; Olsson, M. Multiple paternity in reptiles: Patterns and processes. Mol. Ecol. 2008, 17, 2566–2580. [Google Scholar] [CrossRef]
- Lee, P.L.M.; Schofield, G.; Haughey, R.I.; Mazaris, A.D.; Hays, G.C. A Review of Patterns of Multiple Paternity across Sea Turtle Rookeries. Adv Mar Biol. 2018, 79, 1–31. [Google Scholar] [CrossRef]
- Valenzuela, N. Multiple paternity in side-neck turtles Podocnemis expansa: Evidence from microsatellite DNA data. Mol. Ecol. 2000, 9, 99–105. [Google Scholar] [CrossRef]
- Cai, W.; Chen, W.; Wang, Y.; Bu, X.; Xia, X.; Nie, L. Sperm storage in the oviduct of the Chinese pond turtle Mauremys reevesii depends on oestrogen-based suppression of the TLR2/4 immune pathway. Reprod. Fertil. Dev. 2021, 33, 736–745. [Google Scholar] [CrossRef]
- Kitayama, C.; Tomiyasu, J.; Bochimoto, H.; Kondo, S.; Tokuda, K.; Ogawa, R.; Okubo, S.; Kondoh, D. Histological findings of sperm storage in green turtle (Chelonia mydas) oviduct. Sci. Rep. 2021, 11, 1–9. [Google Scholar] [CrossRef]
- Chen, S.; Zhang, L.; Le, Y.; Waqas, Y.; Chen, W.; Zhang, Q.; Ullah, S.; Liu, T.; Hu, L.; Li, Q.; et al. Sperm storage and spermatozoa interaction with epithelial cells in oviduct of Chinese soft-shelled turtle, Pelodiscus sinensis. Ecol. Evol. 2015, 5, 3023–3030. [Google Scholar] [CrossRef]
- Chen, H.; Liu, T.; Holt, W.V.; Yang, P.; Zhang, L.; Zhang, L.; Han, X.; Bian, X.; Chen, Q. Advances in Understanding Mech-anisms of Long-Term Sperm Storage-the Soft-Shelled Turtle Model. Histol. Histopathol. 2020, 35, 1–23. [Google Scholar] [CrossRef]
- Hall, T.A. BioEdit: A User-Friendly Biological Sequence Alignment Editor and Analysis Program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Hoffberg, S.L.; Kieran, T.J.; Catchen, J.M.; Devault, A.; Faircloth, B.C.; Mauricio, R.; Glenn, T.C. RADcap: Sequence capture of dual-digest RADseq libraries with identifiable duplicates and reduced missing data. Mol. Ecol. Resour. 2016, 16, 1264–1278. [Google Scholar] [CrossRef]
- Wingett, S.W.; Andrews, S. FastQ Screen: A tool for multi-genome mapping and quality control. F1000Research 2018, 7, 1338. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Dodt, M.; Roehr, J.T.; Ahmed, R.; Dieterich, C. FLEXBAR—Flexible Barcode and Adapter Processing for Next-Generation Sequencing Platforms. Biolog. 2012, 1, 895–905. [Google Scholar] [CrossRef]
- Zhang, J.; Kobert, K.; Flouri, T.; Stamatakis, A. PEAR: A fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics 2013, 30, 614–620. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Catchen, J.; Hohenlohe, P.A.; Bassham, S.; Amores, A.; Cresko, W.A. Stacks: An analysis tool set for population genomics. Mol. Ecol. 2013, 22, 3124–3140. [Google Scholar] [CrossRef] [PubMed]
- Danecek, P.; Auton, A.; Abecasis, G.; Albers, C.A.; Banks, E.; DePristo, M.A.; Handsaker, R.E.; Lunter, G.; Marth, G.T.; Sherry, S.T.; et al. The variant call format and VCFtools. Bioinformatics 2011, 27, 2156–2158. [Google Scholar] [CrossRef] [PubMed]
- Wickham, H.; Averick, M.; Bryan, J.; Chang, W.; McGowan, L.D.A.; François, R.; Grolemund, G.; Hayes, A.; Henry, L.; Hester, J.; et al. Welcome to the Tidyverse. J. Open Source Softw. 2019, 4, 1686. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research—An update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef]
- Excoffier, L.; Smouse, P.E.; Quattro, J.M. Analysis of molecular variance inferred from metric distances among DNA haplotypes: Application to human mitochondrial DNA restriction data. Genetics 1992, 131, 479–491. [Google Scholar] [CrossRef] [PubMed]
- Huff, D.R.; Peakall, R.; Smouse, P.E. RAPD variation within and among natural populations of outcrossing buffalograss [Buchloë dactyloides (Nutt.) Engelm.]. Theor. Appl. Genet. 1993, 86, 927–934. [Google Scholar] [CrossRef] [PubMed]
- Weir, B.S.; Cockerham, C.C. Estimating F-Statistics for the Analysis of Population Structure. Evolution 1984, 38, 1358–1370. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.W.; Daly, M.J.; et al. PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef]
- Chang, C.C.; Chow, C.C.; Tellier, L.C.; Vattikuti, S.; Purcell, S.M.; Lee, J.J. Second-generation PLINK: Rising to the challenge of larger and richer datasets. GigaScience 2015, 4, 7. [Google Scholar] [CrossRef] [PubMed]

| Alexander (n = 89) | Taninim (n = 8) | Hula (n = 34) | Türkiye (n = 8) | |
|---|---|---|---|---|
| Alexander | 0.131 | 0.288 | 0.251 | |
| Tanninim | 0.084 | 0.395 | 0.202 | |
| Hula | 0.187 | 0.242 | 0.462 | |
| Türkiye | 0.170 | 0.144 | 0.302 |
| Alexander | Taninim | Hula | |
|---|---|---|---|
| Tanninim | 0.12 | ||
| Hula | 0.06 | 0.04 | |
| Türkiye | 0.00 | −0.01 | −0.04 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gaspar, A.; Arantes, L.S.; Ohana, T.; Bodenheimer, Y.E.; Tikochinski, G.; Levy, O.; Mor, B.J.; Vainberg, M.; Gat, T.; Mbedi, S.; et al. The Mediterranean Habitat of the Nile Soft-Shelled Turtle (Trionyx triunguis): Genomic and Reproductive Insights into an Endangered Population. Int. J. Mol. Sci. 2025, 26, 8822. https://doi.org/10.3390/ijms26188822
Gaspar A, Arantes LS, Ohana T, Bodenheimer YE, Tikochinski G, Levy O, Mor BJ, Vainberg M, Gat T, Mbedi S, et al. The Mediterranean Habitat of the Nile Soft-Shelled Turtle (Trionyx triunguis): Genomic and Reproductive Insights into an Endangered Population. International Journal of Molecular Sciences. 2025; 26(18):8822. https://doi.org/10.3390/ijms26188822
Chicago/Turabian StyleGaspar, Adi, Larissa S. Arantes, Talya Ohana, Yair E. Bodenheimer, Gili Tikochinski, Opal Levy, Bar J. Mor, Muriel Vainberg, Tomer Gat, Susan Mbedi, and et al. 2025. "The Mediterranean Habitat of the Nile Soft-Shelled Turtle (Trionyx triunguis): Genomic and Reproductive Insights into an Endangered Population" International Journal of Molecular Sciences 26, no. 18: 8822. https://doi.org/10.3390/ijms26188822
APA StyleGaspar, A., Arantes, L. S., Ohana, T., Bodenheimer, Y. E., Tikochinski, G., Levy, O., Mor, B. J., Vainberg, M., Gat, T., Mbedi, S., Sparmann, S., Türkozan, O., Levy, Y., Leader, N., Milstein, D., Mazzoni, C. J., & Tikochinski, Y. (2025). The Mediterranean Habitat of the Nile Soft-Shelled Turtle (Trionyx triunguis): Genomic and Reproductive Insights into an Endangered Population. International Journal of Molecular Sciences, 26(18), 8822. https://doi.org/10.3390/ijms26188822






