Correlation Between the Proportion of Senescence-Associated β-Galactosidase-Stained CD8+ T Cells and Age: A Cross-Sectional Study in Japan
Abstract
1. Introduction
2. Results
3. Discussion
4. Materials and Methods
4.1. Participants and Analysis
4.2. Clinical Features
4.3. PBMC Collection and Preservation
4.4. Flow Cytometric Analysis of PBMCs
- Naïve T cells: CD197 (CCR7)+CD45RA+;
- TCM: CD197 (CCR7)+CD45RA-;
- TEM: CD197 (CCR7)-CD45RA-;
- TEMRA: CD197 (CCR7)-CD45RA+;
- PD-1-positive cells: CD279+;
- T cells with SA-βGalhigh expression.
4.5. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- UN. World Population Prospects: The 2024 Revision. Available online: https://www.un.org/development/desa/pd/world-population-prospects-2024 (accessed on 22 July 2025).
- De Lepeleire, J.; Iliffe, S.; Mann, E.; Degryse, J.M. Frailty: An emerging concept for general practice. Br. J. Gen. Pract. 2009, 59, e177–e182. [Google Scholar] [CrossRef]
- Clegg, A.; Young, J.; Iliffe, S.; Rikkert, M.O.; Rockwood, K. Frailty in elderly people. Lancet 2013, 381, 752–762. [Google Scholar] [CrossRef]
- Beard, J.R.; Officer, A.; de Carvalho, I.A.; Sadana, R.; Pot, A.M.; Michel, J.P.; Lloyd-Sherlock, P.; Epping-Jordan, J.E.; Peeters, G.M.E.E.G.; Mahanani, W.R.; et al. The World report on ageing and health: A policy framework for healthy ageing. Lancet 2016, 387, 2145–2154. [Google Scholar] [CrossRef]
- Majnarić, L.T.; Bosnić, Z.; Guljaš, S.; Vučić, D.; Kurevija, T.; Volarić, M.; Martinović, I.; Wittlinger, T. Low Psychological Resilience in Older Individuals: An Association with Increased Inflammation, Oxidative Stress and the Presence of Chronic Medical Conditions. Int. J. Mol. Sci. 2021, 22, 8970. [Google Scholar] [CrossRef] [PubMed]
- Martínez de Toda, I.; Ceprián, N.; Díaz-Del Cerro, E.; De la Fuente, M. The Role of Immune Cells in Oxi-Inflamm-Aging. Cells 2021, 10, 2974. [Google Scholar] [CrossRef]
- Bektas, A.; Schurman, S.H.; Sen, R.; Ferrucci, L. Aging, inflammation and the environment. Exp. Gerontol. 2018, 105, 10–18. [Google Scholar] [CrossRef] [PubMed]
- Moqri, M.; Herzog, C.; Poganik, J.R.; Ying, K.; Justice, J.N.; Belsky, D.W.; Higgins-Chen, A.T.; Chen, B.H.; Cohen, A.A.; Fuellen, G.; et al. Validation of biomarkers of aging. Nat. Med. 2024, 30, 360–372. [Google Scholar] [CrossRef]
- Levine, M.E. Modeling the rate of senescence: Can estimated biological age predict mortality more accurately than chronological age? J. Gerontol. A Biol. Sci. Med. Sci. 2013, 68, 667–674. [Google Scholar] [CrossRef] [PubMed]
- Moqri, M.; Herzog, C.; Poganik, J.R.; Justice, J.; Belsky, D.W.; Higgins-Chen, A.; Moskalev, A.; Fuellen, G.; Cohen, A.A.; Bautmans, I.; et al. Biomarkers of aging for the identification and evaluation of longevity interventions. Cell 2023, 186, 3758–3775. [Google Scholar] [CrossRef]
- Gorgoulis, V.; Adams, P.D.; Alimonti, A.; Bennett, D.C.; Bischof, O.; Bishop, C.; Campisi, J.; Collado, M.; Evangelou, K.; Ferbeyre, G.; et al. Cellular senescence: Defining a path forward. Cell 2019, 179, 813–827. [Google Scholar] [CrossRef]
- He, S.; Sharpless, N.E. Senescence in health and disease. Cell 2017, 169, 1000–1011. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.Y.; Han, J.A.; Im, J.S.; Morrone, A.; Johung, K.; Goodwin, E.C.; Kleijer, W.J.; DiMaio, D.; Hwang, E.S. Senescence-associated β-galactosidase is lysosomal β-galactosidase. Aging Cell 2006, 5, 187–195. [Google Scholar] [CrossRef]
- Hooten, N.N.; Evans, M.K. Techniques to induce and quantify cellular senescence. J. Vis. Exp. 2017, 123, 55533. [Google Scholar]
- González-Gualda, E.; Baker, A.G.; Fruk, L.; Muñoz-Espín, D. A guide to assessing cellular senescence in vitro and in vivo. FEBS J. 2021, 288, 56–80. [Google Scholar] [CrossRef]
- Hickson, L.J.; Prata, L.G.; Bobart, S.A.; Evans, T.K.; Giorgadze, N.; Hashmi, S.K.; Herrmann, S.M.; Jensen, M.D.; Jia, Q.; Jordan, K.L.; et al. Senolytics decrease senescent cells in humans: Preliminary report from a clinical trial of Dasatinib plus Quercetin in individuals with diabetic kidney disease. EBioMedicine 2019, 47, 446–456, Erratum in EBioMedicine 2020, 52, 102595. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Zamudio, R.I.; Dewald, H.K.; Vasilopoulos, T.; Gittens-Williams, L.; Fitzgerald-Bocarsly, P.; Herbig, U. Senescence-associated β-Galactosidase reveals the abundance of senescent CD8+ T cells in aging humans. Aging Cell 2021, 20, e13344. [Google Scholar] [CrossRef]
- Liu, Z.; Liang, Q.; Ren, Y.; Guo, C.; Ge, X.; Wang, L.; Cheng, Q.; Luo, P.; Zhang, Y.; Han, X. Immunosenescence: Molecular mechanisms and diseases. Signal Transduct. Target. Ther. 2023, 8, 200. [Google Scholar] [CrossRef]
- Nga, H.T.; Nguyen, T.L.; Yi, H.S. T-Cell Senescence in Human Metabolic Diseases. Diabetes Metab J. 2024, 48, 864–881. [Google Scholar] [CrossRef]
- Laphanuwat, P.; Gomes, D.C.; Akbar, A.N. Senescent T cells: Beneficial and detrimental roles. Immunol. Rev. 2023, 316, 160–175. [Google Scholar] [CrossRef]
- Lee, Y.H.; Kim, S.R.; Han, D.H.; Yu, H.T.; Han, Y.D.; Kim, J.H.; Kim, S.H.; Lee, C.J.; Min, B.H.; Kim, D.H.; et al. Senescent T Cells Predict the Development of Hyperglycemia in Humans. Diabetes 2019, 68, 156–162. [Google Scholar] [CrossRef]
- Ju, S.H.; Lim, J.Y.; Song, M.; Kim, J.M.; Kang, Y.E.; Yi, H.S.; Joung, K.H.; Lee, J.H.; Kim, H.J.; Ku, B.J. Distinct effects of rosuvastatin and rosuvastatin/ezetimibe on senescence markers of CD8+ T cells in patients with type 2 diabetes mellitus: A randomized controlled trial. Front. Endocrinol. 2024, 15, 1336357. [Google Scholar] [CrossRef]
- Shimizu, Y.; Shimodan, S.; Hayashida, M.; Yazaki, M.; Sakurada, T.; Watanabe, T.; Ishii, Y.; Hirose, Y.; Saito, J.; Teramoto, S. Preliminary data on the senolytic effects of Agrimonia pilosa Ledeb. Extract containing agrimols for immunosenescence in middle-aged humans: A randomized, double-blind, placebo-controlled, parallel-group comparison study. Nutrients 2025, 17, 667. [Google Scholar] [CrossRef] [PubMed]
- Quan, X.Q.; Ruan, L.; Zhou, H.R.; Gao, W.L.; Zhang, Q.; Zhang, C.T. Age-related changes in peripheral T-cell subpopulations in elderly individuals: An observational study. Open Life Sci. 2023, 18, 20220557. [Google Scholar] [CrossRef]
- Chang, S.T.; Chuang, Y.F.; Li, A.H.; Fan, Y.T.; Liao, M.R.; Chen, I.Y.; Hung, R.W.; Yang, T.O.; Chiu, Y.L. Age-dependent immune profile in healthy individuals: An original study, systematic review and meta-analysis. Immun. Ageing 2024, 21, 75. [Google Scholar] [CrossRef] [PubMed]
- Goronzy, J.J.; Fang, F.; Cavanagh, M.M.; Qi, Q.; Weyand, C.M. Naive T cell maintenance and function in human aging. J. Immunol. 2015, 194, 4073–4080. [Google Scholar] [CrossRef]
- Briceño, O.; Lissina, A.; Wanke, K.; Afonso, G.; von Braun, A.; Ragon, K.; Miquel, T.; Gostick, E.; Papagno, L.; Stiasny, K.; et al. Reduced naïve CD8(+) T-cell priming efficacy in elderly adults. Aging Cell 2016, 15, 14–21. [Google Scholar] [CrossRef]
- Moskowitz, D.M.; Zhang, D.W.; Hu, B.; Le Saux, S.; Yanes, R.E.; Ye, Z.; Buenrostro, J.D.; Weyand, C.M.; Greenleaf, W.J.; Goronzy, J.J. Epigenomics of human CD8 T cell differentiation and aging. Sci. Immunol. 2017, 2, eaag0192. [Google Scholar] [CrossRef]
- Turano, P.S.; Akbulut, E.; Dewald, H.K.; Vasilopoulos, T.; Fitzgerald-Bocarsly, P.; Herbig, U.; Martínez-Zamudio, R.I. Epigenetic mechanisms regulating CD8+ T cell senescence in aging humans. bioRxiv 2025. [Google Scholar] [CrossRef]
- Salumets, A.; Tserel, L.; Rumm, A.P.; Türk, L.; Kingo, K.; Saks, K.; Oras, A.; Uibo, R.; Tamm, R.; Peterson, H.; et al. Epigenetic quantification of immunosenescent CD8+ TEMRA cells in human blood. Aging Cell 2022, 21, e13607. [Google Scholar] [CrossRef] [PubMed]
- Serra, S.; Chetty, R. p16. J. Clin. Pathol. 2018, 71, 853–858. [Google Scholar] [CrossRef]
- Safwan-Zaiter, H.; Wagner, N.; Wagner, K.-D. P16INK4A—More Than a Senescence Marker. Life 2022, 12, 1332. [Google Scholar] [CrossRef]
- Di Micco, R.; Krizhanovsky, V.; Baker, D.; d’Adda di Fagagna, F. Cellular senescence in ageing: From mechanisms to therapeutic opportunities. Nat. Rev. Mol. Cell Biol. 2021, 22, 75–95. [Google Scholar] [CrossRef]
- Baixauli, F.; Acín-Pérez, R.; Villarroya-Beltrí, C.; Mazzeo, C.; Nuñez-Andrade, N.; Gabandé-Rodriguez, E.; Ledesma, M.D.; Blázquez, A.; Martin, M.A.; Falcón-Pérez, J.M.; et al. Mitochondrial respiration controls lysosomal function during inflammatory T cell responses. Cell Metab. 2015, 22, 485–498. [Google Scholar] [CrossRef] [PubMed]
- Desdín-Micó, G.; Soto-Heredero, G.; Aranda, J.F.; Oller, J.; Carrasco, E.; Gabandé-Rodríguez, E.; Blanco, E.M.; Alfranca, A.; Cussó, L.; Desco, M.; et al. T cells with dysfunctional mitochondria induce multimorbidity and premature senescence. Science 2020, 368, 1371–1376. [Google Scholar] [CrossRef] [PubMed]
- Fazeli, P.; Kalani, M.; Hosseini, M. T memory stem cell characteristics in autoimmune diseases and their promising therapeutic values. Front. Immunol. 2023, 14, 1204231. [Google Scholar] [CrossRef]
- Fang, Y.; Doyle, M.F.; Chen, J.; Mez, J.; Satizabal, C.L.; Alosco, M.L.; Qiu, W.Q.; Lunetta, K.L.; Murabito, J.M. Circulating immune cell phenotypes are associated with age, sex, CMV, and smoking status in the Framingham Heart Study offspring participants. Aging 2023, 15, 3939–3966, Correction in Aging 2023, 15, 7855–7856.. [Google Scholar] [CrossRef]
- Ramasubramanian, R.; Meier, H.C.; Vivek, S.; Klopack, E.; Crimmins, E.M.; Faul, J.; Nikolich-Žugich, J.; Thyagarajan, B. Evaluation of T-cell aging-related immune phenotypes in the context of biological aging and multimorbidity in the Health and Retirement Study. Immun. Ageing 2022, 19, 33. [Google Scholar] [CrossRef] [PubMed]
- Tomusiak, A.; Floro, A.; Tiwari, R.; Riley, R.; Matsui, H.; Andrews, N.; Kasler, H.G.; Verdin, E. Development of an epigenetic clock resistant to changes in immune cell composition. Commun. Biol. 2024, 7, 934. [Google Scholar] [CrossRef]
- Xu, W.; Larbi, A. Markers of T Cell Senescence in Humans. Int. J. Mol. Sci. 2017, 18, 1742. [Google Scholar] [CrossRef]
- Zhao, X.; Zhang, Y.; Gao, Z.; Han, Y. Prognostic value of peripheral naive CD8+ T cells in oligometastatic non-small-cell lung cancer. Future Oncol. 2022, 18, 55–65. [Google Scholar] [CrossRef]
- Lee, T.H.; Chen, J.J.; Wu, C.Y.; Lin, T.Y.; Hung, S.C.; Yang, H.Y. Immunosenescence, gut dysbiosis, and chronic kidney disease: Interplay and implications for clinical management. Biomed. J. 2024, 47, 100638. [Google Scholar] [CrossRef] [PubMed]
- Valieva, Y.; Ivanova, E.; Fayzullin, A.; Kurkov, A.; Igrunkova, A. Senescence-associated β-galactosidase detection in pathology. Diagnostics 2022, 12, 2309. [Google Scholar] [CrossRef] [PubMed]
- Verzola, D.; Gandolfo, M.T.; Gaetani, G.; Ferraris, A.; Mangerini, R.; Ferrario, F.; Villaggio, B.; Gianiorio, F.; Tosetti, F.; Weiss, U.; et al. Accelerated senescence in the kidneys of patients with type 2 diabetic nephropathy. Am. J. Physiol. Renal Physiol. 2008, 295, F1563–F1573. [Google Scholar] [CrossRef]
- Esposito, P.; Picciotto, D.; Verzola, D.; Garibotto, G.; Parodi, E.L.; Sofia, A.; Costigliolo, F.; Gaggero, G.; Zanetti, V.; Saio, M.; et al. SA-β-Gal in kidney tubules as a predictor of renal outcome in patients with chronic kidney disease. J. Clin. Med. 2024, 13, 322. [Google Scholar] [CrossRef]
- Katsuumi, G.; Shimizu, I.; Suda, M.; Yoshida, Y.; Furihata, T.; Joki, Y.; Hsiao, C.L.; Jiaqi, L.; Fujiki, S.; Abe, M.; et al. SGLT2 inhibition eliminates senescent cells and alleviates pathological aging. Nat. Aging 2024, 4, 926–938. [Google Scholar] [CrossRef]
- Watanabe, T.; Yazaki, M.; Yazaki, T.; Furukawa, M.; Izumo, N. Senotherapeutic effect of Agrimonia pilosa Ledeb. in targeting senescent cells in naturally aged mice. Food Biosci. 2024, 59, 103903. [Google Scholar] [CrossRef]
- Kared, H.; Martelli, S.; Ng, T.P.; Pender, S.L.; Larbi, A. CD57 in human natural killer cells and T-lymphocytes. Cancer Immunol. Immunother. 2016, 65, 441–452. [Google Scholar] [CrossRef]
- Chou, J.P.; Effros, R.B. T cell replicative senescence in human aging. Curr. Pharm. Des. 2013, 19, 1680–1698. [Google Scholar]
- Panossian, L.A.; Porter, V.R.; Valenzuela, H.F.; Zhu, X.; Reback, E.; Masterman, D.; Cummings, J.L.; Effros, R.B. Telomere shortening in T cells correlates with Alzheimer’s disease status. Neurobiol. Aging 2003, 24, 77–84. [Google Scholar] [CrossRef]
- Winford, E.; Lutshumba, J.; Martin, B.J.; Wilcock, D.M.; Jicha, G.A.; Nikolajczyk, B.S.; Stowe, A.M.; Bachstetter, A.D. Terminally differentiated effector memory T cells associate with cognitive and AD-related biomarkers in an aging-based community cohort. Immun. Ageing 2024, 21, 36. [Google Scholar] [CrossRef] [PubMed]
- Kouli, A.; Jensen, M.; Papastavrou, V.; Scott, K.M.; Kolenda, C.; Parker, C.; Solim, I.H.; Camacho, M.; Martin-Ruiz, C.; Williams-Gray, C.H. T lymphocyte senescence is attenuated in Parkinson’s disease. J. Neuroinflammation 2021, 18, 228. [Google Scholar] [CrossRef]
- Capelle, C.M.; Ciré, S.; Hedin, F.; Hansen, M.; Pavelka, L.; Grzyb, K.; Kyriakis, D.; Hunewald, O.; Konstantinou, M.; Revets, D.; et al. Early-to-mid stage idiopathic Parkinson’s disease shows enhanced cytotoxicity and differentiation in CD8 T-cells in females. Nat. Commun. 2023, 14, 7461. [Google Scholar] [CrossRef]
- Kronzer, V.L.; Bridges, S.L., Jr.; Davis, J.M., 3rd. Why women have more autoimmune diseases than men: An evolutionary perspective. Evol. Appl. 2020, 14, 629–633. [Google Scholar] [CrossRef]
- Rubtsov, A.V.; Rubtsova, K.; Kappler, J.W.; Marrack, P. Genetic and hormonal factors in female-biased autoimmunity. Autoimmun. Rev. 2010, 9, 494–498. [Google Scholar] [CrossRef]
- Márquez, E.J.; Chung, C.H.; Marches, R.; Rossi, R.J.; Nehar-Belaid, D.; Eroglu, A.; Mellert, D.J.; Kuchel, G.A.; Banchereau, J.; Ucar, D. Sexual-dimorphism in human immune system aging. Nat. Commun. 2020, 11, 751. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, J.R.; Pinto, T.N.; Arruda, L.B.; da Silva, C.C.; de Carvalho, C.R.; Pinto, R.M.C.; da Silva Duarte, A.J.; Benard, G. Age-associated phenotypic imbalance in TCD4 and TCD8 cell subsets: Comparison between healthy aged, smokers, COPD patients and young adults. Immun. Ageing 2022, 19, 9. [Google Scholar] [CrossRef] [PubMed]
- Park, T.-I.; Yang, A.H.; Kanth, B.K.; Pack, S.P. Aptamers as Diagnostic and Therapeutic Agents for Aging and Age-Related Diseases. Biosensors 2025, 15, 232. [Google Scholar] [CrossRef]
- Furrer, R.; Handschin, C. Biomarkers of aging: From molecules and surrogates to physiology and function. Physiol. Rev. 2025, 105, 1609–1694. [Google Scholar] [CrossRef] [PubMed]
- Cummings, S.R.; Kritchevsky, S.B. Endpoints for geroscience clinical trials: Health outcomes, biomarkers, and biologic age. Geroscience 2022, 44, 2925–2931. [Google Scholar] [CrossRef]
- Ogrodnik, M.; Acosta, J.C.; Adams, P.D.; di Fagagna, F.D.; Baker, D.J.; Bishop, C.L.; Chandra, T.; Collado, M.; Gil, J.; Gorgoulis, V.; et al. Guidelines for minimal information on cellular senescence experimentation in vivo. Cell 2024, 187, 4150–4175. [Google Scholar] [CrossRef]
| All | (N = 632) | Male | (N = 303) | Female | (N = 329) | t-Test | Wilcoxon | ||
|---|---|---|---|---|---|---|---|---|---|
| Variables | (Unit) | Mean | (SD) | Mean | (SD) | Mean | (SD) | p | p |
| Age | (years) | 50.2 | (5.22) | 50.2 | (5.22) | 50.2 | (5.23) | 0.987 | 0.912 |
| BMI | (kg/m2) | 21.9 | (3.22) | 22.9 | (3.19) | 20.9 | (2.92) | <0.001 | <0.001 |
| HbA1c | (%) | 5.4 | (0.31) | 5.4 | (0.34) | 5.4 | (0.28) | 0.350 | 0.965 |
| Blood glucose | (mg/dL) | 87.0 | (8.31) | 87.7 | (8.79) | 86.5 | (7.81) | 0.071 | 0.016 |
| Triglyceride | (mg/dL) | 101.4 | (65.99) | 114.4 | (75.50) | 89.5 | (53.21) | <0.001 | <0.001 |
| Total cholesterol | (mg/dL) | 218.9 | (35.69) | 213.1 | (34.17) | 224.2 | (36.27) | <0.001 | <0.001 |
| HDL cholesterol | (mg/dL) | 71.6 | (19.10) | 64.2 | (17.30) | 78.4 | (18.16) | <0.001 | <0.001 |
| LDL cholesterol | (mg/dL) | 123.9 | (30.85) | 123.4 | (29.90) | 124.4 | (31.74) | 0.694 | 0.868 |
| ALT | (U/L) | 19.9 | (13.13) | 22.8 | (13.88) | 17.3 | (11.81) | 0.197 | 0.010 |
| AST | (U/L) | 21.7 | (9.37) | 22.2 | (8.17) | 21.3 | (10.34) | <0.001 | <0.001 |
| γ-GTP | (U/L) | 32.0 | (49.89) | 38.8 | (40.52) | 25.8 | (56.52) | 0.001 | <0.001 |
| Urea nitrogen | (mg/dL) | 13.6 | (3.70) | 14.1 | (3.62) | 13.1 | (3.71) | 0.001 | 0.001 |
| Creatinine | (mg/dL) | 0.8 | (0.15) | 0.9 | (0.13) | 0.7 | (0.09) | <0.001 | <0.001 |
| SBP | (mmHg) | 116.6 | (13.88) | 119.2 | (13.45) | 114.1 | (13.84) | <0.001 | <0.001 |
| DBP | (mmHg) | 72.7 | (11.20) | 76.0 | (10.64) | 69.6 | (10.81) | <0.001 | <0.001 |
| Pulse rate | (bpm) | 70.6 | (10.29) | 70.8 | (10.68) | 70.3 | (9.93) | 0.548 | 0.704 |
| CD4 cells | (count) | 16,609.5 | (6208.88) | 16,433.2 | (6301.24) | 16,771.8 | (6127.70) | 0.494 | 0.159 |
| CD8 cells | (count) | 6092.7 | (3185.58) | 6125.7 | (3176.89) | 6062.2 | (3198.10) | 0.802 | 0.334 |
| CD4/CD8 | (ratio) | 3.2 | (1.51) | 3.2 | (1.60) | 3.2 | (1.41) | 0.767 | 0.154 |
| All | (N = 632) | Male | (N = 303) | Female | (N = 329) | t-Test | Wilcoxon | |
|---|---|---|---|---|---|---|---|---|
| Variables | Mean | (SD) | Mean | (SD) | Mean | (SD) | p | p |
| Total CD8+ | 19.1 | (6.59) | 18.9 | (6.58) | 19.3 | (6.60) | 0.487 | 0.488 |
| Naïve in all subsets | 34.6 | (17.50) | 34.3 | (17.77) | 34.8 | (17.28) | 0.721 | 0.468 |
| TCM in all subsets | 21.5 | (9.37) | 21.5 | (9.74) | 21.5 | (9.03) | 0.925 | 0.689 |
| TEM in all subsets | 30.1 | (12.52) | 31.2 | (12.88) | 29.1 | (12.11) | 0.036 | 0.031 |
| TEMRA in all subsets | 13.9 | (11.69) | 13.1 | (10.40) | 14.6 | (12.74) | 0.102 | 0.178 |
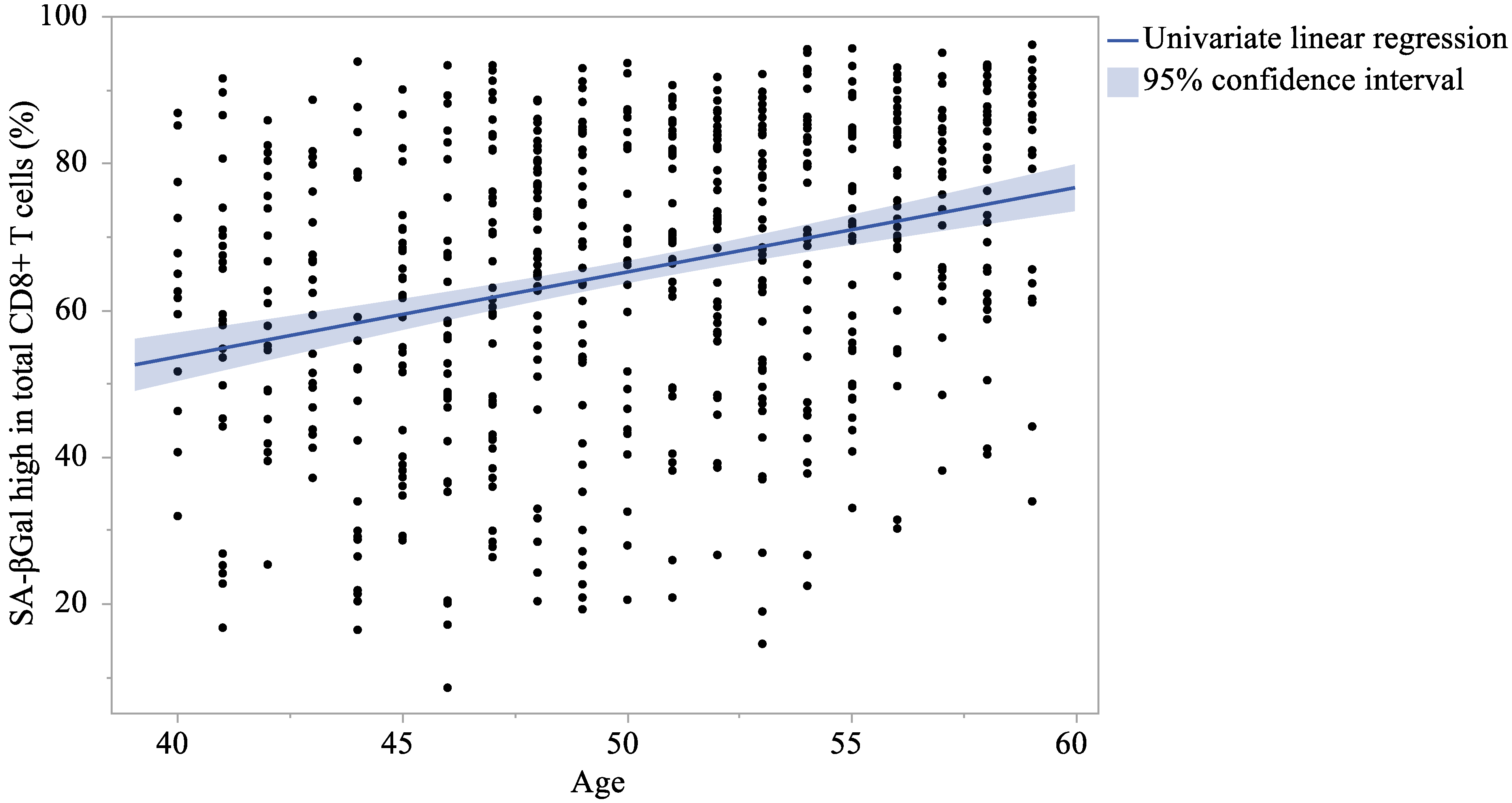
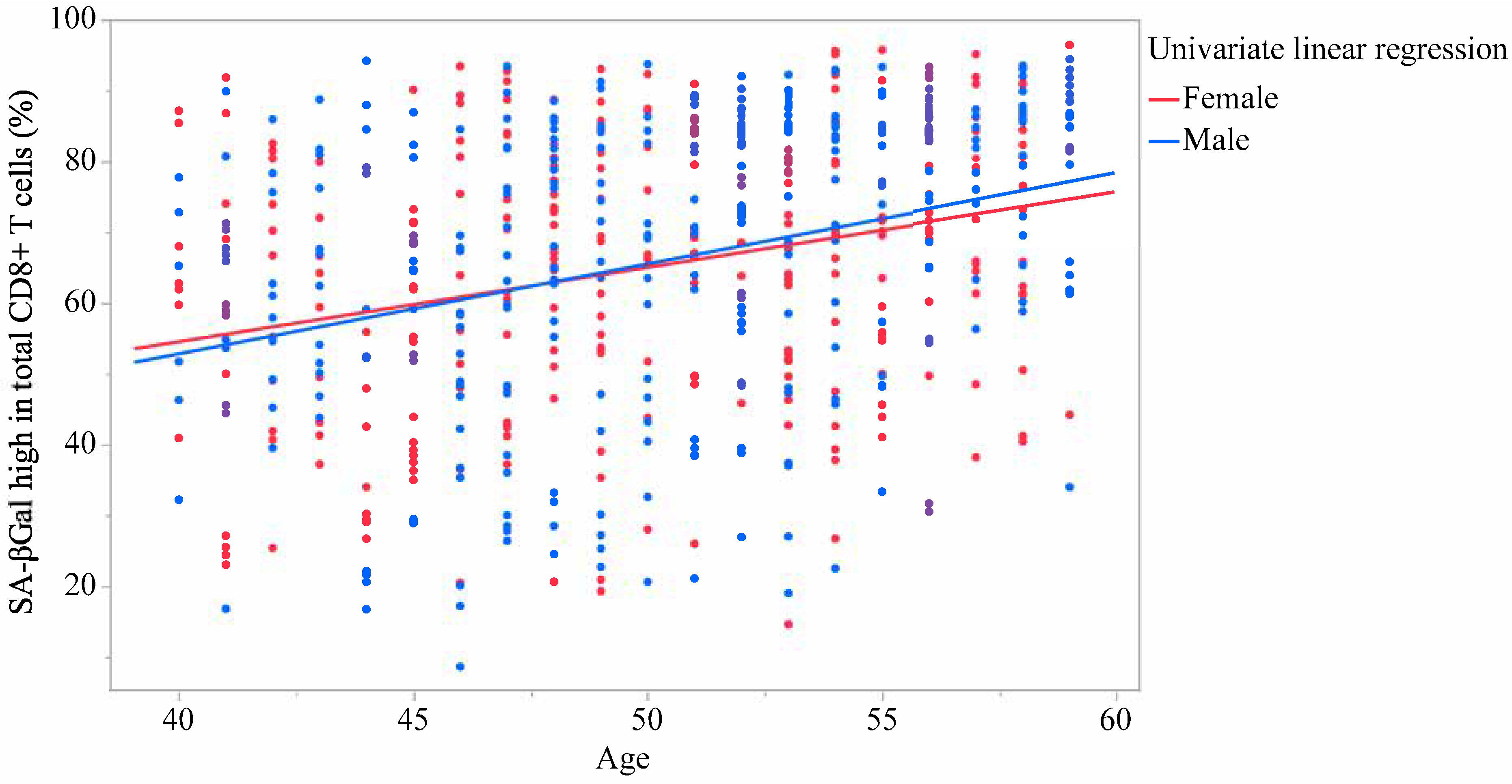
| SA-βGalhigh in total CD8+ | 65.5 | (20.07) | 65.7 | (20.91) | 65.2 | (19.30) | 0.737 | 0.456 |
| SA-βGalhigh in naïve | 10.1 | (9.27) | 10.1 | (9.55) | 10.1 | (9.02) | 0.994 | 0.443 |
| SA-βGalhigh in TCM | 53.1 | (22.62) | 53.0 | (22.93) | 53.2 | (22.36) | 0.908 | 0.882 |
| SA-βGalhigh in TEM | 93.2 | (7.72) | 93.0 | (8.21) | 93.5 | (7.24) | 0.384 | 0.679 |
| SA-βGalhigh in TEMRA | 96.3 | (5.33) | 96.3 | (5.47) | 96.3 | (5.21) | 0.918 | 0.617 |
| PD-1+ | 9.8 | (5.83) | 10.5 | (6.53) | 9.2 | (5.05) | 0.007 | 0.031 |
| All | Male | Female | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Characteristics | Pearson | p | Spearman | p | Pearson | p | Spearman | p | Pearson | p | Spearman | p |
| Total CD8+ | −0.119 | 0.003 | −0.117 | 0.003 | −0.157 | 0.006 | −0.148 | 0.010 | −0.084 | 0.126 | −0.084 | 0.127 |
| Naïve in all subsets | −0.308 | <0.001 | −0.320 | <0.001 | −0.326 | <0.001 | −0.348 | <0.001 | −0.291 | <0.001 | −0.297 | <0.001 |
| TCM in all subsets | 0.098 | 0.014 | 0.085 | 0.033 | 0.148 | 0.010 | 0.122 | 0.034 | 0.048 | 0.388 | 0.046 | 0.406 |
| TEM in all subsets | 0.263 | <0.001 | 0.259 | <0.001 | 0.296 | <0.001 | 0.287 | <0.001 | 0.233 | <0.001 | 0.233 | <0.001 |
| TEMRA in all subsets | 0.102 | 0.010 | 0.114 | 0.004 | 0.053 | 0.362 | 0.112 | 0.052 | 0.140 | 0.011 | 0.117 | 0.034 |
| SA-βGalhigh in total CD8+ | 0.300 | <0.001 | 0.314 | <0.001 | 0.317 | <0.001 | 0.348 | <0.001 | 0.284 | <0.001 | 0.282 | <0.001 |
| SA-βGalhigh in naïve | 0.254 | <0.001 | 0.275 | <0.001 | 0.300 | <0.001 | 0.328 | <0.001 | 0.210 | <0.001 | 0.222 | <0.001 |
| SA-βGalhigh in TCM | 0.269 | <0.001 | 0.272 | <0.001 | 0.346 | <0.001 | 0.350 | <0.001 | 0.197 | <0.001 | 0.196 | <0.001 |
| SA-βGalhigh in TEM | 0.169 | <0.001 | 0.202 | <0.001 | 0.175 | 0.002 | 0.228 | <0.001 | 0.162 | 0.003 | 0.170 | 0.002 |
| SA-βGalhigh in TEMRA | 0.148 | <0.001 | 0.199 | <0.001 | 0.131 | 0.023 | 0.184 | 0.001 | 0.164 | 0.003 | 0.211 | <0.001 |
| Univairate | Multivariate | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Characteristics | β | 95% CI | p | β | 95% CI | p | ||||
| Total CD8+ | −0.151 | −0.249 | to | −0.053 | 0.003 | −0.150 | −0.248 | to | −0.051 | 0.003 |
| Naïve in all subsets | −1.034 | −1.284 | to | −0.784 | <0.001 | −1.038 | −1.288 | to | −0.788 | <0.001 |
| TCM in all subsets | 0.175 | 0.036 | to | 0.315 | 0.014 | 0.176 | 0.036 | to | 0.316 | 0.014 |
| TEM in all subsets | 0.630 | 0.449 | to | 0.811 | <0.001 | 0.633 | 0.453 | to | 0.814 | <0.001 |
| TEMRA in all subsets | 0.228 | 0.054 | to | 0.403 | 0.010 | 0.229 | 0.054 | to | 0.403 | 0.010 |
| SA-βGalhigh in total CD8+ | 1.154 | 0.867 | to | 1.441 | <0.001 | 1.156 | 0.869 | to | 1.444 | <0.001 |
| SA-βGalhigh in naïve | 0.452 | 0.317 | to | 0.586 | <0.001 | 0.450 | 0.315 | to | 0.584 | <0.001 |
| SA-βGalhigh in TCM | 1.166 | 0.840 | to | 1.493 | <0.001 | 1.163 | 0.836 | to | 1.490 | <0.001 |
| SA-βGalhigh in TEM | 0.249 | 0.135 | to | 0.363 | <0.001 | 0.249 | 0.135 | to | 0.364 | <0.001 |
| SA-βGalhigh in TEMRA | 0.151 | 0.072 | to | 0.230 | <0.001 | 0.151 | 0.072 | to | 0.230 | <0.001 |
| All | Male | Female | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Characteristics | Pearson | p | Spearman | p | Pearson | p | Spearman | p | Pearson | p | Spearman | p |
| Naïve in all subsets | −0.915 | <0.001 | −0.928 | <0.001 | −0.925 | <0.001 | −0.937 | <0.001 | −0.906 | <0.001 | −0.919 | <0.001 |
| TCM in all subsets | −0.049 | 0.218 | −0.092 | 0.021 | −0.020 | 0.725 | −0.062 | 0.281 | −0.080 | 0.148 | −0.113 | 0.041 |
| TEM in all subsets | 0.789 | <0.001 | 0.797 | <0.001 | 0.822 | <0.001 | 0.828 | <0.001 | 0.758 | <0.001 | 0.773 | <0.001 |
| TEMRA in all subsets | 0.565 | <0.001 | 0.618 | <0.001 | 0.582 | <0.001 | 0.624 | <0.001 | 0.565 | <0.001 | 0.608 | <0.001 |
| Univairate | Multivariate | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Characteristics | β | 95% CI | p | β | 95% CI | p | ||||
| Naïve in all subsets | −1.050 | −1.081 | to | −1.005 | <0.001 | −1.044 | −1.082 | to | −1.006 | <0.001 |
| TCM in all subsets | −0.105 | −0.273 | to | −0.062 | 0.218 | −0.171 | −0.331 | to | −0.010 | 0.037 |
| TEM in all subsets | 1.265 | 1.188 | to | 1.342 | <0.001 | 1.231 | 1.152 | to | 1.310 | <0.001 |
| TEMRA in all subsets | 0.970 | 0.859 | to | 1.081 | <0.001 | 0.932 | 0.826 | to | 1.039 | <0.001 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tsubokawa, M.; Shimizu, Y.; Yazaki, M.; Shimodan, S.; Noguchi, M.; Yamazaki, A.; Watanabe, T.; Ocho, M.; Sakurada, T.; Hirose, Y.; et al. Correlation Between the Proportion of Senescence-Associated β-Galactosidase-Stained CD8+ T Cells and Age: A Cross-Sectional Study in Japan. Int. J. Mol. Sci. 2025, 26, 8799. https://doi.org/10.3390/ijms26188799
Tsubokawa M, Shimizu Y, Yazaki M, Shimodan S, Noguchi M, Yamazaki A, Watanabe T, Ocho M, Sakurada T, Hirose Y, et al. Correlation Between the Proportion of Senescence-Associated β-Galactosidase-Stained CD8+ T Cells and Age: A Cross-Sectional Study in Japan. International Journal of Molecular Sciences. 2025; 26(18):8799. https://doi.org/10.3390/ijms26188799
Chicago/Turabian StyleTsubokawa, Masaya, Yoshiki Shimizu, Misato Yazaki, Shieri Shimodan, Masayuki Noguchi, Arisa Yamazaki, Tomomichi Watanabe, Makoto Ocho, Tsuyoshi Sakurada, Yoshie Hirose, and et al. 2025. "Correlation Between the Proportion of Senescence-Associated β-Galactosidase-Stained CD8+ T Cells and Age: A Cross-Sectional Study in Japan" International Journal of Molecular Sciences 26, no. 18: 8799. https://doi.org/10.3390/ijms26188799
APA StyleTsubokawa, M., Shimizu, Y., Yazaki, M., Shimodan, S., Noguchi, M., Yamazaki, A., Watanabe, T., Ocho, M., Sakurada, T., Hirose, Y., Saito, J., & Ishii, Y. (2025). Correlation Between the Proportion of Senescence-Associated β-Galactosidase-Stained CD8+ T Cells and Age: A Cross-Sectional Study in Japan. International Journal of Molecular Sciences, 26(18), 8799. https://doi.org/10.3390/ijms26188799






