Ole-e-1 Interacts with FWL Genes to Modulate Cell Division and Determine Fruit Size in Pears
Abstract
1. Introduction
2. Results
2.1. Tissue Fluorescence In Situ Hybridization Analysis of FWL1 and FWL5
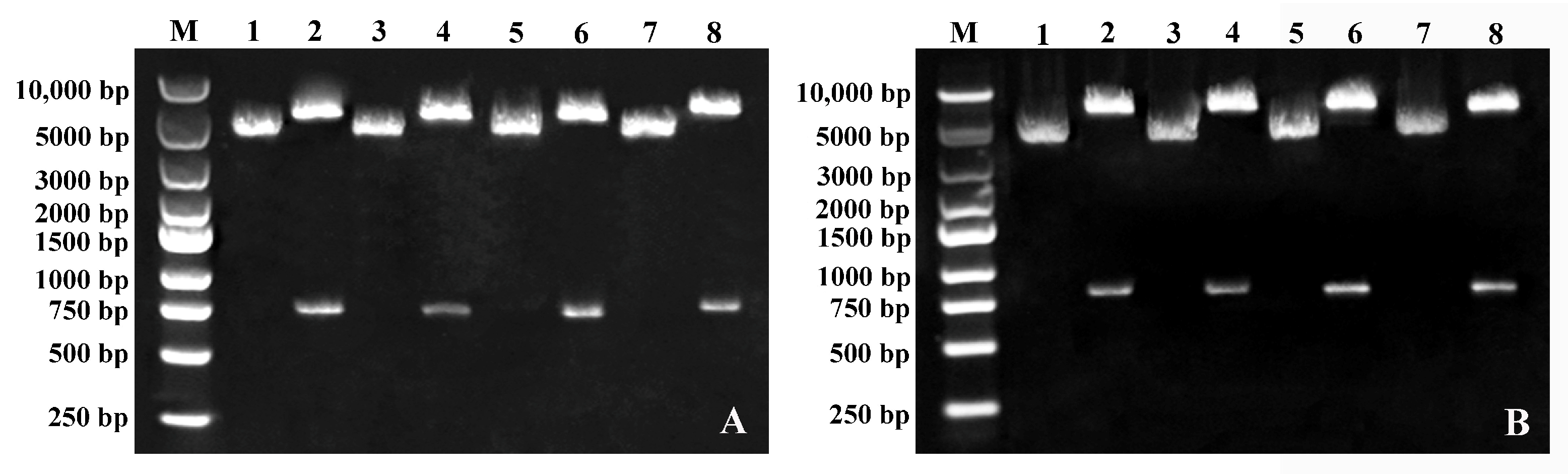
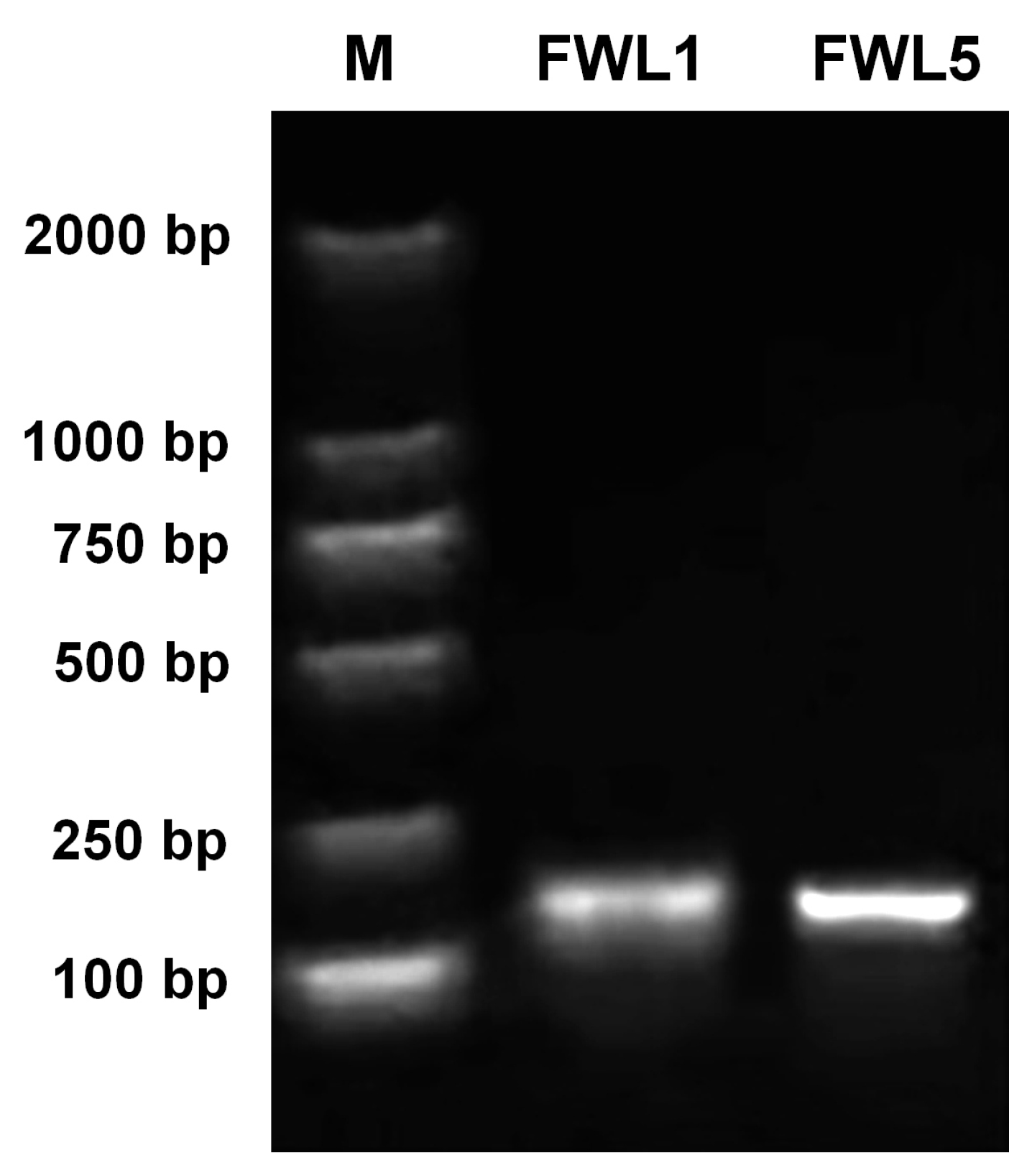
2.2. Silencing of FWL Genes via TRV-Based VIGS
2.3. Effects of FWL Gene Silencing on Fruit Size, Cell Size, and Cell Number
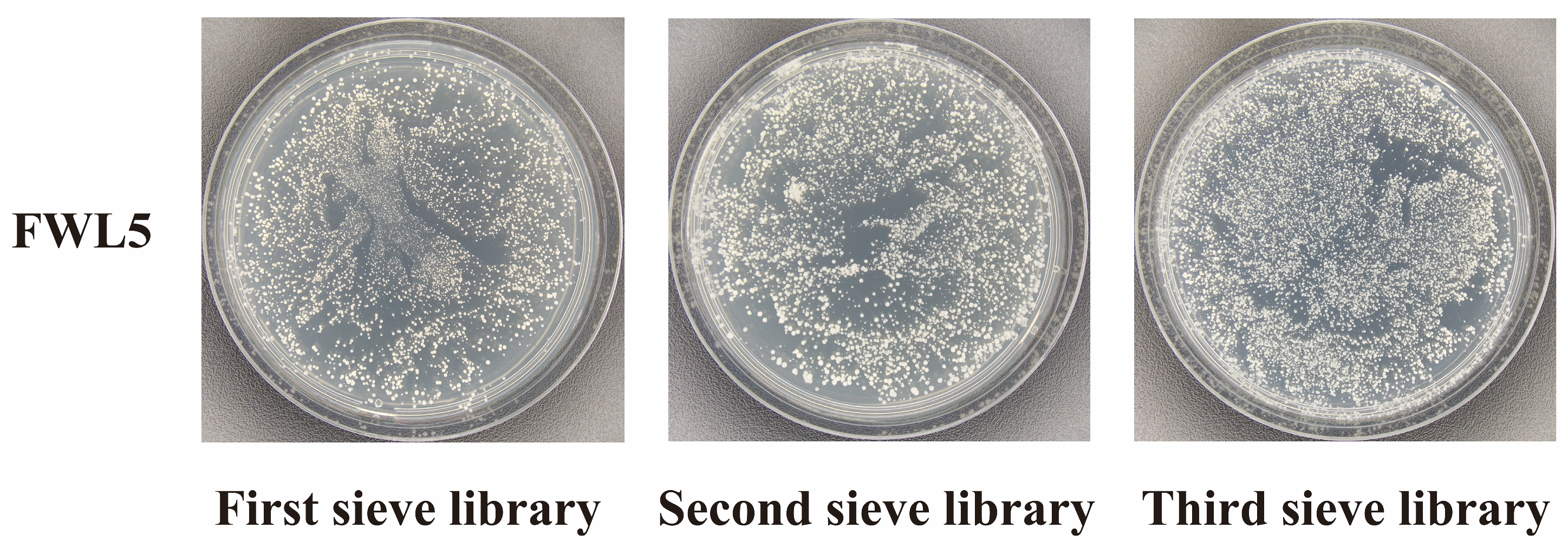
2.4. Library Screening and Identification of Candidate Interacting Proteins
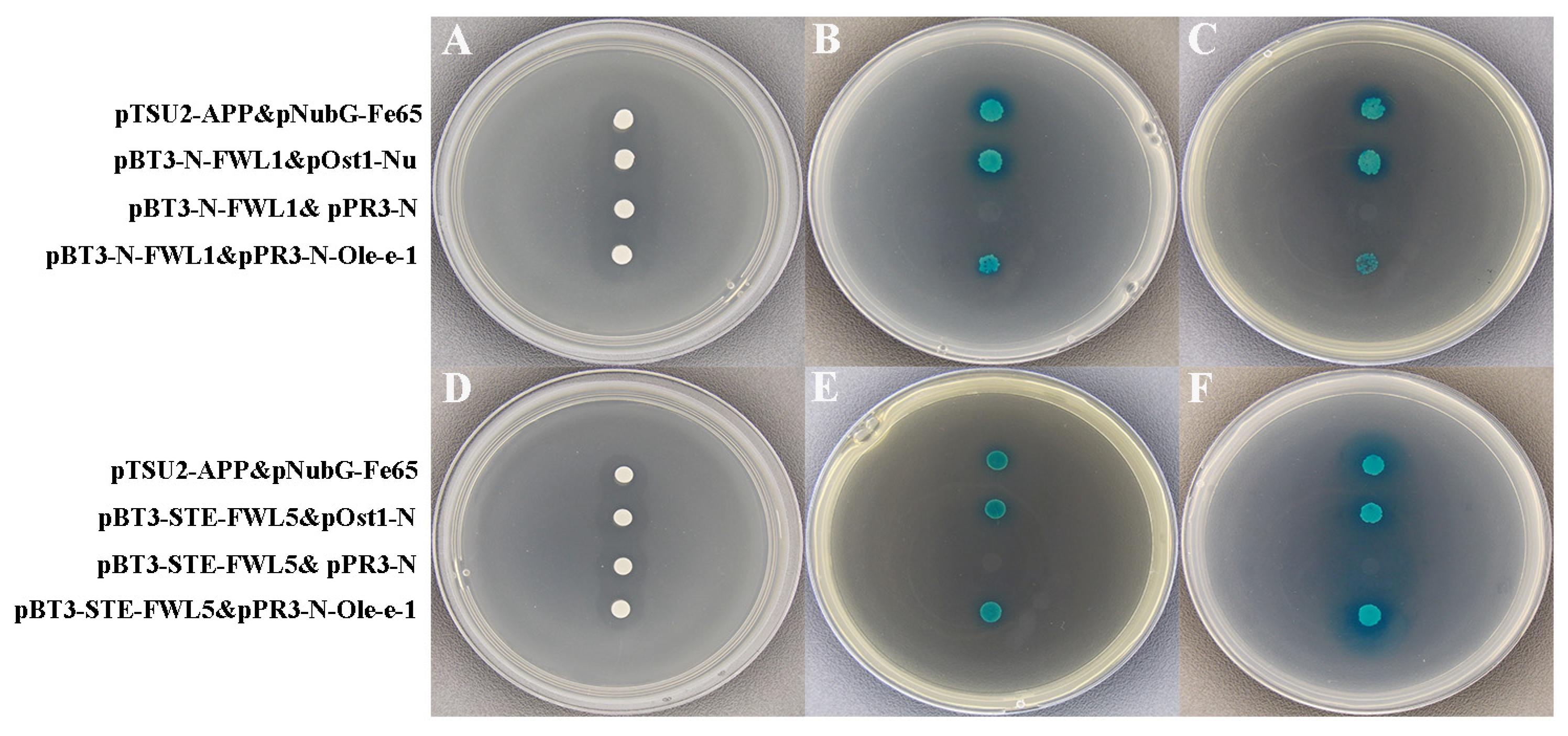
2.5. Self-Activation and Functional Assay of Decoy Expression Vectors
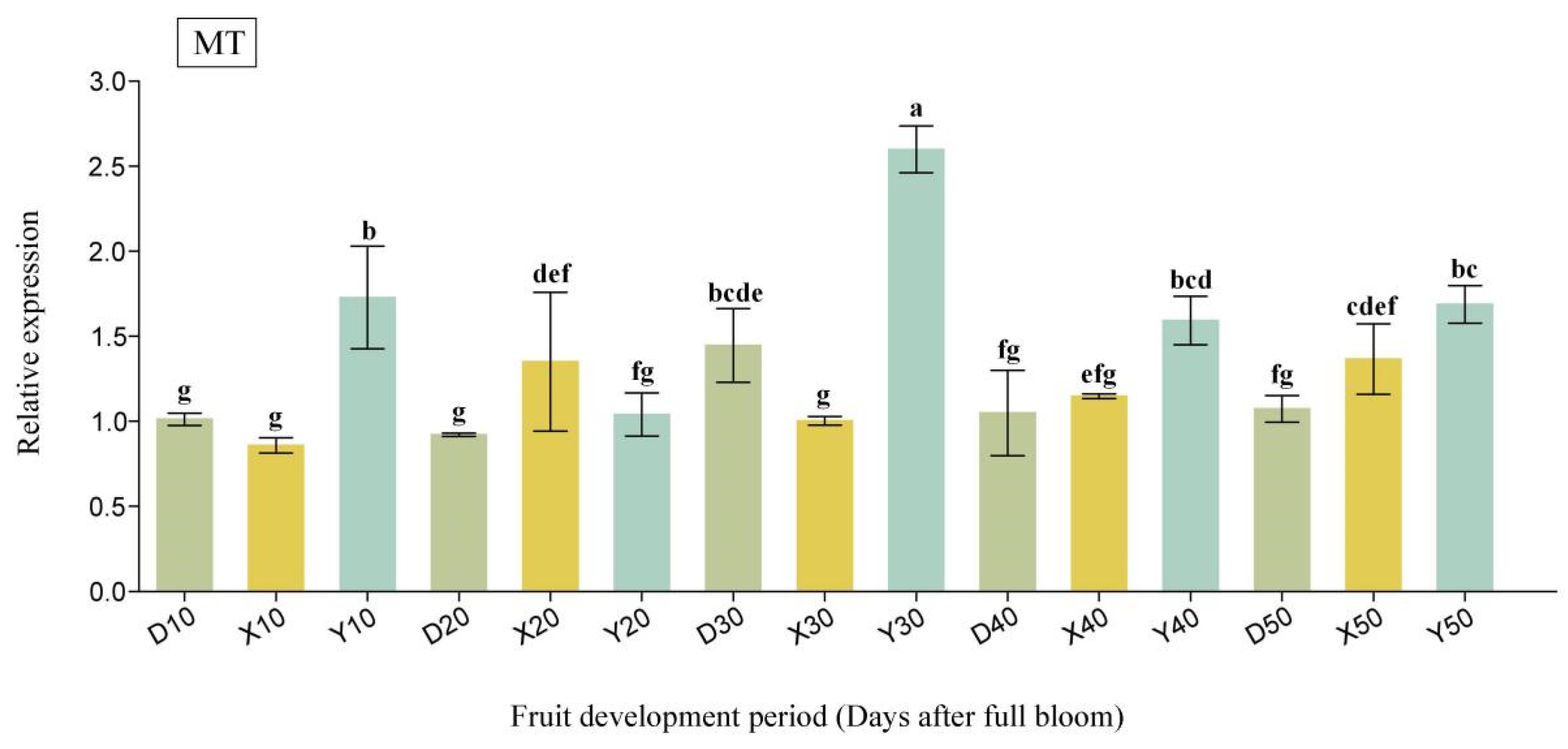
2.6. Expression Analysis of Candidate Interactions Protein-Related Genes in Pear Fruit During Critical Cell Division Stages
2.7. Validation of FWL1/5 Interactions with Ole-e-1
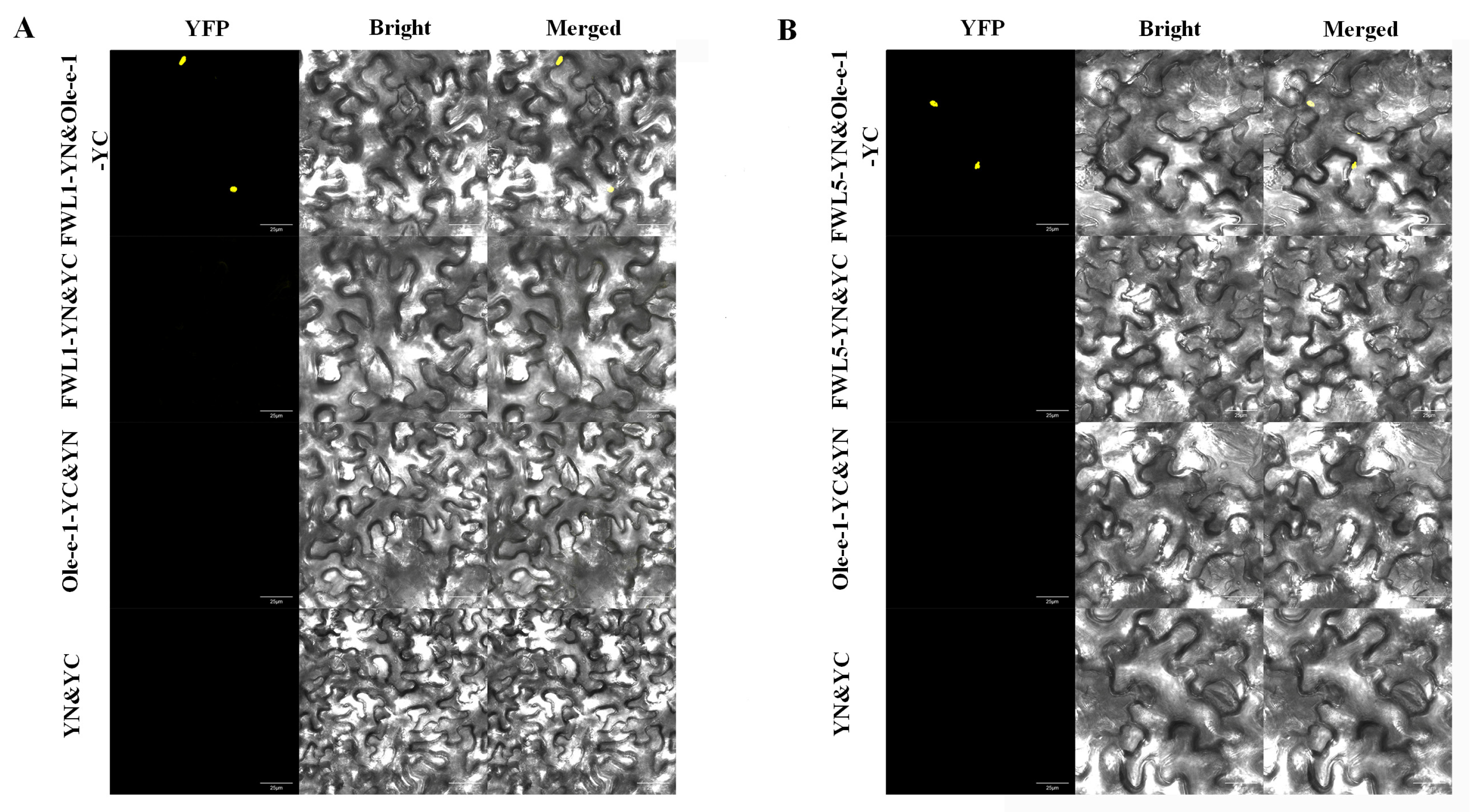
2.8. BiFC Demonstrates That FWL1/5 Interacts with Ole-e-1 Protein
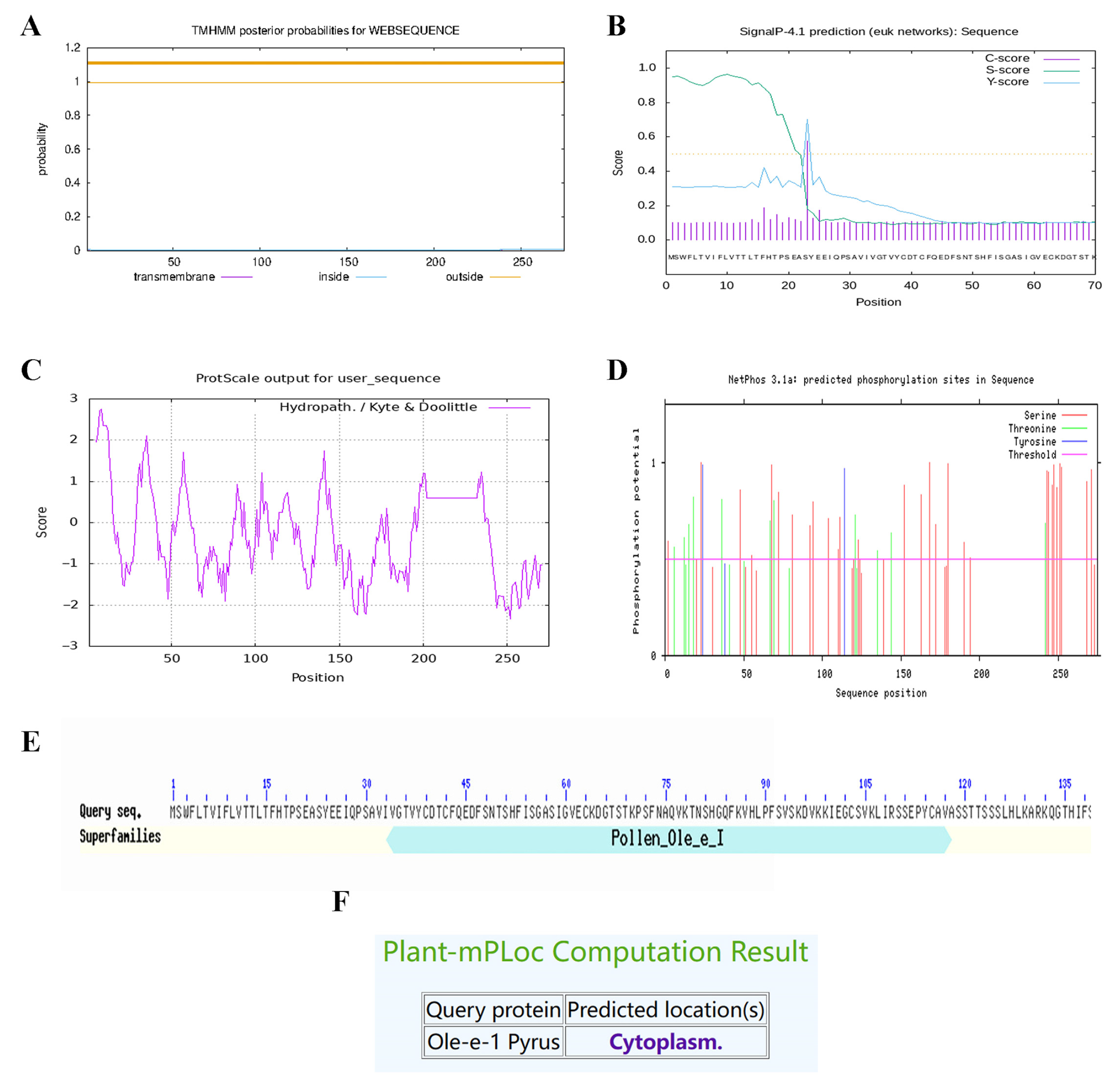
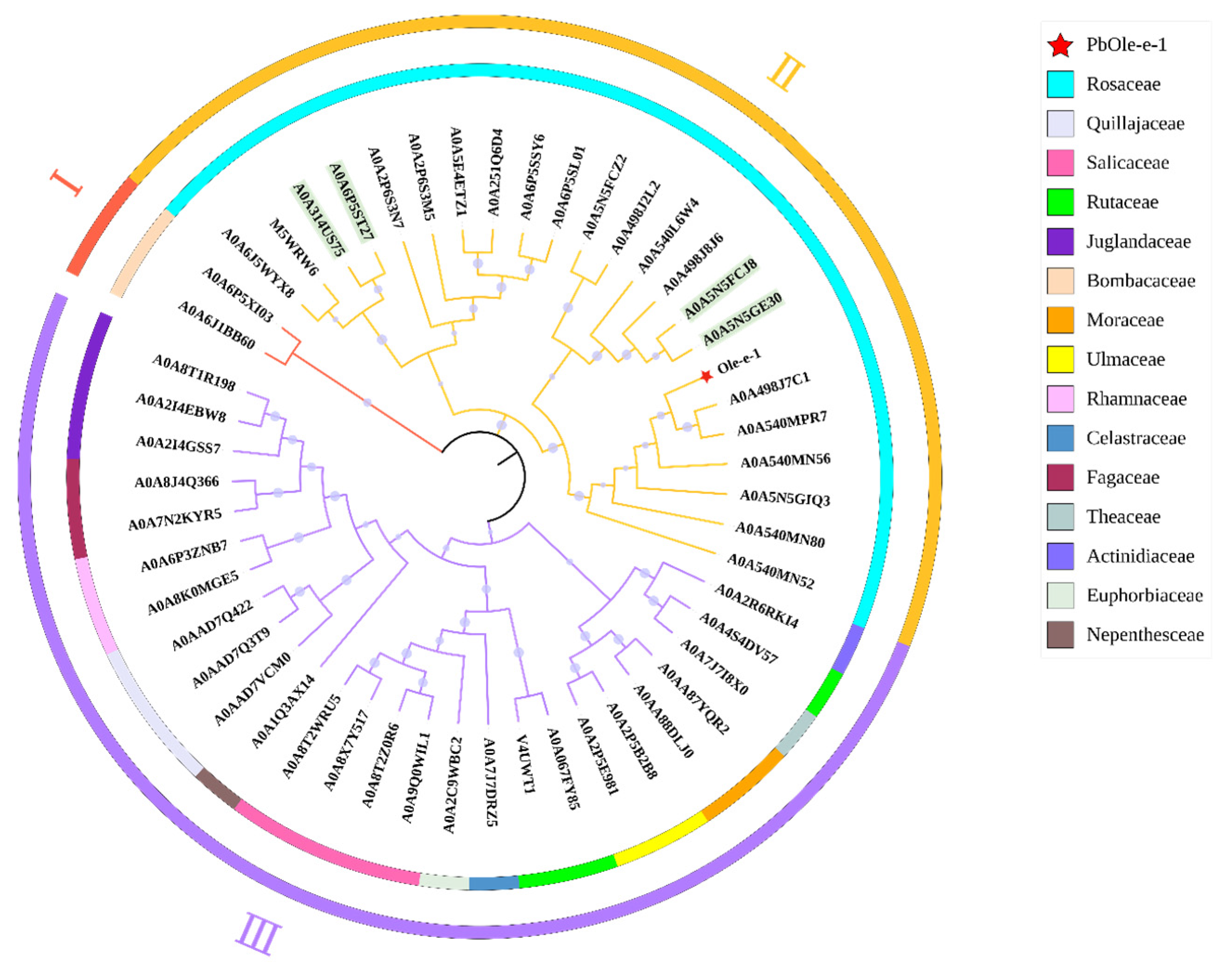
2.9. Ole-e-1 Bioinformatics Analysis
3. Discussion
4. Materials and Methods
4.1. Test Materials and Growth Conditions
4.2. FWL1 and FWL5 Tissue Fluorescence In Situ Hybridization (FISH)
4.3. Construction of VIGS Vectors for Pear FWL Genes
4.4. VIGS of FWL Genes
4.5. Library Construction and Protein Screening
4.6. Construction of Decoy Expression Vectors and Self-Activation Assay
4.7. Fluorescence Quantification
4.8. Yeast Two-Hybrid Validation
4.9. Bimolecular Fluorescence Complementation (BiFC)
4.10. Candidate Gene Biosignature Analysis
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Appendix A
| Gene Name | Sequence (5′-3′) |
|---|---|
| FWL1 | CATTTTGGAGCGGTAGAAGCATGAGTAGCAGCATGGACAAGCTGT |
| FWL5 | CAACGACTGCCTAAACAGACCTGTGTAGATTGCACACATCCACCA |
| Primers | Primers Sequence (5′-3′) |
|---|---|
| FWL1-F | CGGAATTCGACAGGTATCCCGGTCAGCT |
| FWL1-R | CGGGATCCCCAAGGGATGAGAGCACGA |
| FWL5-F | CGGAATTCGAGAGTTGCTGGACTGGACT |
| FWL5-R | CGGGATCCGCAATTCCTCCTTCAACGCA |
| FWL1-F (pBT3-N) | GAATTCCTGCAGGGCCATTACGGCCATGTACTCGTCACACCCAAGGG |
| FWL1-R (pBT3-N) | TACTTACCATGGGGCCGAGGCGGCTTTATCTCGACTCATGCCTTCCTC |
| FWL1-F (pBT3-C) | CACTAATCTAGACGGCCATTACGGCCATGTACTCGTCACACCCAAGGG |
| FWL1-R (pBT3-C) | ATGGAGGCCTTTGGCCGAGGCGGCTTTATCTCGACTCATGCCTTCCTC |
| FWL1-F (pBT3-SUC) | AATATCTGCAATGGCCATTACGGCCATGTACTCGTCACACCCAAGGG |
| FWL1-R (pBT3-SUC) | ATTCCTGCAGATGGCCGAGGCGGCTTTATCTCGACTCATGCCTTCCTC |
| FWL1-F (pBT3-STE) | TATTTTATGTAATGGCCATTACGGCCATGTACTCGTCACACCCAAGGG |
| FWL1-R (pBT3-STE) | ATTCCTGCAGATGGCCGAGGCGGCTTTATCTCGACTCATGCCTTCCTC |
| FWL5-F (pBT3-N) | GAATTCCTGCAGGGCCATTACGGCCATGGCAGAAGGGAATCCGCA |
| FWL5-R (pBT3-N) | TACTTACCATGGGGCCGAGGCGGCCACAGGCTGCAGAGCCAG |
| FWL5-F (pBT3-C) | CACTAATCTAGACGGCCATTACGGCCATGGCAGAAGGGAATCCGCA |
| FWL5-R (pBT3-C) | ATGGAGGCCTTTGGCCGAGGCGGCCACAGGCTGCAGAGCCAG |
| FWL5-F (pBT3-SUC) | AATATCTGCAATGGCCATTACGGCCATGGCAGAAGGGAATCCGCA |
| FWL5-R (pBT3-SUC) | ATTCCTGCAGATGGCCGAGGCGGCCACAGGCTGCAGAGCCAG |
| FWL5-F (pBT3-STE) | TATTTTATGTAATGGCCATTACGGCCATGGCAGAAGGGAATCCGCA |
| FWL5-R (pBT3-STE) | ATTCCTGCAGATGGCCGAGGCGGCCACAGGCTGCAGAGCCAG |
| pPR3-N-Ole-e-1-F | TGTTCCAGATTACGCTGGATCCATGTCTTGGTTTCTTACAGTTATCTTTCTGG |
| pPR3-N-Ole-e-1-R | ACTAATTACATGACTCGAGGTCGACTCATGGTGCTGAACCCGAT |
| MT-F | GTGGTAAATGTGGTTGCGGGTC |
| MT-R | CCGCACTTGCATCCGTTCTC |
| Ole-e-1-F | GCCTTCTGCAGTAATTGTTGGC |
| Ole-e-1-R | GTCCCATCTTTGCATTCCACACC |
| UBI-F | GCTCGCAGTGCTCCAGTTCTAC |
| UBI-R | AACATAGGTCAACCCGCACTT |
| pEarleyGate201-YN-FWL1-F | ACAAGTTTGTACAAAAAAATGTACTCGTCACACCCAAGG |
| pEarleyGate201-YN-FWL1-R | CACCACTTTGTACAAGAATTTATCTCGACTCATGCCTTCC |
| pEarleyGate201-YN-FWL5-F | ACAAGTTTGTACAAAAAAATGGCAGAAGGGAATCCG |
| pEarleyGate201-YN-FWL5-R | CACCACTTTGTACAAGAACACAGGCTGCAGAGCCAGCT |
| pEarleyGate202-YC-Ole-e-1-F | ACAAGTTTGTACAAAAAAATGTCTTGGTTTCTTACAGTTATCTTTCTGG |
| pEarleyGate202-YC-Ole-e-1-R | CACCACTTTGTACAAGAATGGTGCTGAACCCGATGGT |
| Combination | Plasmid Combination | Function |
|---|---|---|
| 1 | pBT3-N-FWL5&pOst1-NubI | Functional testing |
| 2 | pBT3-C-FWL5&pOst1-NubI | Functional testing |
| 3 | pBT3-SUC-FWL5&pOst1-NubI | Functional testing |
| 4 | pBT3-STE-FWL5&pOst1-NubI | Functional testing |
| 5 | pBT3-N-FWL5&pPR3-N | Self-activation detection |
| 6 | pBT3-C-FWL5&pPR3-N | Self-activation detection |
| 7 | pBT3-SUC-FWL5&pPR3-N | Self-activation detection |
| 8 | pBT3-STE-FWL5&pPR3-N | Self-activation detection |
References
- Camarero, M.C.; Briegas, B.; Corbacho, J.; Labrador, J.; Román, Á.C.; Verde, A.; Gallardo, M.; Gomez-Jimenez, M.C. Variations in Fruit Ploidy Level and Cell Size Between Small- and Large-Fruited Olive Cultivars During Fruit Ontogeny. Plants 2024, 13, 990. [Google Scholar] [CrossRef] [PubMed]
- Sha, G.; Cheng, J.; Wang, X.; Xue, Q.; Zhang, H.; Zhai, R.; Yang, C.; Wang, Z.; Xu, L. PbbHLH137 interacts with PbGIF1 to regulate pear fruit development by promoting cell expansion to increase fruit size. Physiol. Plant. 2024, 176, e14451. [Google Scholar] [CrossRef]
- Tian, Z.; Wu, B.; Liu, J.; Zhang, L.; Wu, T.; Wang, Y.; Han, Z.; Zhang, X. Genetic variations in MdSAUR36 participate in the negative regulation of mesocarp cell division and fruit size in Malus species. Mol. Breed. 2024, 44, 1. [Google Scholar] [CrossRef]
- Zhao, M.; Huang, Q.; Zhang, B.; Guo, W.; Lv, G.; Wang, D.; Yang, Y.; Wang, C.; Wu, X.; Zhang, Z.; et al. Histological and transcriptomic analysis reveals the cell number and plant hormone related to fruit size in Litchi chinensis Sonn. Sci. Hortic. 2025, 341, 114007. [Google Scholar] [CrossRef]
- Causse, M.; Chaïb, J.; Lecomte, L.; Buret, M.; Hospital, F. Both additivity and epistasis control the genetic variation for fruit quality traits in tomato. Theor. Appl. Genet. 2007, 115, 429–442. [Google Scholar] [CrossRef]
- Grandillo, S.; Ku, H.M.; Tanksley, S.D. Identifying the loci responsible for natural variation in fruit size and shape in tomato. Theor. Appl. Genet. 1999, 99, 978–987. [Google Scholar] [CrossRef]
- Rodríguez, G.R.; Muños, S.; Anderson, C.; Sim, S.C.; Michel, A.; Causse, M.; McSpadden Gardener, B.B.; Francis, D.; van der Knaap, E. Distribution of SUN, OVATE, LC, and FAS in the tomato germplasm and the relationship to fruit shape diversity. Plant Physiol. 2011, 156, 275–285. [Google Scholar] [CrossRef]
- Alpert, K.B.; Tanksley, S.D. High-resolution mapping and isolation of a yeast artificial chromosome contig containing fw2.2: A major fruit weight quantitative trait locus in tomato. Proc. Natl. Acad. Sci. USA 1996, 93, 15503–15507. [Google Scholar] [CrossRef]
- Frary, A.; Nesbitt, T.C.; Frary, A.; Grandillo, S.; van der Knaap, E.; Cong, B.; Liu, J.; Meller, J.; Elber, R.; Alpert, K.B.; et al. Fw2.2: A Quantitative Trait Locus Key to the Evolution of Tomato Fruit Size. Science 2000, 289, 85–88. [Google Scholar] [CrossRef]
- Liu, J.; Cong, B.; Tanksley, S.D. Generation and analysis of an artificial gene dosage series in tomato to study the mechanisms by which the cloned quantitative trait locus fw2.2 Controls fruit size. Plant Physiol. 2003, 132, 292–299. [Google Scholar] [CrossRef]
- Tanksley, S.D. The Genetic, Developmental, and Molecular Bases of Fruit Size and Shape Variation in Tomato. Plant Cell Online 2004, 16, S181–S189. [Google Scholar] [CrossRef]
- Kortstee, A.J.; Appeldoorn, N.J.G.; Oortwijn, M.E.P.; Visser, R.G.F. Differences in regulation of carbohydrate metabolism during early fruit development between domesticated tomato and two wild relatives. Planta 2007, 226, 929–939. [Google Scholar] [CrossRef]
- Dahan, Y.; Rosenfeld, R.; Zadiranov, V.; Irihimovitch, V. A proposed conserved role for an avocado fw2.2-like gene as a negative regulator of fruit cell division. Planta 2010, 232, 663–676. [Google Scholar] [CrossRef]
- Li, Z.; He, C. Physalis floridana Cell Number Regulator1 encodes a cell membrane-anchored modulator of cell cycle and negatively controls fruit size. J. Exp. Bot. 2015, 66, 257–270. [Google Scholar] [CrossRef] [PubMed]
- Tian, J.; Zeng, B.; Luo, S.; Li, X.; Wu, B.; Li, J. Cloning, localization and expression analysis of two fw2.2-like genes in small- and large-fruited pear species. J. Integr. Agric. 2016, 15, 282–294. [Google Scholar] [CrossRef]
- Guo, M.; Rupe, M.A.; Dieter, J.A.; Zou, J.; Spielbauer, D.; Duncan, K.E.; Howard, R.J.; Hou, Z.; Simmons, C.R. Cell Number Regulator1 Affects Plant and Organ Size in Maize: Implications for Crop Yield Enhancement and Heterosis. Plant Cell 2010, 22, 1057–1073. [Google Scholar] [CrossRef]
- Xu, J.; Xiong, W.; Cao, B.; Yan, T.; Luo, T.; Fan, T.; Luo, M. Molecular characterization and functional analysis of “fruit-weight2.2-like” gene family in rice. Planta 2013, 238, 643–655. [Google Scholar] [CrossRef] [PubMed]
- Gao, Q.; Li, G.; Sun, H.; Xu, M.; Wang, H.; Ji, J.; Wang, D.; Yuan, C.; Zhao, X. Targeted Mutagenesis of the Rice FW 2.2-Like Gene Family Using the CRISPR/Cas9 System Reveals OsFWL4 as a Regulator of Tiller Number and Plant Yield in Rice. Int. J. Mol. Sci. 2020, 21, 809. [Google Scholar] [CrossRef]
- Libault, M.; Zhang, X.C.; Govindarajulu, M.; Qiu, J.; Ong, Y.T.; Brechenmacher, L.; Berg, R.H.; Hurley-Sommer, A.; Taylor, C.G.; Stacey, G. A member of the highly conserved FWL (tomato FW2.2-like) gene family is essential for soybean nodule organogenesis. Plant J. 2010, 62, 852–864. [Google Scholar] [CrossRef]
- Wang, G.-L.; Zhang, C.-L.; Huo, H.-Q.; Sun, X.-S.; Zhang, Y.-L.; Hao, Y.-J.; You, C.-X. The SUMO E3 Ligase MdSIZ1 Sumoylates a Cell Number Regulator MdCNR8 to Control Organ Size. Front. Plant Sci. 2022, 13, 836935. [Google Scholar] [CrossRef] [PubMed]
- Cong, B.; Liu, J.; Tanksley, S.D. Natural alleles at a tomato fruit size quantitative trait locus differ by heterochronic regulatory mutations. Proc. Natl. Acad. Sci. USA 2002, 99, 13606–13611. [Google Scholar] [CrossRef]
- Su, W. The Cellular and Molecular Mechanisms of Pome Size Domestication in Eriobotrya japonica; South China: Guangzhou, China, 2017. [Google Scholar]
- Beauchet, A.; Gévaudant, F.; Gonzalez, N.; Chevalier, C. In search of the still unknown function of FW2.2/CELL NUMBER REGULATOR, a major regulator of fruit size in tomato. J. Exp. Bot. 2021, 72, 5300–5311. [Google Scholar] [CrossRef]
- Beauchet, A.; Bollier, N.; Grison, M.; Rofidal, V.; Gévaudant, F.; Bayer, E.; Gonzalez, N.; Chevalier, C. The CELL NUMBER REGULATOR FW2.2 protein regulates cell-to-cell communication in tomato by modulating callose deposition at plasmodesmata. Plant Physiol. 2024, 196, 883–901. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Wang, Z.; Shi, Z.; Zhang, S.; Ming, R.; Zhu, S.; Khan, M.A.; Tao, S.; Korban, S.S.; Wang, H.; et al. The genome of the pear (Pyrus bretschneideri Rehd.). Genome Res. 2013, 23, 396–408. [Google Scholar] [CrossRef]
- Zhang, Y.; Tian, J.; Zhang, F.; Wen, Y. Cloning and expression analysis of FWL5 gene in pear. Mol. Plant Breed. 2022, 20, 4940–4947. [Google Scholar] [CrossRef]
- Sai, J.; Wen, Y.; Hao, Z.; Tian, J. Construction of Yeast Two-Hybrid Three-Frame cDNA Library and Screening of Interacting Proteins in FWL1 Membrane System of Pear Young Fruit. Xinjiang Agric. Sci. 2022, 59, 1877–1888. [Google Scholar] [CrossRef]
- Yuan, J.; Chen, D.; Ren, Y.; Zhang, X.; Zhao, J. Characteristic and expression analysis of a metallothionein gene, OsMT2b, down-regulated by cytokinin suggests functions in root development and seed embryo germination of rice. Plant Physiol. 2008, 146, 1637–1650. [Google Scholar] [CrossRef]
- Fonseca-García, C.; López-García, C.M.; Pacheco, R.; Armada, E.; Nava, N.; Pérez-Aguilar, R.; Solis-Miranda, J.; Quinto, C. Metallothionein1A Regulates Rhizobial Infection and Nodulation in Phaseolus vulgaris. Int. J. Mol. Sci. 2022, 23, 1491. [Google Scholar] [CrossRef] [PubMed]
- Nakagawa, Y.; Katagiri, T.; Shinozaki, K.; Qi, Z.; Tatsumi, H.; Furuichi, T.; Kishigami, A.; Sokabe, M.; Kojima, I.; Sato, S.; et al. Arabidopsis plasma membrane protein crucial for Ca2+ influx and touch sensing in roots. Proc. Natl. Acad. Sci. USA 2007, 104, 3639–3644. [Google Scholar] [CrossRef]
- Song, W.-Y.; Hörtensteiner, S.; Tomioka, R.; Lee, Y.; Martinoia, E. Common Functions or Only Phylogenetically Related? The Large Family of PLAC8 Motif-Containing/PCR Genes. Mol. Cells 2011, 31, 1–8. [Google Scholar] [CrossRef]
- Tsyganov, V.E.; Tsyganova, A.V.; Gorshkov, A.P.; Seliverstova, E.V.; Kim, V.E.; Chizhevskaya, E.P.; Belimov, A.A.; Serova, T.A.; Ivanova, K.A.; Kulaeva, O.A.; et al. Efficacy of a Plant-Microbe System: Pisum sativum (L.) Cadmium-Tolerant Mutant and Rhizobium leguminosarum Strains, Expressing Pea Metallothionein Genes PsMT1 and PsMT2, for Cadmium Phytoremediation. Front. Microbiol. 2020, 11, 15. [Google Scholar] [CrossRef]
- Larson, J.D.; Hoskins, A.A. Dynamics and consequences of spliceosome E complex formation. eLife 2017, 6, e27592. [Google Scholar] [CrossRef] [PubMed]
- Bateman, A.; Coin, L.; Durbin, R.; Finn, R.D.; Hollich, V.; Griffiths-Jones, S.; Khanna, A.; Marshall, M.; Moxon, S.; Sonnhammer, E.L.L.; et al. The Pfam protein families database. Nucleic Acids Res. 2004, 32, 138D–141D. [Google Scholar] [CrossRef]
- Carlos, J.; Isabel, M.; de Dios Alche, J. Systematic and Phylogenetic Analysis of the Ole e 1 Pollen Protein Family Members in Plants. In Systems and Computational Biology-Bioinformatics and Computational Modeling; InTech: London, UK, 2011; pp. 245–260. [Google Scholar]
- Rodríguez, R.; Villalba, M.; Batanero, E.; González, E.M.; Monsalve, R.I.; Huecas, S.; Tejera, M.L.; Ledesma, A. Allergenic diversity of the olive pollen. Allergy 2002, 57, 6–16. [Google Scholar] [CrossRef] [PubMed]
- Muschietti, J.; Dircks, L.; Vancanneyt, G.; McCormick, S. LAT52 protein is essential for tomato pollen development: Pollen expressing antisense LAT52 RNA hydrates and germinates abnormally and cannot achieve fertilization. Plant J. 1994, 6, 321–338. [Google Scholar] [CrossRef]
- Alché, J.D.D.; M’rani-Alaoui, M.; Castro, A.J.; Rodríguez-García, M.I. Ole e 1, the Major Allergen from Olive (Olea europaea L.) Pollen, Increases Its Expression and Is Released to the Culture Medium During In Vitro Germination. Plant Cell Physiol. 2004, 45, 1149–1157. [Google Scholar] [CrossRef] [PubMed]
- Tang, W.; Newton, R.J. Polyamines reduce salt-induced oxidative damage by increasing the activities of antioxidant enzymes and decreasing lipid peroxidation in Virginia pine. Plant Growth Regul. 2005, 46, 31–43. [Google Scholar] [CrossRef]
- Bruex, A.; Kainkaryam, R.M.; Wieckowski, Y.; Kang, Y.H.; Bernhardt, C.; Xia, Y.; Zheng, X.; Wang, J.Y.; Lee, M.M.; Benfey, P.; et al. A Gene Regulatory Network for Root Epidermis Cell Differentiation in Arabidopsis. PLoS Genet. 2012, 8, e1002446. [Google Scholar] [CrossRef]
- Jiang, S.Y.; Jasmin, P.X.H.; Ting, Y.Y.; Ramachandran, S. Genome-Wide Identification and Molecular Characterization of Ole_e_I, Allerg_1 and Allerg_2 Domain-containing Pollen-Allergen-like Genes in Oryza sativa. DNA Res. 2005, 12, 167–179. [Google Scholar] [CrossRef]
- Hu, B.; Liu, B.; Liu, L.; Liu, C.; Xu, L.; Ruan, Y. Epigenetic control of Pollen Ole e 1 allergen and extensin family gene expression in Arabidopsis thaliana. Acta Physiol. Plant. 2014, 36, 2203–2209. [Google Scholar] [CrossRef]
- Jackson, P.K. The Hunt for Cyclin. Cell 2008, 134, 199–202. [Google Scholar] [CrossRef]
- Rutschmann, F.; Stalder, U.; Piotrowski, M.; Oecking, C.; Schaller, A. LeCPK1, a calcium-dependent protein kinase from tomato. Plasma membrane targeting and biochemical characterization. Plant Physiol. 2002, 129, 156–168. [Google Scholar] [CrossRef]
- Singh, D.; Verma, N.; Rengasamy, B.; Banerjee, G.; Sinha, A.K. The small RNA biogenesis in rice is regulated by MAP kinase-mediated OsCDKD phosphorylation. New Phytol. 2024, 244, 1482–1497. [Google Scholar] [CrossRef]
- Kollárová, E.; Baquero Forero, A.; Cvrčková, F. The Arabidopsis thaliana Class II Formin FH13 Modulates Pollen Tube Growth. Front. Plant Sci. 2021, 12, 599961. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Yang, X.; Zhang, X.; Song, Y.; Liang, W.; Zhang, D. Rice morphology determinant-mediated actin filament organization contributes to pollen tube growth. Plant Physiol. 2018, 177, 255–270. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zhang, Y.; Wu, J.; Meng, L.; Ren, H. At FH16, an Arabidopsis Type II Formin, Binds and Bundles Both Microfilaments and Microtubules, and Preferentially Binds to Microtubules. J. Integr. Plant Biol. 2013, 55, 1002–1015. [Google Scholar] [CrossRef] [PubMed]
- Pu, X.; Tian, J.; Li, J.; Zhang, Y.; Li, P.; Qin, W.; Jing, C. Analysis on expression stability of internal reference genes at cell division stage of pear fruits. Non-Wood For. Res. 2020, 38, 66–74. [Google Scholar] [CrossRef]

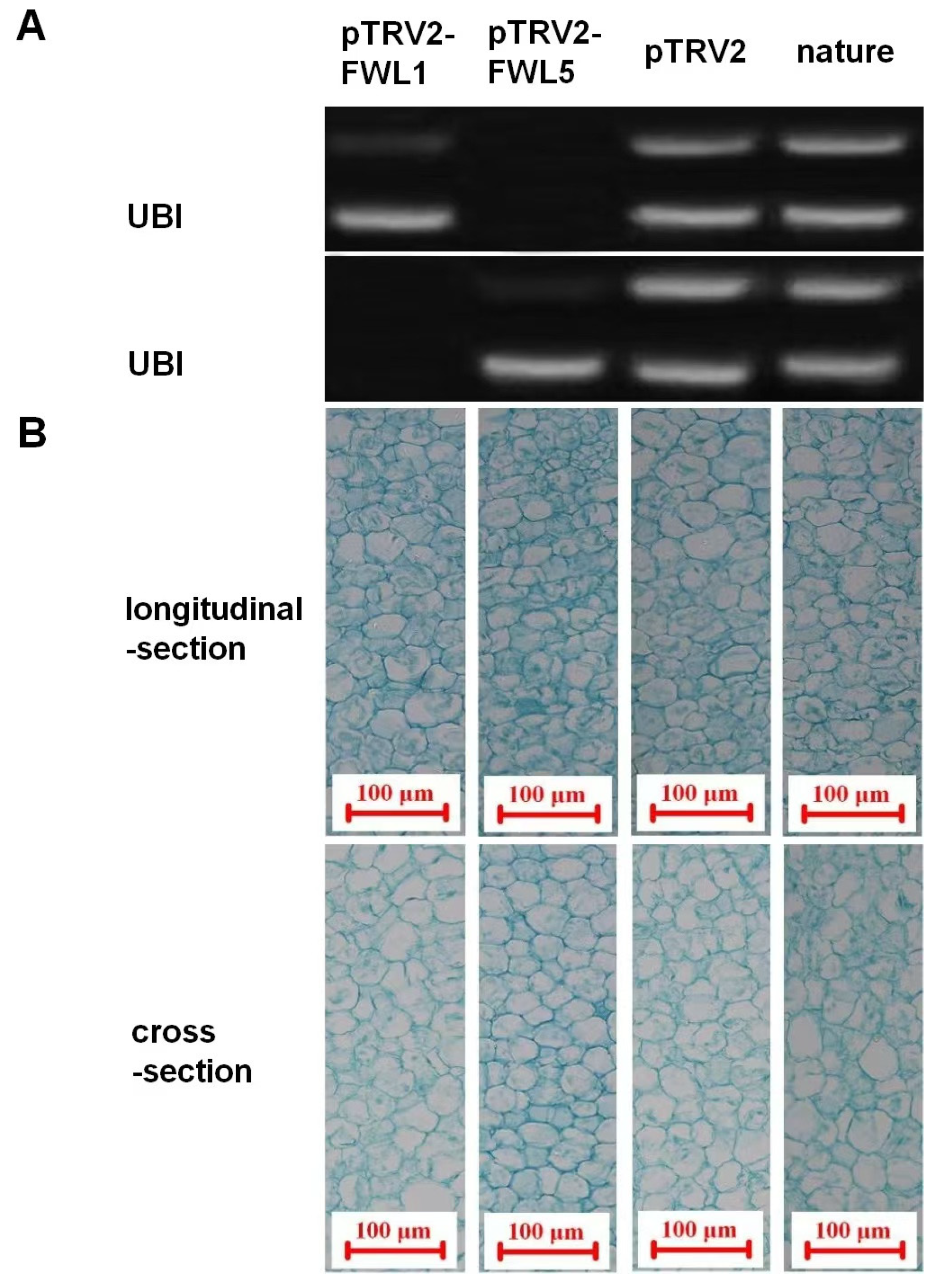


| Treatment | Fruit Weight (g) | Fruit Volume (mm3) | Cell Volume (×10−6 mm3) | Cell Numbers (×104) |
|---|---|---|---|---|
| pTRV2-FWL1 | 1.13 ± 0.04 b | 596.98 ± 30.54 b | 20.65 ± 0.62 a | 2890.94 ± 75.78 b |
| pTRV2-FWL5 | 1.58 ± 0.56 a | 663.33 ± 34.65 a | 18.94 ± 0.39 b | 3502.27 ± 83.44 a |
| pTRV2 | 1.18 ± 0.09 b | 608.82 ± 31.70 b | 20.43 ± 0.60 a | 2980.03 ± 73.45 b |
| nature | 1.12 ± 0.06 b | 589.65 ± 28.36 b | 20.75 ± 0.53 a | 2841.69 ± 69.78 b |
| Number | Accession | Marginal Notes | Frequency |
|---|---|---|---|
| 1 | XM_009352003.2 | metallothionein-like protein type 2 | 7 |
| 2 | XM_009367135.2 | uncharacterized | 6 |
| 3 | XM_009379993.2 | collagen and calcium-binding EGF domain-containing protein 1-like | 5 |
| 4 | XM_009379993.3 | branchpoint-bridging protein-like | 5 |
| 5 | BX640903.1 | Homo sapiens mRNA | 3 |
| 6 | GQ906589.1 | metallothionein-like protein (Met1) | 3 |
| 7 | NM_001302326.1 | auxin-repressed 12.5 kDa protein-like (ARP) | 3 |
| 8 | XM_008343913.3 | small ubiquitin-related modifier 1-like | 3 |
| 9 | XM_009337215.2 | V-type proton ATPase 16 kDa proteolipid subunit-like | 3 |
| 10 | XM_009340070.2 | aquaporin TIP1-3-like | 3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sai, J.; Wen, Y.; Zhang, Y.; Pu, X.; Chen, C.; Wang, L.; Zhu, M.; Tian, J. Ole-e-1 Interacts with FWL Genes to Modulate Cell Division and Determine Fruit Size in Pears. Int. J. Mol. Sci. 2025, 26, 8804. https://doi.org/10.3390/ijms26188804
Sai J, Wen Y, Zhang Y, Pu X, Chen C, Wang L, Zhu M, Tian J. Ole-e-1 Interacts with FWL Genes to Modulate Cell Division and Determine Fruit Size in Pears. International Journal of Molecular Sciences. 2025; 26(18):8804. https://doi.org/10.3390/ijms26188804
Chicago/Turabian StyleSai, Jingyi, Yue Wen, Yan Zhang, Xiaoqiu Pu, Chen Chen, Lei Wang, Mengli Zhu, and Jia Tian. 2025. "Ole-e-1 Interacts with FWL Genes to Modulate Cell Division and Determine Fruit Size in Pears" International Journal of Molecular Sciences 26, no. 18: 8804. https://doi.org/10.3390/ijms26188804
APA StyleSai, J., Wen, Y., Zhang, Y., Pu, X., Chen, C., Wang, L., Zhu, M., & Tian, J. (2025). Ole-e-1 Interacts with FWL Genes to Modulate Cell Division and Determine Fruit Size in Pears. International Journal of Molecular Sciences, 26(18), 8804. https://doi.org/10.3390/ijms26188804






