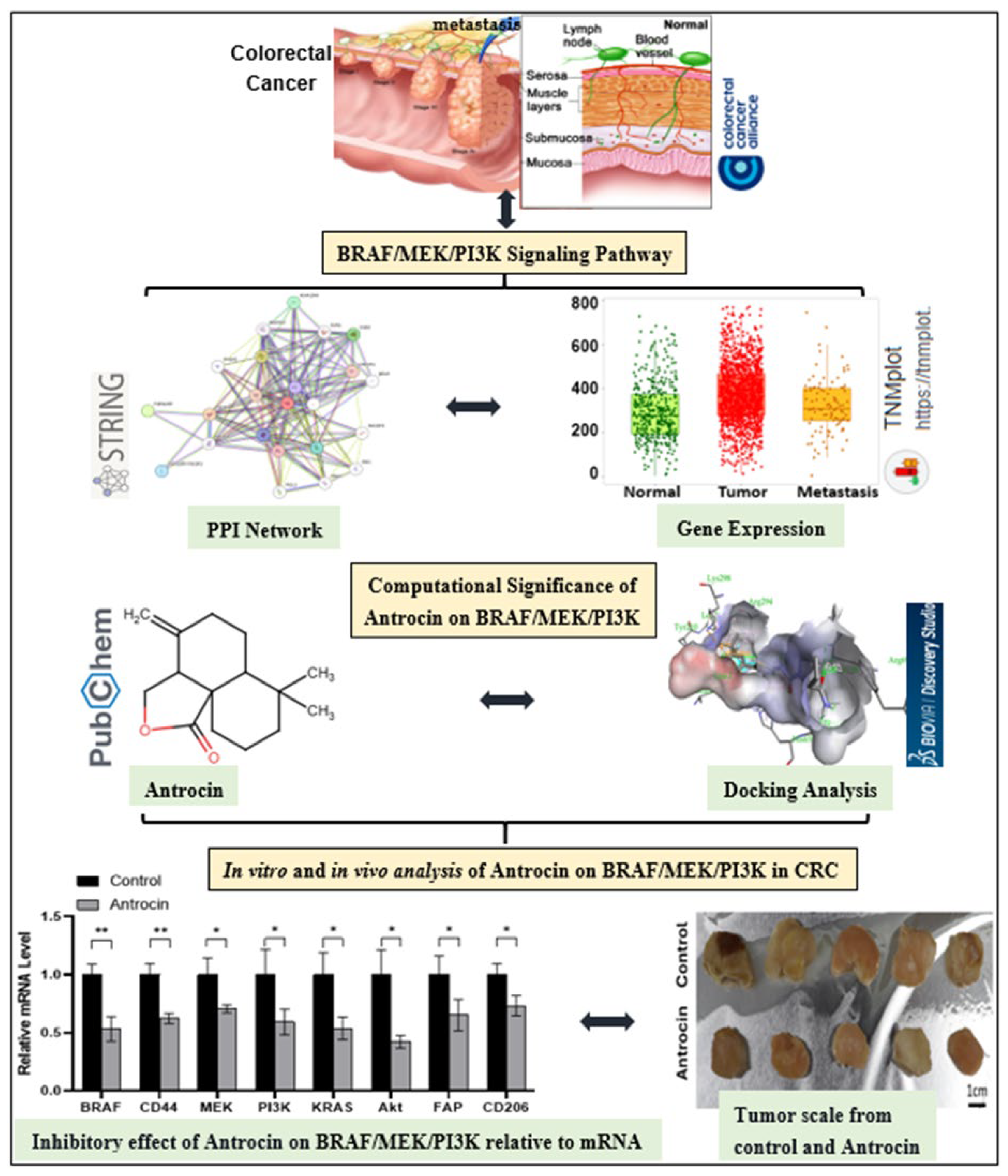
Integrated In Silico and Experimental Validation of Antrocin as a Plant-Derived Multi-Target Therapeutic for BRAF/MEK/PI3K-Driven Colorectal Cancer
Abstract
1. Introduction
2. Results
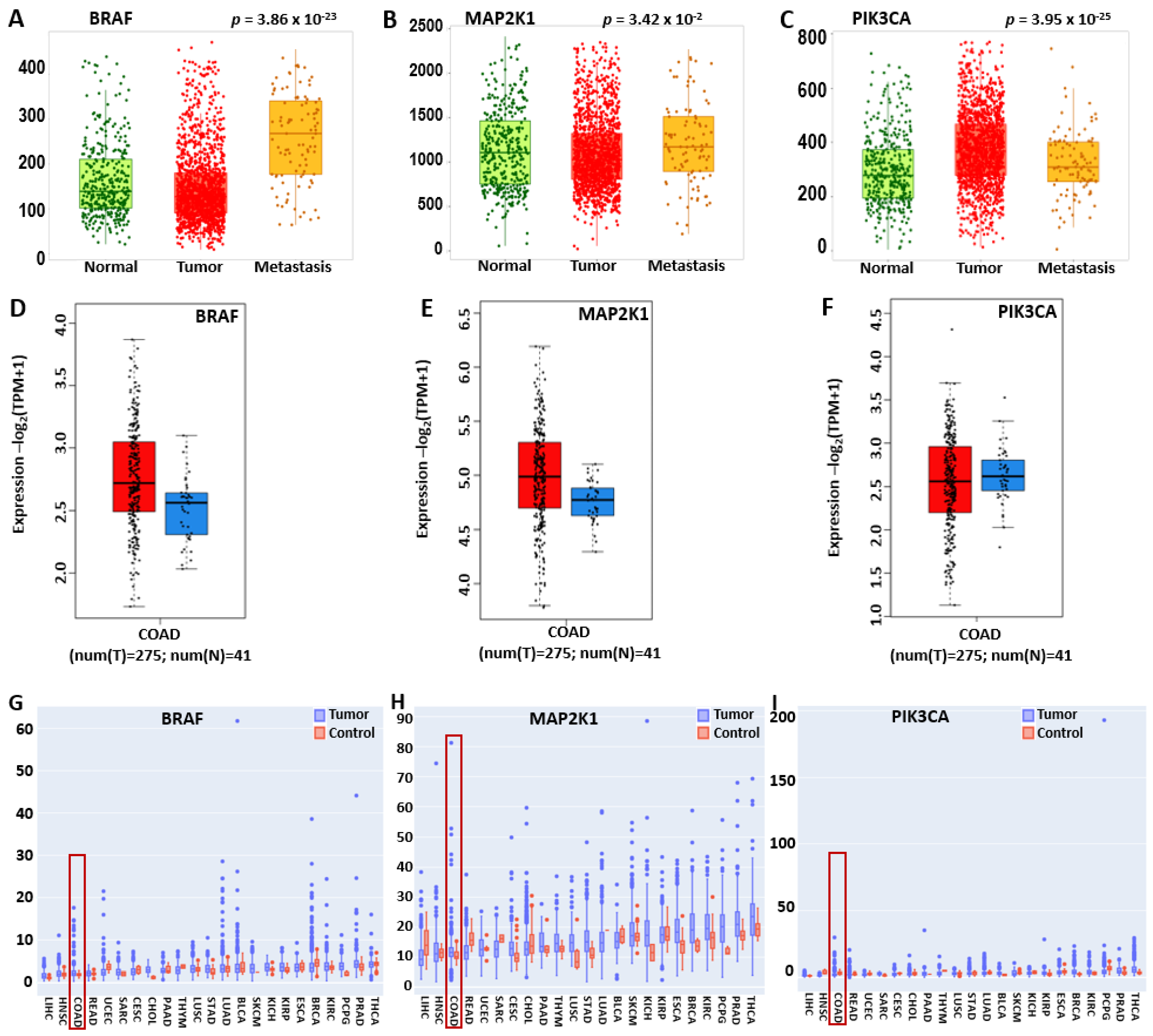
2.1. BRAF/MEK/PI3K Is Overexpressed in the Pan-Cancer and Promotes Tumor Proliferation and Metastasis in CRC
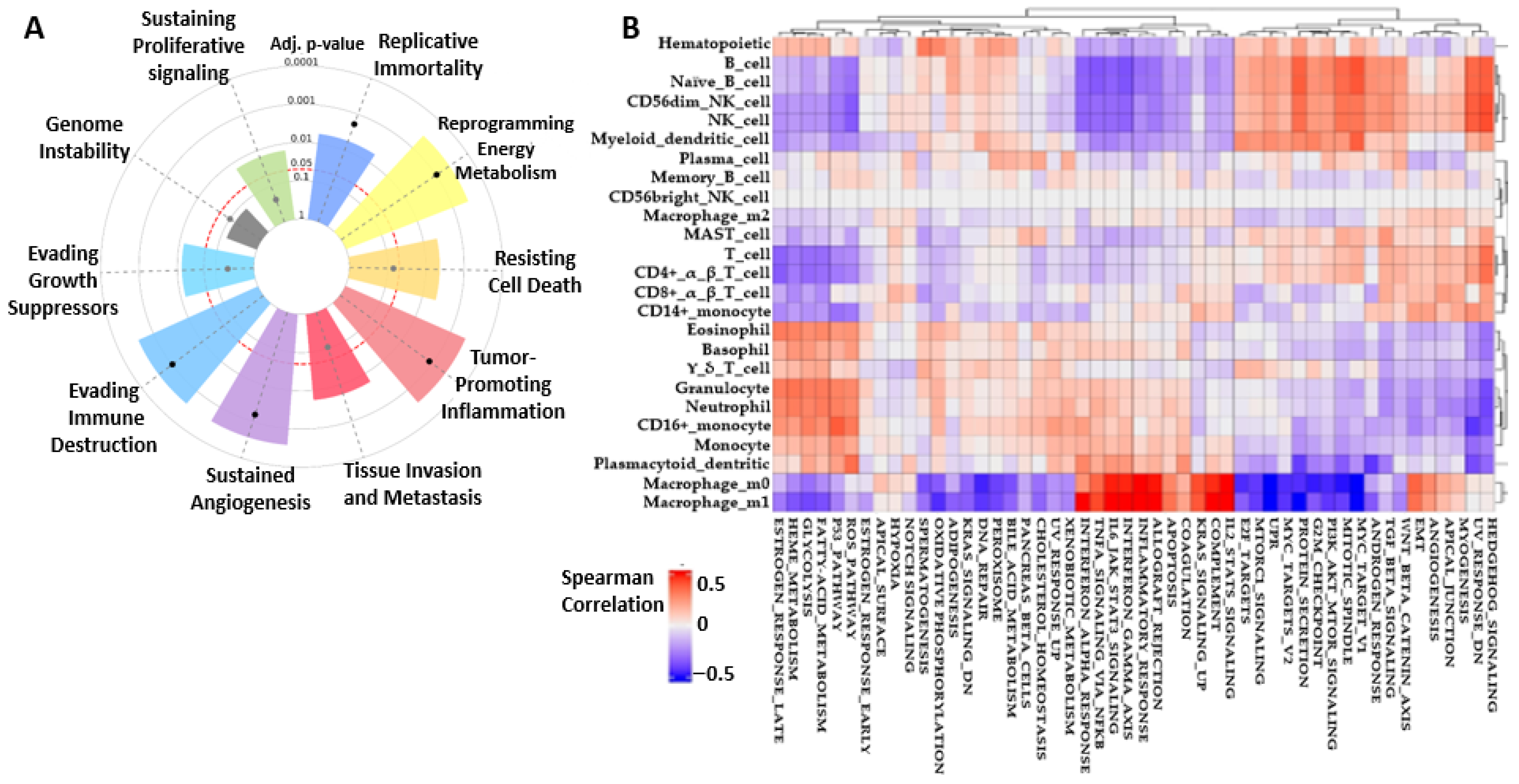
2.2. BRAF/MEK/PI3K Cancer Hallmark and CRC Correlation Cell Type Activities
2.3. BRAF/MAP2K1/PIK3CA Gene Ontology (GO) and KEGG Pathway
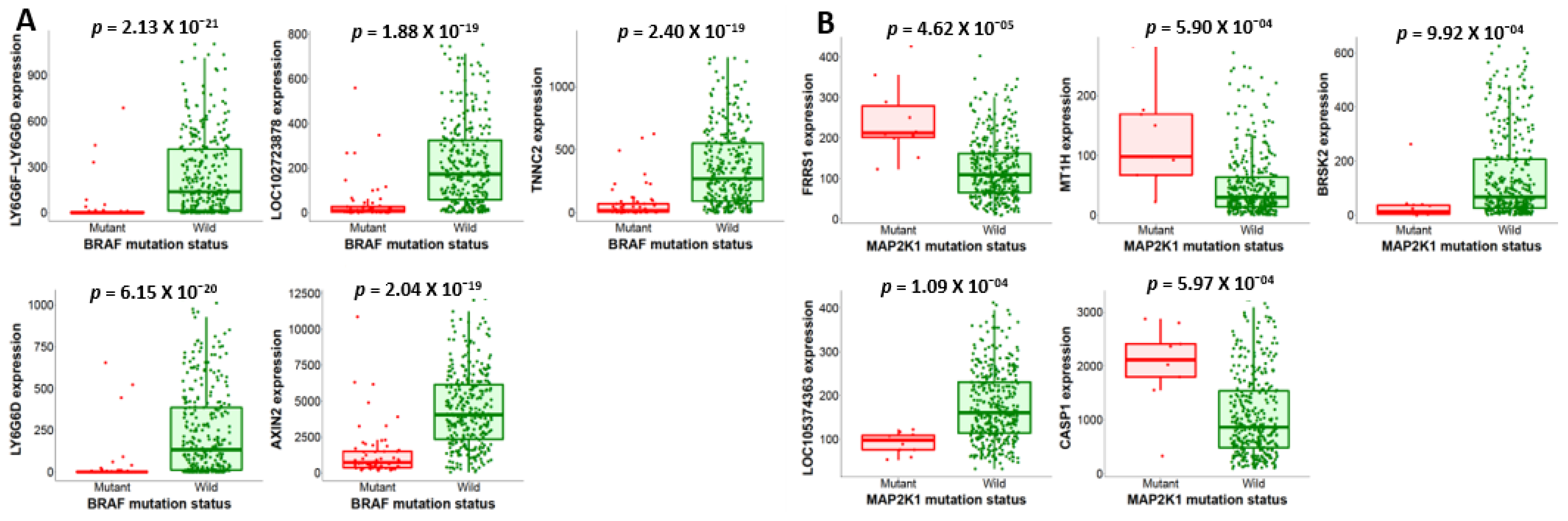
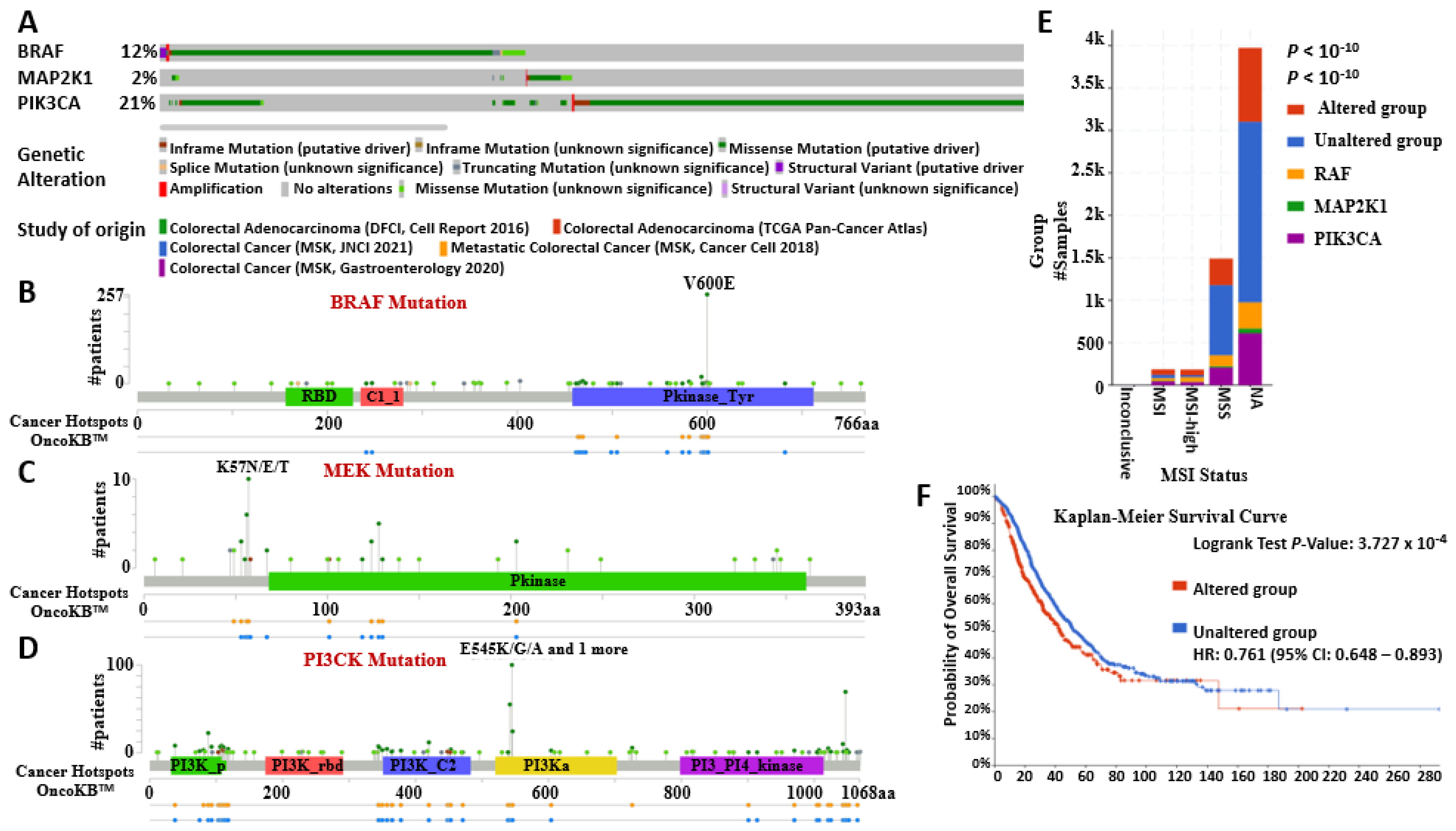
2.4. Mutations in BRAF/MEK/PI3K and Their Expression Changes Promote CRC Tumor Growth
2.5. BRAF/MEK/PI3K Oncogenic Signature Promotes Tumor Aggressiveness, Therapy Resistance, and Poor Overall Survival
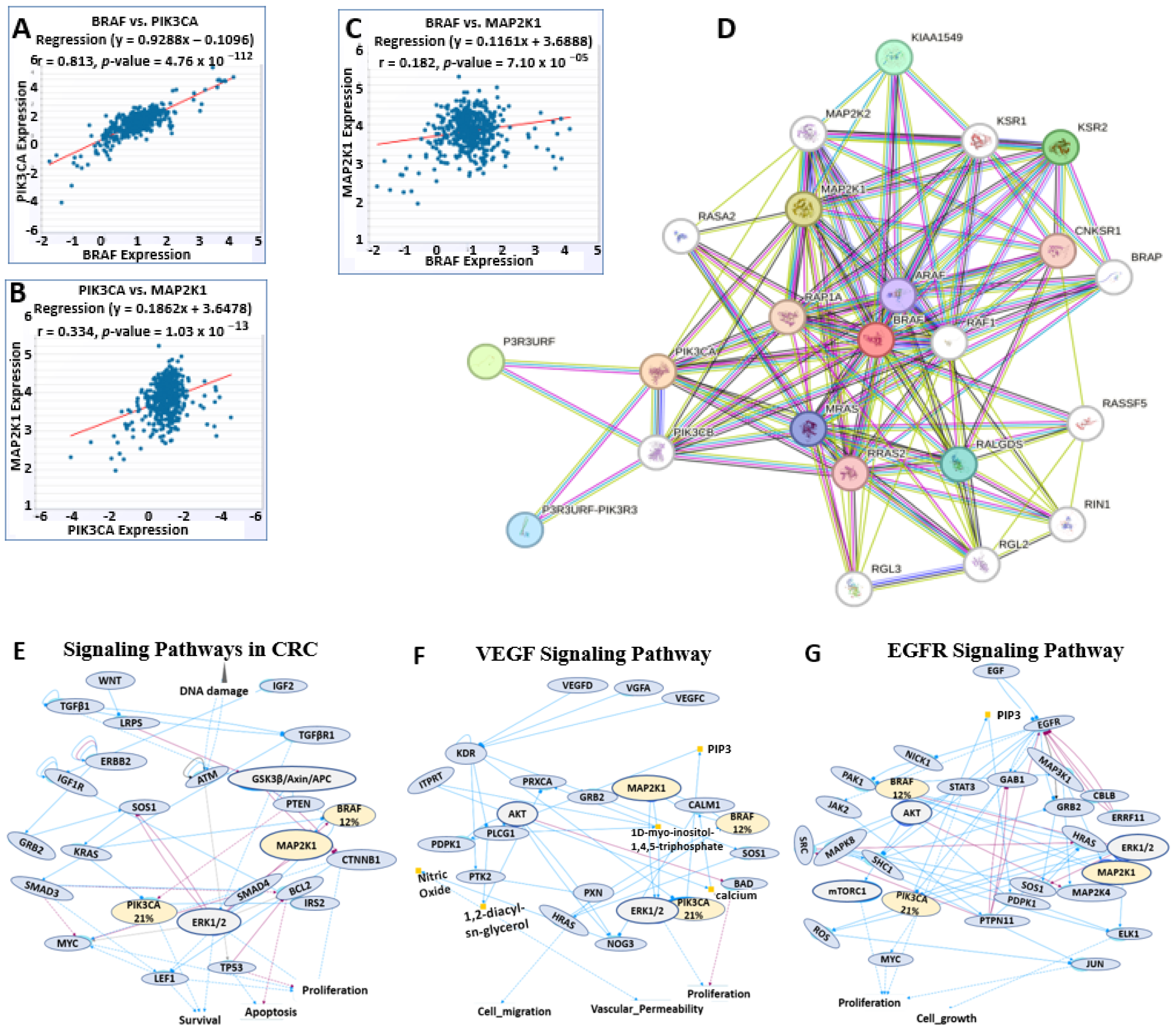
2.6. BRAF/MEK/PI3K Correlation and Functional Enrichment in Colorectal Cancer
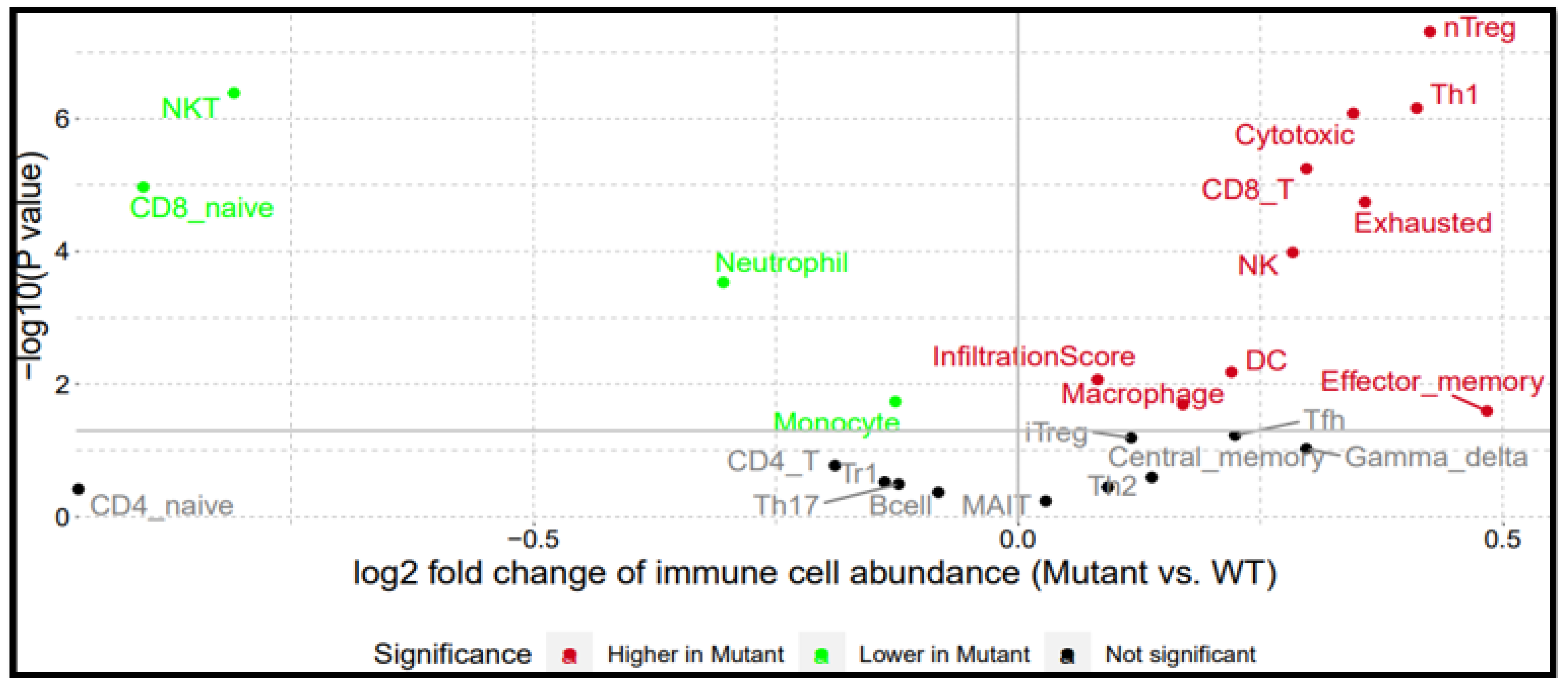
2.7. BRAF/MEK/PI3K Overexpression Promotes the Recruitment of Pro-Tumor Immune Cells in the CRC Tumor Microenvironment
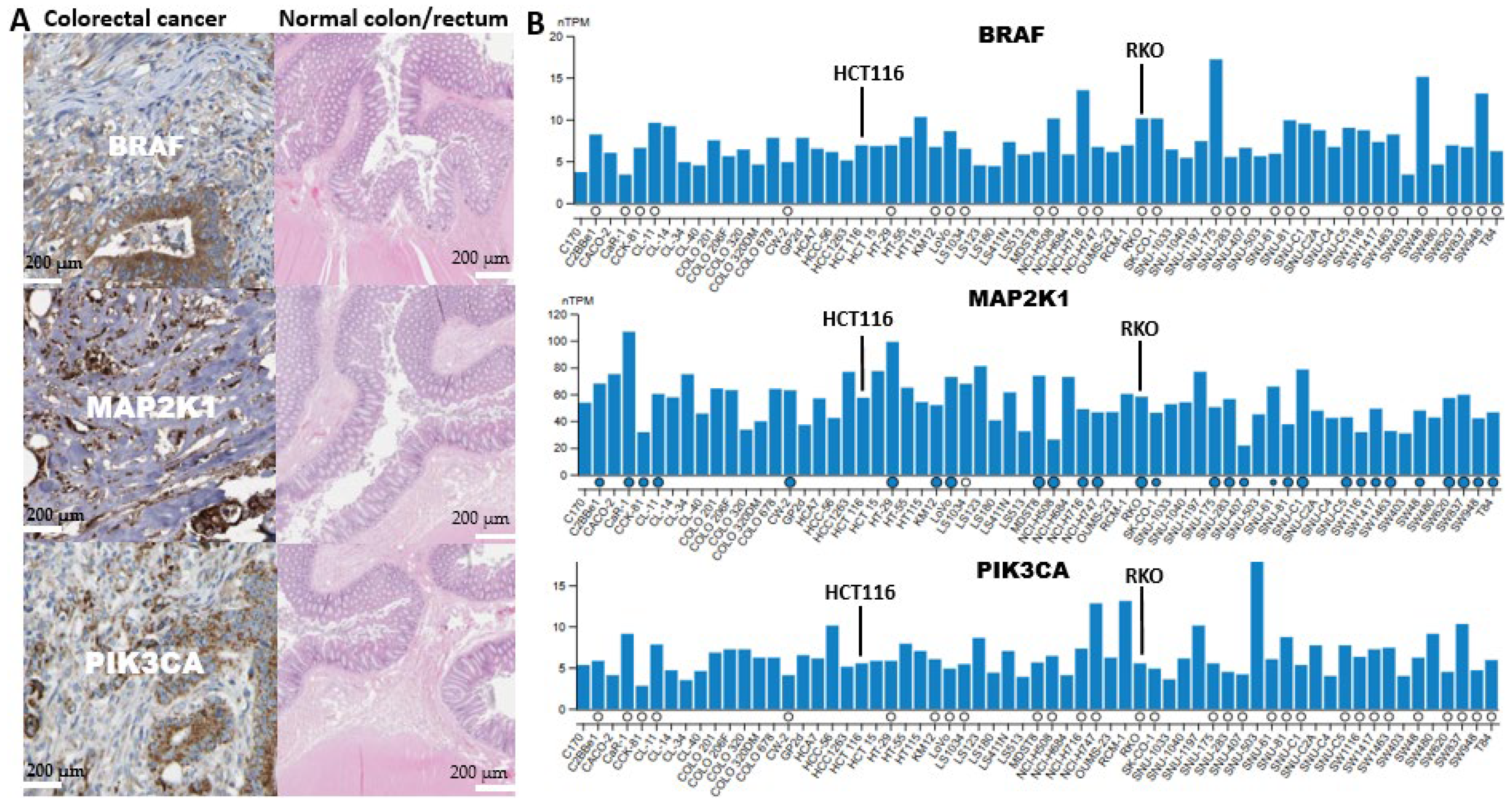
2.8. Analysis of the Presence of BRAF/MEK/PI3K in a Cancerous Colon and Its Expression in CRC Cell Lines
2.9. High BRAF/MEK/PI3K Expression Is Associated with Multi-Drug Resistance in CRC
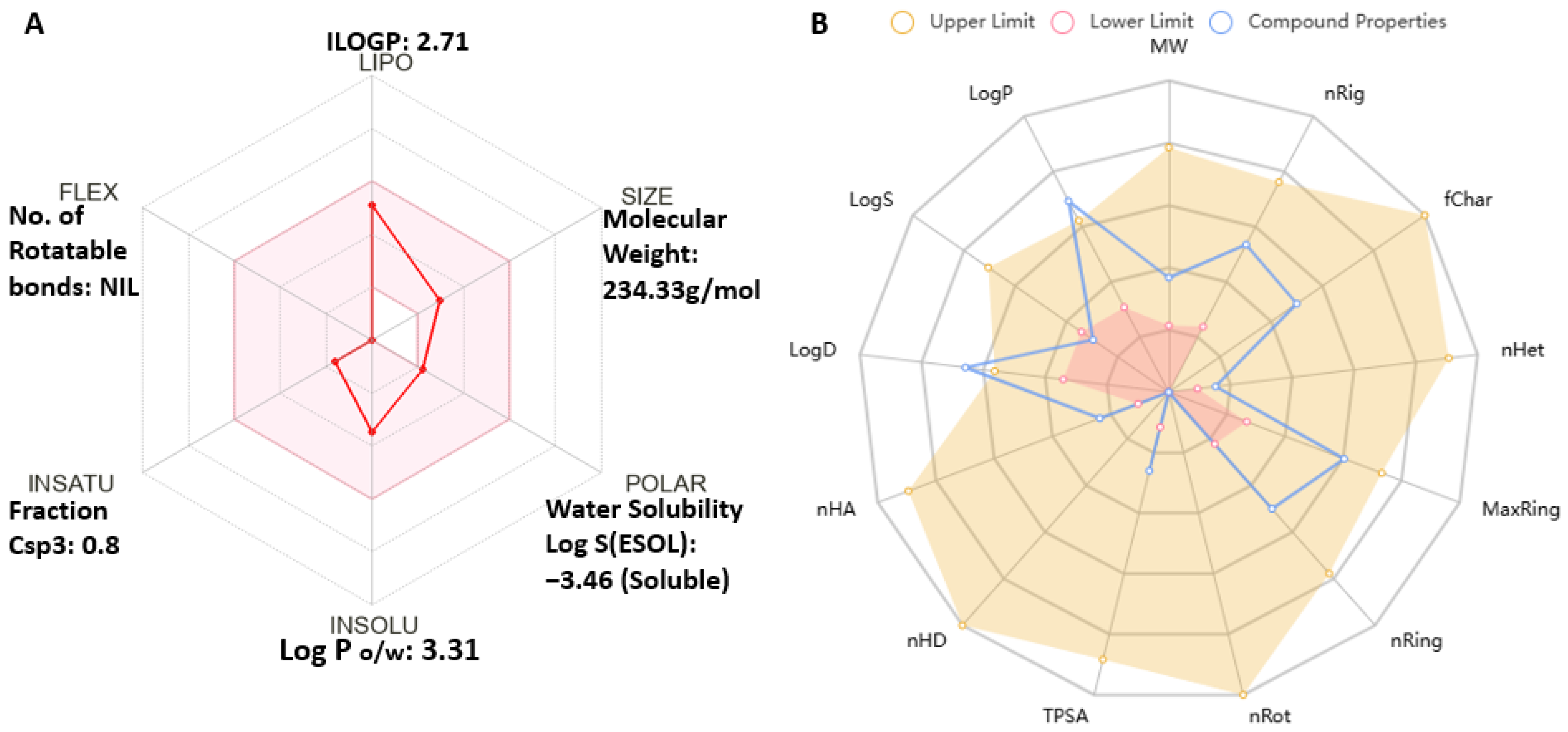
2.10. Antrocin Meets All Drug-Likeness, Absorption, Distribution, Metabolism, and Excretion (ADME) Properties of Small-Molecule Drugs
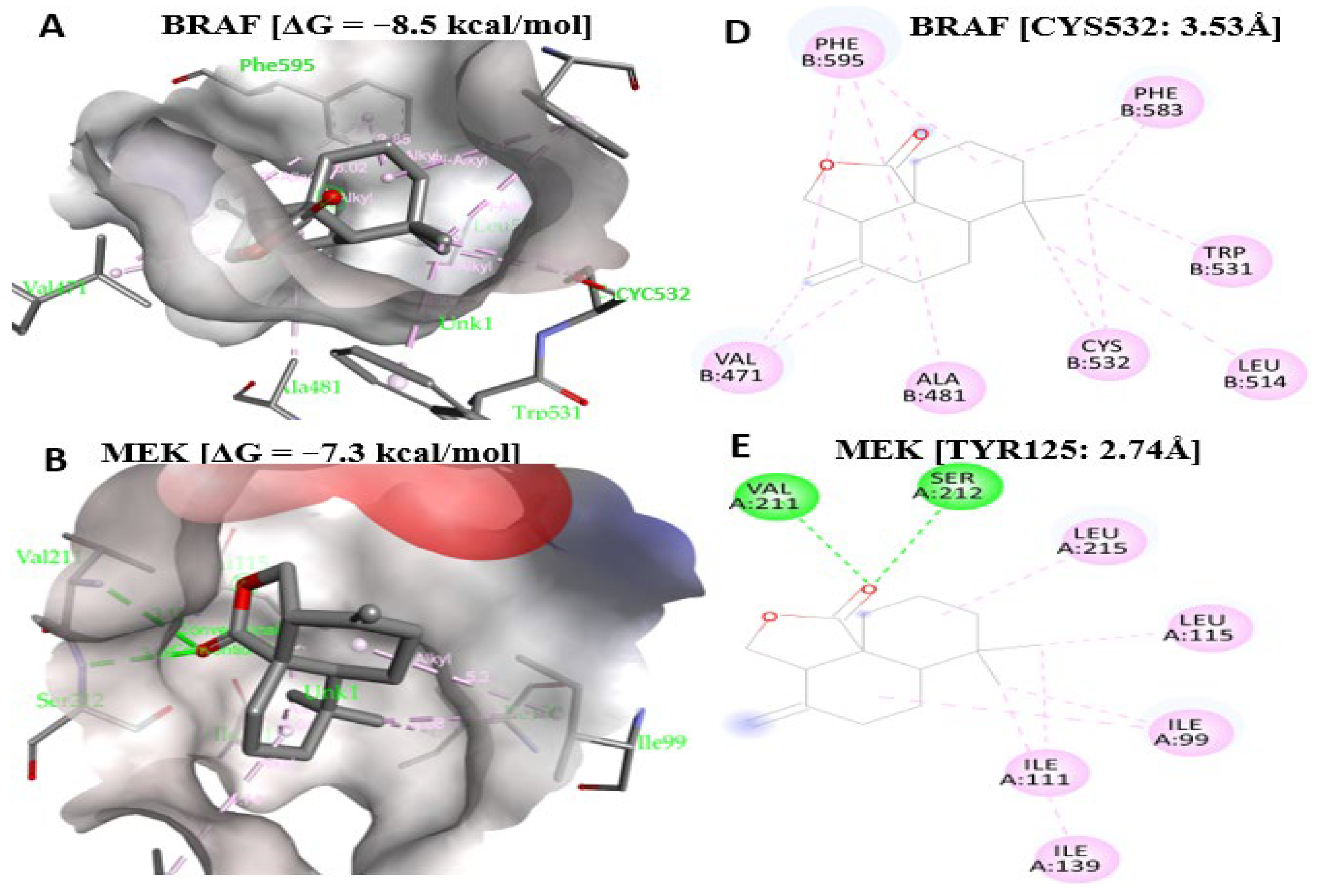
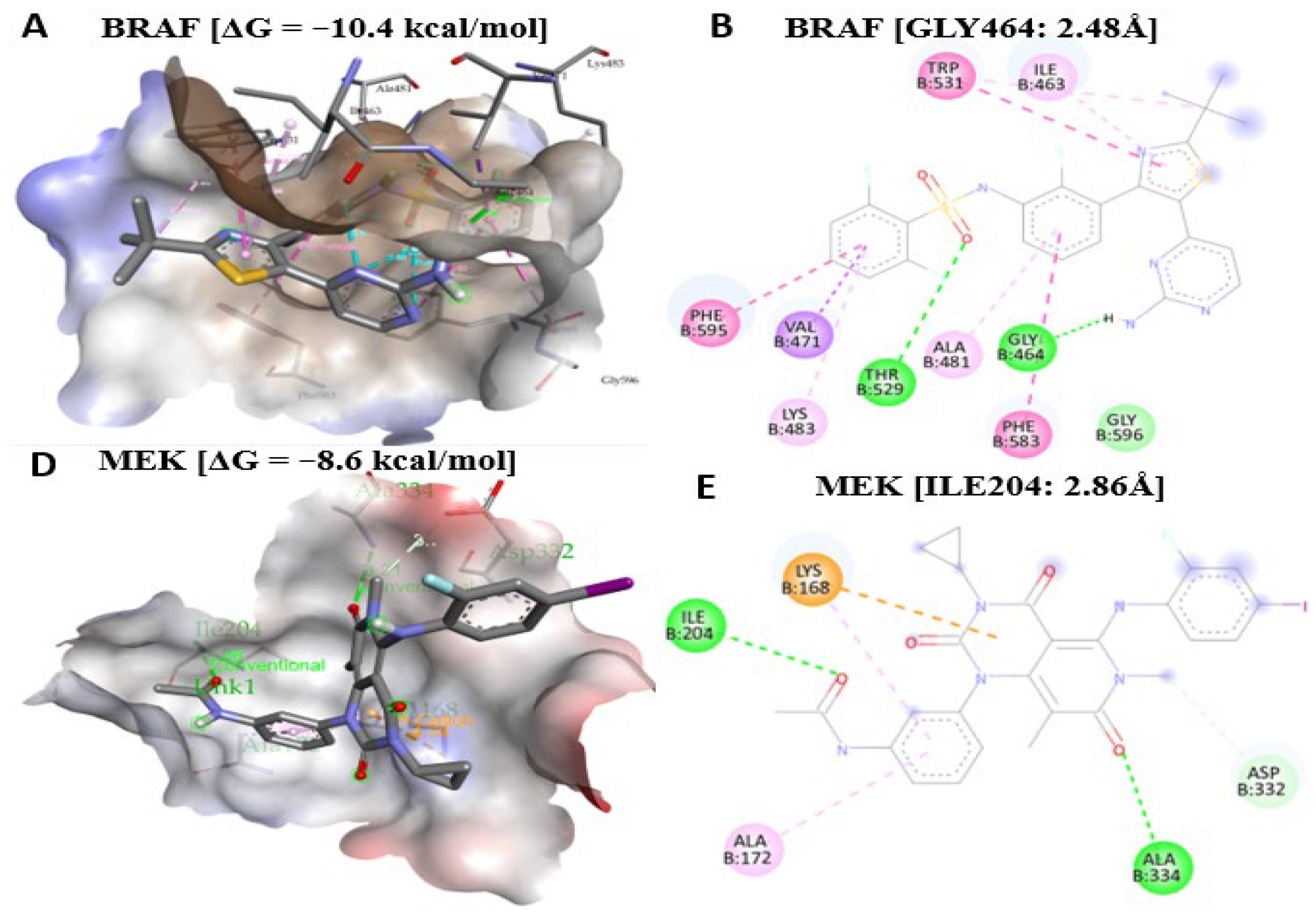
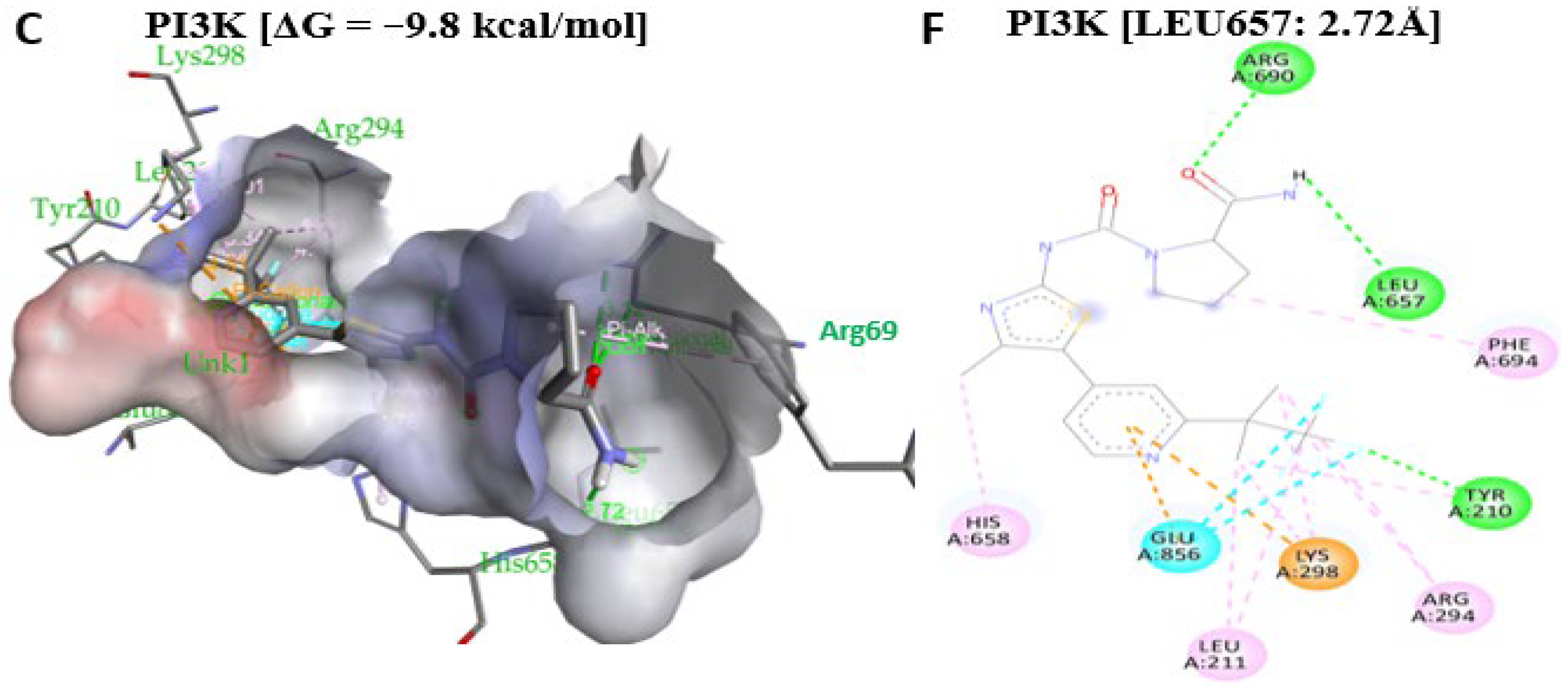
2.11. Antrocin Is a Potential Drug for Targeting the BRAF/MEK/PI3K Oncogenic Signature
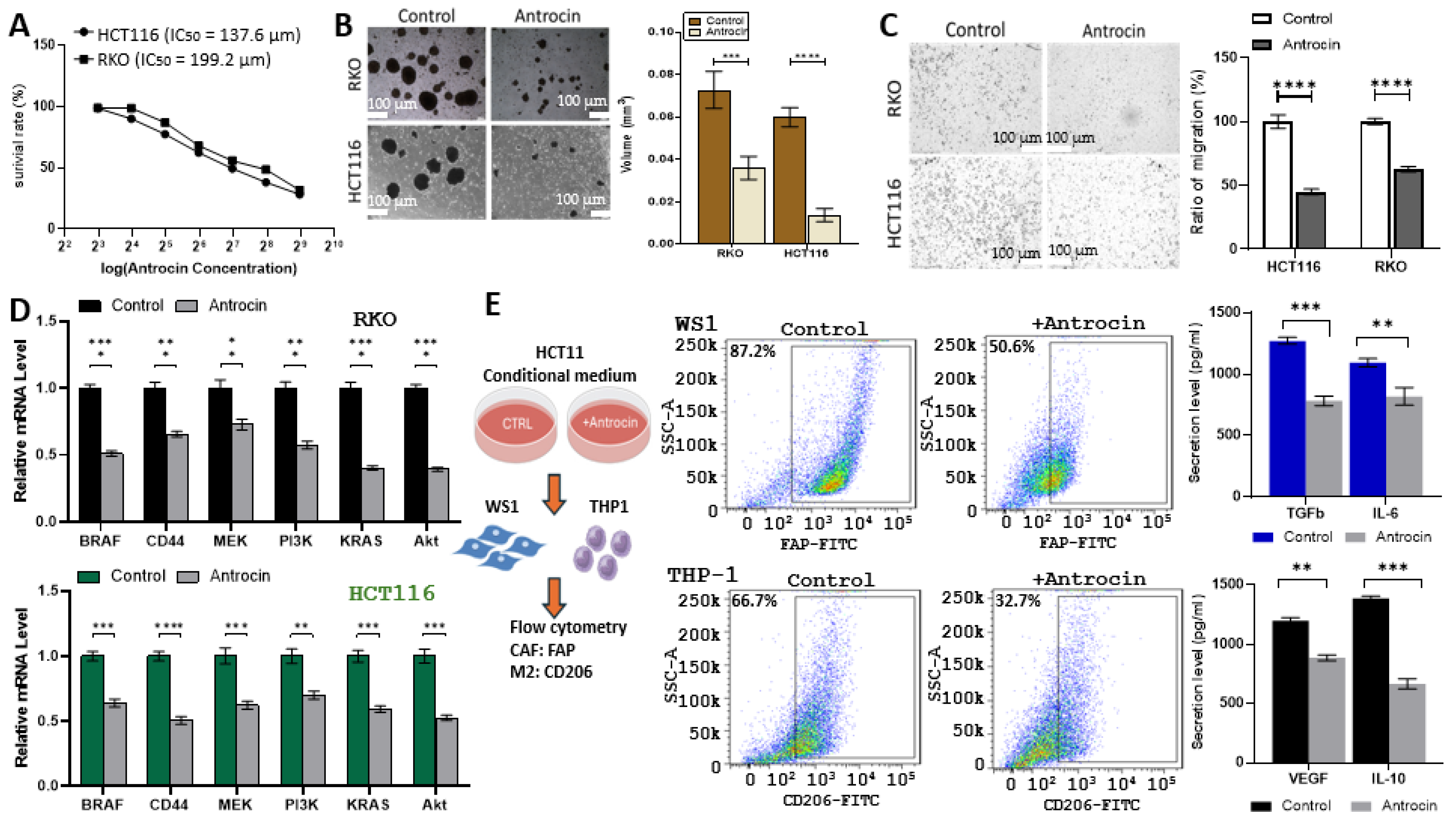
2.12. In Vitro Validation of Antrocin’s CRC Inhibitory Potential Through Inhibition of Multiple Oncogenic Targets
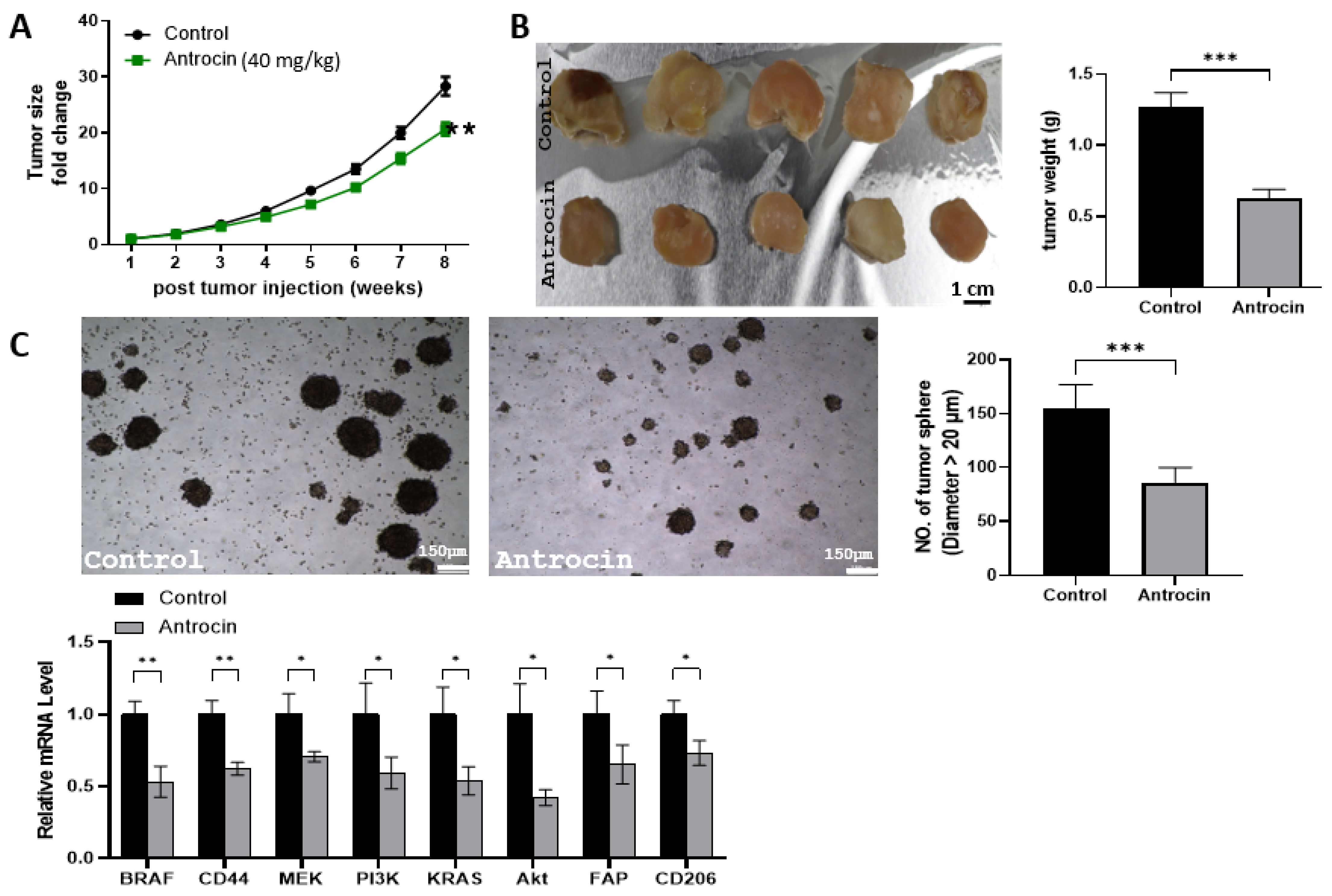
2.13. Antrocin Treatment Significantly Delays Tumor Growth of HCT116 Tumoroids
3. Discussion
4. Materials and Methods
4.1. BRAF/MEK/PI3K Differential Expression in Colorectal Cancer and Pan-Cancers
4.2. Evaluating BRAF/MEK/PI3K Mutations and Associated Gene Expression Changes in CRC
4.3. Network Construction and Pathways Enrichment Analysis in CRC
4.4. Correlation Between BRAF/MEK/PI3K Expression and Immune Cell Infiltration Levels
4.5. Correlation Between Drug Sensitivity and BRAF/MEK/PI3K Expression in Colorectal Cancer
4.6. Chemical Origin and Assessment of the Drug-Likeness, Absorption, Distribution, Metabolism, and Excretion (ADME) Properties of Antrocin
4.7. Molecular Docking Analysis
4.8. Cell Culture
4.9. Antrocin Preparation and Treatment
4.10. Tumor Spheroid Formation Assay
4.11. Migration Assay
4.12. Quantitative Real-Time PCR (qPCR) Analysis
4.13. Conditioned Medium Preparation and Flow Cytometry
4.14. ELISA Cytokine Profiling
4.15. In Vivo Evaluation of Antrocin’s Therapeutic Potential
4.16. Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CRC | Colorectal cancer |
| TME | Tumor microenvironment |
| BRAF | B-Raf proto-oncogene, serine/threonine kinase |
| MEK | Mitogen-activated protein kinase kinase (MAPK/ERK kinase) |
| PI3K | Phosphoinositide 3-kinase |
| MAP2K1 | Mitogen-activated protein kinase kinase 1 (MEK1) |
| PIK3CA | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha |
| GSCA | Gene Set Cancer Analysis (integrated cancer genomics platform) |
References
- Bray, F.; Laversanne, M.; Sung, H.; Ferlay, J.; Siegel, R.L.; Soerjomataram, I.; Jemal, A. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2024, 74, 229–263. [Google Scholar] [CrossRef]
- Dekker, E.; Tanis, P.J.; Vleugels, J.L.A.; Kasi, P.M.; Wallace, M.B. Colorectal cancer. Lancet 2019, 394, 1467–1480. [Google Scholar] [CrossRef] [PubMed]
- Stefani, C.; Miricescu, D.; Stanescu-Spinu, I.-I.; Nica, R.I.; Greabu, M.; Totan, A.R.; Jinga, M. Growth Factors, PI3K/AKT/mTOR and MAPK Signaling Pathways in Colorectal Cancer Pathogenesis: Where Are We Now? Int. J. Mol. Sci. 2021, 22, 10260. [Google Scholar] [CrossRef] [PubMed]
- Voutsadakis, I.A. KRAS mutated colorectal cancers with or without PIK3CA mutations: Clinical and molecular profiles inform current and future therapeutics. Crit. Rev. Oncol. Hematol. 2023, 186, 103987. [Google Scholar] [CrossRef] [PubMed]
- Nors, J.; Iversen, L.H.; Erichsen, R.; Gotschalck, K.A.; Andersen, C.L. Incidence of recurrence and time to recurrence in stage I to III colorectal cancer: A nationwide Danish cohort study. JAMA Oncol. 2024, 10, 54–62. [Google Scholar] [CrossRef]
- Siegel, R.L.; Giaquinto, A.N.; Jemal, A. Cancer statistics, 2024. CA Cancer J. Clin. 2024, 74, 12–49. [Google Scholar] [CrossRef]
- Jafari, A.; Hosseini, F.A.; Jalali, F.S. A systematic review of the economic burden of colorectal cancer. Health Sci. Rep. 2024, 7, e70002. [Google Scholar] [CrossRef]
- Barbosa, R.; Acevedo, L.A.; Marmorstein, R. The MEK/ERK Network as a Therapeutic Target in Human Cancer. Mol. Cancer Res. 2021, 19, 361–374. [Google Scholar] [CrossRef]
- Bye, B.; Jack, J.; Pierce, A.; Walsh, R.M.; Eades, A.; Olou, A.; Chalise, P.; VanSaun, M. Combined PI3K and MAPK inhibition synergizes to suppress PDAC. bioRxiv 2023. [Google Scholar] [CrossRef]
- Corrales, E.; Levit-Zerdoun, E.; Metzger, P.; Mertes, R.; Lehmann, A.; Münch, J.; Lemke, S.; Kowar, S.; Boerries, M. PI3K/AKT signaling allows for MAPK/ERK pathway independency mediating dedifferentiation-driven treatment resistance in melanoma. Cell Commun. Signal. 2022, 20, 187. [Google Scholar] [CrossRef]
- Ciombor, K.K.; Strickler, J.H.; Bekaii-Saab, T.S.; Yaeger, R. BRAF-Mutated Advanced Colorectal Cancer: A Rapidly Changing Therapeutic Landscape. J. Clin. Oncol. 2022, 40, 2706–2715. [Google Scholar] [CrossRef]
- Temraz, S.; Mukherji, D.; Shamseddine, A. Dual inhibition of MEK and PI3K pathway in KRAS and BRAF mutated colorectal cancers. Int. J. Mol. Sci. 2015, 16, 22976–22988. [Google Scholar] [CrossRef]
- Kopetz, S.; Murphy, D.A.; Pu, J.; Ciardiello, F.; Desai, J.; Van Cutsem, E.; Wasan, H.S.; Yoshino, T.; Saffari, H.; Zhang, X. Molecular profiling of BRAF-V600E-mutant metastatic colorectal cancer in the phase 3 BEACON CRC trial. Nat. Med. 2024, 30, 3261–3271. [Google Scholar] [CrossRef]
- Muradi Muhar, A.; Velaro, A.J.; Prananda, A.T.; Nugraha, S.E.; Halim, P.; Syahputra, R.A. Precision medicine in colorectal cancer: Genomics profiling and targeted treatment. Front. Pharmacol. 2025, 16, 1532971. [Google Scholar] [CrossRef]
- Grob, J.J.; Amonkar, M.M.; Karaszewska, B.; Schachter, J.; Dummer, R.; Mackiewicz, A.; Stroyakovskiy, D.; Drucis, K.; Grange, F.; Chiarion-Sileni, V.; et al. Comparison of dabrafenib and trametinib combination therapy with vemurafenib monotherapy on health-related quality of life in patients with unresectable or metastatic cutaneous BRAF Val600-mutation-positive melanoma (COMBI-v): Results of a phase 3, open-label, randomised trial. Lancet Oncol 2015, 16, 1389–1398. [Google Scholar]
- Xu, T.; Wang, X.; Changsong, Q.; Wang, Z.; Li, J.; Shen, L.; Wang, J.; Lin, X.; Zhang, L.; Lin, J. Genomic profiles of BRAF inhibitor resistance mechanisms in metastatic colorectal cancer. J. Clin. Oncol. 2021, 39, e15527. [Google Scholar] [CrossRef]
- Wang, H.; Tang, R.; Jiang, L.; Jia, Y. The role of PIK3CA gene mutations in colorectal cancer and the selection of treatment strategies. Front. Pharmacol. 2024, 15, 1494802. [Google Scholar] [CrossRef] [PubMed]
- Dankner, M.; Rousselle, E.; Petrecca, S.; Fabi, F.; Nowakowski, A.; Lazaratos, A.-M.; Rajadurai, C.V.; Stein, A.J.; Bian, D.; Tai, P. Clinical Activity of Mitogen-Activated Protein Kinase Inhibitors in Patients With MAP2K1 (MEK1)-Mutated Metastatic Cancers. JCO Precis. Oncol. 2025, 9, e2400199. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Tzeng, D.T.; Huang, Y.P.; Lin, C.J.; Lo, U.G.; Wu, C.L.; Lin, H.; Hsieh, J.T.; Tang, C.H.; Lai, C.H. Antrocin Sensitizes Prostate Cancer Cells to Radiotherapy Through Inhibiting PI3K/AKT and MAPK Signaling Pathways. Cancers 2018, 11, 34. [Google Scholar] [CrossRef] [PubMed]
- Rao, Y.K.; Wu, A.T.; Geethangili, M.; Huang, M.T.; Chao, W.-J.; Wu, C.H.; Deng, W.P.; Yeh, C.T.; Tzeng, Y.M. Identification of Antrocin From Antrodia Camphorata as a Selective and Novel Class of Small Molecule Inhibitor of Akt/mTOR Signaling in Metastatic Breast Cancer MDA-MB-231 Cells. Chem. Res. Toxicol. 2010, 24, 238–245. [Google Scholar]
- Greger, J.G.; Eastman, S.D.; Zhang, V.; Bleam, M.R.; Hughes, A.M.; Smitheman, K.N.; Dickerson, S.H.; Laquerre, S.G.; Liu, L.; Gilmer, T.M. Combinations of BRAF, MEK, and PI3K/mTOR inhibitors overcome acquired resistance to the BRAF inhibitor GSK2118436 dabrafenib, mediated by NRAS or MEK mutations. Mol. Cancer Ther. 2012, 11, 909–920. [Google Scholar] [CrossRef] [PubMed]
- Irvine, M.; Stewart, A.; Pedersen, B.; Boyd, S.; Kefford, R.; Rizos, H. Oncogenic PI3K/AKT promotes the step-wise evolution of combination BRAF/MEK inhibitor resistance in melanoma. Oncogenesis 2018, 7, 72. [Google Scholar] [CrossRef] [PubMed]
- Wee, S.; Jagani, Z.; Xiang, K.X.; Loo, A.; Dorsch, M.; Yao, Y.-M.; Sellers, W.R.; Lengauer, C.; Stegmeier, F. PI3K pathway activation mediates resistance to MEK inhibitors in KRAS mutant cancers. Cancer Res. 2009, 69, 4286–4293. [Google Scholar] [CrossRef] [PubMed]
- Sedgwick, P. Spearman’s rank correlation coefficient. Bmj 2014, 349, g7327. [Google Scholar] [CrossRef]
- Nunes, L.; Li, F.; Wu, M.; Luo, T.; Hammarström, K.; Torell, E.; Ljuslinder, I.; Mezheyeuski, A.; Edqvist, P.-H.; Löfgren-Burström, A. Prognostic genome and transcriptome signatures in colorectal cancers. Nature 2024, 633, 137–146. [Google Scholar] [CrossRef]
- Gao, S.; Tibiche, C.; Zou, J.; Zaman, N.; Trifiro, M.; O’Connor-McCourt, M.; Wang, E. Identification and construction of combinatory cancer hallmark–based gene signature sets to predict recurrence and chemotherapy benefit in stage II colorectal cancer. JAMA Oncol. 2016, 2, 37–45. [Google Scholar] [CrossRef]
- Cercek, A.; Chatila, W.K.; Yaeger, R.; Walch, H.; Fernandes, G.D.S.; Krishnan, A.; Palmaira, L.; Maio, A.; Kemel, Y.; Srinivasan, P.; et al. A Comprehensive Comparison of Early-Onset and Average-Onset Colorectal Cancers. J. Natl. Cancer Inst. 2021, 113, 1683–1692. [Google Scholar] [CrossRef]
- Yu, J.; Liu, D.; Sun, X.; Yang, K.; Yao, J.; Cheng, C.; Wang, C.; Zheng, J. CDX2 inhibits the proliferation and tumor formation of colon cancer cells by suppressing Wnt/β-catenin signaling via transactivation of GSK-3β and Axin2 expression. Cell Death Dis. 2019, 10, 26. [Google Scholar] [CrossRef]
- Zhai, J.; Chen, H.; Wong, C.C.; Peng, Y.; Gou, H.; Zhang, J.; Pan, Y.; Chen, D.; Lin, Y.; Wang, S.; et al. ALKBH5 Drives Immune Suppression Via Targeting AXIN2 to Promote Colorectal Cancer and Is a Target for Boosting Immunotherapy. Gastroenterology 2023, 165, 445–462. [Google Scholar] [CrossRef]
- Cheng, L.Z.; Huang, D.L.; Tang, Z.R.; Zhang, J.H.; Xiong, T.; Zhou, C.; Zhang, N.X.; Fu, R.; Cheng, Y.X.; Wu, Z.Q. Pharmacological targeting of Axin2 suppresses cell growth and metastasis in colorectal cancer. Br. J. Pharmacol. 2023, 180, 3071–3091. [Google Scholar] [CrossRef]
- Liu, H.; Liu, L.; Liu, Q.; He, F.; Zhu, H. LncRNA HOXD-AS1 affects proliferation and apoptosis of cervical cancer cells by promoting FRRS1 expression via transcription factor ELF1. Cell Cycle 2022, 21, 416–426. [Google Scholar] [CrossRef] [PubMed]
- Saiyin, H.; Na, N.; Han, X.; Fang, Y.; Wu, Y.; Lou, W.; Yang, X. BRSK2 induced by nutrient deprivation promotes Akt activity in pancreatic cancer via downregulation of mTOR activity. Oncotarget 2017, 8, 44669–44681. [Google Scholar] [CrossRef] [PubMed]
- Tamir, T.Y.; Drewry, D.H.; Wells, C.; Major, M.B.; Axtman, A.D. PKIS deep dive yields a chemical starting point for dark kinases and a cell active BRSK2 inhibitor. Sci. Rep. 2020, 10, 15826. [Google Scholar] [CrossRef] [PubMed]
- Neyazi, M.; Bharadwaj, S.S.; Bullers, S.; Varenyiova, Z.; Travis, S.; Arancibia-Cárcamo, C.V.; Powrie, F.; Geremia, A. Overexpression of Cancer-Associated Stem Cell Gene OLFM4 in the Colonic Epithelium of Patients With Primary Sclerosing Cholangitis. Inflamm. Bowel Dis. 2021, 27, 1316–1327. [Google Scholar] [CrossRef]
- Liu, W.; Li, H.; Aerbajinai, W.; Botos, I.; Rodgers, G.P. OLFM4-RET fusion is an oncogenic driver in small intestine adenocarcinoma. Oncogene 2022, 41, 72–82. [Google Scholar] [CrossRef]
- Zhi, L.; Zhao, L.; Zhang, X.; Liu, W.; Gao, B.; Wang, F.; Wang, X.; Wang, G. SLCO1B3 promotes colorectal cancer tumorigenesis and metastasis through STAT3. Aging 2021, 13, 22164–22175. [Google Scholar] [CrossRef]
- Sun, R.; Ying, Y.; Tang, Z.; Liu, T.; Shi, F.; Li, H.; Guo, T.; Huang, S.; Lai, R. The Emerging Role of the SLCO1B3 Protein in Cancer Resistance. Protein Pept. Lett. 2020, 27, 17–29. [Google Scholar] [CrossRef]
- Hlaváč, V.; Václavíková, R.; Brynychová, V.; Ostašov, P.; Koževnikovová, R.; Kopečková, K.; Vrána, D.; Gatěk, J.; Souček, P. Role of Genetic Variation in Cytochromes P450 in Breast Cancer Prognosis and Therapy Response. Int. J. Mol. Sci. 2021, 22, 2826. [Google Scholar] [CrossRef]
- Chang, M.T.; Asthana, S.; Gao, S.P.; Lee, B.H.; Chapman, J.S.; Kandoth, C.; Gao, J.; Socci, N.D.; Solit, D.B.; Olshen, A.B.; et al. Identifying recurrent mutations in cancer reveals widespread lineage diversity and mutational specificity. Nat. Biotechnol. 2016, 34, 155–163. [Google Scholar] [CrossRef]
- Gao, J.; Chang, M.T.; Johnsen, H.C.; Gao, S.P.; Sylvester, B.E.; Sumer, S.O.; Zhang, H.; Solit, D.B.; Taylor, B.S.; Schultz, N.; et al. 3D clusters of somatic mutations in cancer reveal numerous rare mutations as functional targets. Genome Med. 2017, 9, 4. [Google Scholar] [CrossRef]
- Müller, M.F.; Ibrahim, A.E.K.; Arends, M.J. Molecular pathological classification of colorectal cancer. Virchows Arch. 2016, 469, 125–134. [Google Scholar] [CrossRef] [PubMed]
- Cohen, R.; Rousseau, B.; Vidal, J.; Colle, R.; Diaz, L.A.; André, T. Immune Checkpoint Inhibition in Colorectal Cancer: Microsatellite Instability and Beyond. Target. Oncol. 2020, 15, 11–24. [Google Scholar] [CrossRef] [PubMed]
- De’ Angelis, G.L.; Bottarelli, L.; Azzoni, C.; De’ Angelis, N.; Leandro, G.; Di Mario, F.; Gaiani, F.; Negri, F. Microsatellite instability in colorectal cancer. Acta Biomed. 2018, 89, 97–101. [Google Scholar] [PubMed]
- Rao, C.; Frodyma, D.E.; Southekal, S.; Svoboda, R.A.; Black, A.R.; Guda, C.; Mizutani, T.; Clevers, H.; Johnson, K.R.; Fisher, K.W. KSR1-and ERK-dependent translational regulation of the epithelial-to-mesenchymal transition. eLife 2021, 10, e66608. [Google Scholar] [CrossRef]
- Neilsen, B.K.; Frodyma, D.E.; Lewis, R.E.; Fisher, K.W. KSR as a therapeutic target for Ras-dependent cancers. Expert. Opin. Ther. Targets 2017, 21, 499–509. [Google Scholar] [CrossRef]
- Zaidh, S.M.; Vengateswaran, H.T.; Habeeb, M.; Aher, K.B.; Bhavar, G.B.; Irfan, N.; Lakshmi, K. Network pharmacology and AI in cancer research uncovering biomarkers and therapeutic targets for RALGDS mutations. Sci. Rep. 2025, 15, 1–13. [Google Scholar] [CrossRef]
- Cui, X.; Shen, K.; Xie, Z.; Liu, T.; Zhang, H. Identification of key genes in colorectal cancer using random walk with restart. Mol. Med. Rep. 2017, 15, 867–872. [Google Scholar] [CrossRef]
- Li, Q.; Geng, S.; Luo, H.; Wang, W.; Mo, Y.Q.; Luo, Q.; Wang, L.; Song, G.B.; Sheng, J.P.; Xu, B. Signaling pathways involved in colorectal cancer: Pathogenesis and targeted therapy. Signal Transduct. Target. Ther. 2024, 9, 266. [Google Scholar] [CrossRef]
- Yang, D.; Liu, J.; Qian, H.; Zhuang, Q. Cancer-associated fibroblasts: From basic science to anticancer therapy. Exp. Mol. Med. 2023, 55, 1322–1332. [Google Scholar] [CrossRef]
- Glabman, R.A.; Choyke, P.L.; Sato, N. Cancer-Associated Fibroblasts: Tumorigenicity and Targeting for Cancer Therapy. Cancers 2022, 14, 3906. [Google Scholar] [CrossRef]
- Christofides, A.; Strauss, L.; Yeo, A.; Cao, C.; Charest, A.; Boussiotis, V.A. The complex role of tumor-infiltrating macrophages. Nat. Immunol. 2022, 23, 1148–1156. [Google Scholar] [CrossRef]
- Cassetta, L.; Pollard, J.W. A timeline of tumour-associated macrophage biology. Nat. Rev. Cancer 2023, 23, 238–257. [Google Scholar] [CrossRef]
- Togashi, Y.; Shitara, K.; Nishikawa, H. Regulatory T cells in cancer immunosuppression—Implications for anticancer therapy. Nat. Rev. Clin. Oncol. 2019, 16, 356–371. [Google Scholar] [CrossRef]
- Li, C.; Jiang, P.; Wei, S.; Xu, X.; Wang, J. Regulatory T cells in tumor microenvironment: New mechanisms, potential therapeutic strategies and future prospects. Mol. Cancer 2020, 19, 116. [Google Scholar] [CrossRef]
- Plitas, G.; Rudensky, A.Y. Regulatory T cells in cancer. Annu. Rev. Cancer Biol. 2020, 4, 459–477. [Google Scholar] [CrossRef]
- Long, N.P.; Lee, W.J.; Huy, N.T.; Lee, S.J.; Park, J.H.; Kwon, S.W. Novel biomarker candidates for colorectal cancer metastasis: A meta-analysis of in vitro studies. Cancer Inform. 2016, 15, CIN.S40301. [Google Scholar] [CrossRef] [PubMed]
- Olejniczak, A.; Szaryńska, M.; Kmieć, Z. In vitro characterization of spheres derived from colorectal cancer cell lines. Int. J. Oncol. 2018, 52, 599–612. [Google Scholar] [CrossRef] [PubMed]
- Ronen, J.; Hayat, S.; Akalin, A. Evaluation of colorectal cancer subtypes and cell lines using deep learning. Life Sci. Alliance 2019, 2, e201900517. [Google Scholar] [CrossRef]
- Deshmukh, R.; Prajapati, M.; Harwansh, R.K. A review on emerging targeted therapies for the management of metastatic colorectal cancers. Med. Oncol. 2023, 40, 159. [Google Scholar] [CrossRef]
- Lee, C.L.; Cremona, M.; Farrelly, A.; Workman, J.A.; Kennedy, S.; Aslam, R.; Carr, A.; Madden, S.; O’Neill, B.; Hennessy, B.T.; et al. Preclinical evaluation of the CDK4/6 inhibitor palbociclib in combination with a PI3K or MEK inhibitor in colorectal cancer. Cancer Biol. Ther. 2023, 24, 2223388. [Google Scholar] [CrossRef]
- Carlos, G.; Anforth, R.; Clements, A.; Menzies, A.M.; Carlino, M.S.; Chou, S.; Fernandez-Peñas, P. Cutaneous Toxic Effects of BRAF Inhibitors Alone and in Combination with MEK Inhibitors for Metastatic Melanoma. JAMA Dermatol. 2015, 151, 1103–1109. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, T.H.; Hsu, C.Y.; Wu, C.W.; Wang, S.H.; Yeh, C.H.; Cheng, K.H.; Tsai, E.M. Vorinostat decrease M2 macrophage polarization through ARID1A(6488delG)/HDAC6/IL-10 signaling pathway in endometriosis-associated ovarian carcinoma. Biomed. Pharmacother. 2023, 161, 114500. [Google Scholar] [CrossRef] [PubMed]
- Berdeja, J.G.; Laubach, J.P.; Richter, J.; Stricker, S.; Spencer, A.; Richardson, P.G.; Chari, A. Panobinostat From Bench to Bedside: Rethinking the Treatment Paradigm for Multiple Myeloma. Clin. Lymphoma Myeloma Leuk. 2021, 21, 752–765. [Google Scholar] [CrossRef] [PubMed]
- Jenner, M.W.; Pawlyn, C.; Davies, F.E.; Menzies, T.; Hockaday, A.; Olivier, C.; Jones, J.R.; Karunanithi, K.; Lindsay, J.; Kishore, B.; et al. The addition of vorinostat to lenalidomide maintenance for patients with newly diagnosed multiple myeloma of all ages: Results from ‘Myeloma XI’, a multicentre, open-label, randomised, phase III trial. Br. J. Haematol. 2023, 201, 267–279. [Google Scholar] [CrossRef]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef]
- Ertl, P.; Rohde, B.; Selzer, P. Fast calculation of molecular polar surface area as a sum of fragment-based contributions and its application to the prediction of drug transport properties. J. Med. Chem. 2000, 43, 3714–3717. [Google Scholar] [CrossRef]
- Yamamoto, M.; Fujiwara, N. Protein phosphatase 6 regulates trametinib sensitivity, a mitogen-activated protein kinase kinase (MEK) inhibitor, by regulating MEK1/2-ERK1/2 signaling in canine melanoma cells. J. Vet. Med. Sci. 2023, 85, 977–984. [Google Scholar] [CrossRef]
- Aaroe, A.; Kurzrock, R.; Goyal, G.; Goodman, A.M.; Patel, H.; Ruan, G.; Ulaner, G.; Young, J.; Li, Z.; Dustin, D. Successful treatment of non-Langerhans cell histiocytosis with the MEK inhibitor trametinib: A multicenter analysis. Blood Adv. 2023, 7, 3984–3992. [Google Scholar] [CrossRef]
- Ballantyne, A.D.; Garnock-Jones, K.P. Dabrafenib: First Global Approval. Drugs 2013, 73, 1367–1376. [Google Scholar] [CrossRef]
- Vasuki, A.; Christy, H.J.; Renugadevi, K.; Dammalli, M. Structure-based pharmacophore modeling and DFT studies of Indian Ocean-derived red algal compounds as PI3Kα inhibitors. Mol. Divers. 2023, 28, 2563–2581. [Google Scholar] [CrossRef]
- Yang, J.; Friedman, R. Combination strategies to overcome drug resistance in FLT(+) acute myeloid leukaemia. Cancer Cell Int. 2023, 23, 161. [Google Scholar] [PubMed]
- Seeliger, D.; de Groot, B.L. Ligand docking and binding site analysis with PyMOL and Autodock/Vina. J. Comput.-Aided Mol. Des. 2010, 24, 417–422. [Google Scholar] [CrossRef] [PubMed]
- Egbuna, C.; Patrick-Iwuanyanwu, K.C.; Onyeike, E.N.; Khan, J.; Alshehri, B. FMS-like tyrosine kinase-3 (FLT3) inhibitors with better binding affinity and ADMET properties than sorafenib and gilteritinib against acute myeloid leukemia: In silico studies. J. Biomol. Struct. Dyn. 2022, 40, 12248–12259. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.M.; Faisal, A.M.; Nila, T.A.; Binti, N.N.; Hosen, M.I.; Shekhar, H.U. A computational in silico approach to predict high-risk coding and non-coding SNPs of human PLCG1 gene. PLoS ONE 2021, 16, e0260054. [Google Scholar] [CrossRef]
- Muzny, D.M.; Bainbridge, M.N.; Chang, K.; Dinh, H.H.; Drummond, J.A.; Fowler, G.; Kovar, C.L.; Lewis, L.R.; Morgan, M.B.; Newsham, I.F.; et al. Comprehensive molecular characterization of human colon and rectal cancer. Nature 2012, 487, 330–337. [Google Scholar] [CrossRef]
- Benmokhtar, S.; Laraqui, A.; Hilali, F.; Bajjou, T.; El Zaitouni, S.; Jafari, M.; Baba, W.; Elannaz, H.; Lahlou, I.A.; Hafsa, C. RAS/RAF/MAPK pathway mutations as predictive biomarkers in middle eastern colorectal cancer: A systematic review. Clin. Med. Insights Oncol. 2024, 18, 11795549241255651. [Google Scholar] [CrossRef]
- Voutsadakis, I.A. PIK3CA mutated colorectal cancers without KRAS, NRAS and BRAF mutations possess common and potentially targetable mutations in epigenetic modifiers and DNA damage response genes. Cancer Genom. Proteom. 2024, 21, 533–548. [Google Scholar] [CrossRef]
- Chuang, J.; Wang, C.; Guo, Y.; Valenzuela, V.; Wu, J.; Fakih, M. MAP2K1 Mutations in Advanced Colorectal Cancer Predict Poor Response to Anti-EGFR Therapy and to Vertical Targeting of MAPK Pathway. Clin. Colorectal. Cancer 2021, 20, 72–78. [Google Scholar] [CrossRef]
- Fennell, L.J.; Kane, A.; Liu, C.; McKeone, D.; Fernando, W.; Su, C.; Bond, C.; Jamieson, S.; Dumenil, T.; Patch, A.-M. APC mutation marks an aggressive subtype of BRAF mutant colorectal cancers. Cancers 2020, 12, 1171. [Google Scholar] [CrossRef]
- Crielaard, B.J.; Lammers, T.; Rivella, S. Targeting iron metabolism in drug discovery and delivery. Nat. Rev. Drug Discov. 2017, 16, 400–423. [Google Scholar] [CrossRef]
- Bu, X.; Wang, L. Iron metabolism and the tumor microenvironment: A new perspective on cancer intervention and therapy. Int. J. Mol. Med. 2024, 55, 39. [Google Scholar] [CrossRef] [PubMed]
- Dong, D.; Zhang, G.; Yang, J.; Zhao, B.; Wang, S.; Wang, L.; Zhang, G.; Shang, P. The role of iron metabolism in cancer therapy focusing on tumor-associated macrophages. J. Cell. Physiol. 2019, 234, 8028–8039. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Qiao, S.; Chen, J.; Wen, X.; Chen, Y.; Song, X.; Xu, J.; Qiao, X.; Yang, J.; Zhang, S. M2-type macrophages and cancer-associated fibroblasts combine to promote colorectal cancer liver metastases. OncoTargets Ther. 2024, 17, 243–260. [Google Scholar] [CrossRef]
- Zhang, R.; Qi, F.; Zhao, F.; Li, G.; Shao, S.; Zhang, X.; Yuan, L.; Feng, Y. Cancer-associated fibroblasts enhance tumor-associated macrophages enrichment and suppress NK cells function in colorectal cancer. Cell Death Dis. 2019, 10, 273. [Google Scholar] [CrossRef] [PubMed]
- Martinelli, E.; Cremolini, C.; Mazard, T.; Vidal, J.; Virchow, I.; Tougeron, D.; Cuyle, P.J.; Chibaudel, B.; Kim, S.; Ghanem, I.; et al. Real-world first-line treatment of patients with BRAF(V600E)-mutant metastatic colorectal cancer: The CAPSTAN CRC study. ESMO Open 2022, 7, 100603. [Google Scholar] [CrossRef]
- Su, Y.-H.; Wu, J.-S.; Dai, Y.-Z.; Chen, Y.-T.; Lin, Y.-X.; Tzeng, Y.-M.; Liao, J.-W. Anti-Oxidant, Anti-Mutagenic Activity and Safety Evaluation of Antrocin. Toxics 2023, 11, 547. [Google Scholar] [CrossRef]
- Bartha, Á.; Győrffy, B. TNMplot.com: A Web Tool for the Comparison of Gene Expression in Normal, Tumor and Metastatic Tissues. Int. J. Mol. Sci. 2021, 22, 2622. [Google Scholar] [CrossRef]
- Tang, Z.; Kang, B.; Li, C.; Chen, T.; Zhang, Z. GEPIA2: An enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019, 47, W556–W560. [Google Scholar] [CrossRef]
- Pepe, G.; Granieri, C.N.; Appierdo, R.; Ausiello, G.; Helmer-Citterich, M.; Gherardini, P. PANDA: PAN cancer Data Analysis web tool. J. Mol. Biol. 2025, 437, 169158. [Google Scholar] [CrossRef]
- Nagy, Á.; Győrffy, B. muTarget: A platform linking gene expression changes and mutation status in solid tumors. Int. J. Cancer 2021, 148, 502–511. [Google Scholar] [CrossRef]
- de Bruijn, I.; Kundra, R.; Mastrogiacomo, B.; Tran, T.N.; Sikina, L.; Mazor, T.; Li, X.; Ochoa, A.; Zhao, G.; Lai, B.; et al. Analysis and Visualization of Longitudinal Genomic and Clinical Data from the AACR Project GENIE Biopharma Collaborative in cBioPortal. Cancer Res. 2023, 83, 3861–3867. [Google Scholar] [CrossRef]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal 2013, 6, pl1. [Google Scholar] [CrossRef] [PubMed]
- Cerami, E. Correction: The cBio Cancer Genomics Portal: An Open Platform for Exploring Multidimensional Cancer Genomics Data. Cancer Discov. 2012, 2, 960. [Google Scholar] [CrossRef] [PubMed]
- Yaeger, R.; Chatila, W.K.; Lipsyc, M.D.; Hechtman, J.F.; Cercek, A.; Sanchez-Vega, F.; Jayakumaran, G.; Middha, S.; Zehir, A.; Donoghue, M.T.A.; et al. Clinical Sequencing Defines the Genomic Landscape of Metastatic Colorectal Cancer. Cancer Cell 2018, 33, 125–136.e3. [Google Scholar] [CrossRef] [PubMed]
- Giannakis, M.; Mu, X.J.; Shukla, S.A.; Qian, Z.R.; Cohen, O.; Nishihara, R.; Bahl, S.; Cao, Y.; Amin-Mansour, A.; Yamauchi, M.; et al. Genomic Correlates of Immune-Cell Infiltrates in Colorectal Carcinoma. Cell Rep. 2016, 15, 857–865. [Google Scholar] [CrossRef]
- Mondaca, S.; Walch, H.; Nandakumar, S.; Chatila, W.K.; Schultz, N.; Yaeger, R. Specific Mutations in APC, but Not Alterations in DNA Damage Response, Associate With Outcomes of Patients With Metastatic Colorectal Cancer. Gastroenterology 2020, 159, 1975–1978.e4. [Google Scholar] [CrossRef]
- Li, J.-H.; Liu, S.; Zhou, H.; Qu, L.-H.; Yang, J.-H. starBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein–RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2013, 42, D92–D97. [Google Scholar] [CrossRef]
- Liu, C.J.; Hu, F.F.; Xie, G.Y.; Miao, Y.R.; Li, X.W.; Zeng, Y.; Guo, A.Y. GSCA: An integrated platform for gene set cancer analysis at genomic, pharmacogenomic and immunogenomic levels. Brief. Bioinform. 2023, 24, bbac558. [Google Scholar] [CrossRef]
- Shi, H.; Fang, L.; Tan, C.; Shi, L.; Zhang, W.; Li, C.-C.; Luo, T.; Yang, Z. Total syntheses of drimane-type sesquiterpenoids enabled by a gold-catalyzed tandem reaction. J. Am. Chem. Soc. 2011, 133, 14944–14947. [Google Scholar] [CrossRef]
- Lipinski, C.A.; Lombardo, F.; Dominy, B.W.; Feeney, P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 2001, 46, 3–26. [Google Scholar] [CrossRef]
- Roskoski, R. Rule of five violations among the FDA-approved small molecule protein kinase inhibitors. Pharmacol. Res. 2023, 191, 106774. [Google Scholar] [CrossRef] [PubMed]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef]
- Ferreira, L.G.; Dos Santos, R.N.; Oliva, G.; Andricopulo, A.D. Molecular Docking and Structure-Based Drug Design Strategies. Molecules 2015, 20, 13384–13421. [Google Scholar] [CrossRef]
- Chen, J.-H.; Wu, A.T.H.; Tzeng, D.T.W.; Huang, C.-C.; Tzeng, Y.-M.; Chao, T.-Y. Antrocin, a bioactive component from Antrodia cinnamomea, suppresses breast carcinogenesis and stemness via downregulation of β-catenin/Notch1/Akt signaling. Phytomedicine 2019, 52, 70–78. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Xu, Z.; Guo, J.; Yang, K.; Zheng, J.; Sun, X. Tumor-associated macrophages (TAMs) depend on MMP1 for their cancer-promoting role. Cell Death Discov. 2021, 7, 343. [Google Scholar] [CrossRef]
- Yin, J.; Zhu, W.; Feng, S.; Yan, P.; Qin, S. The role of cancer-associated fibroblasts in the invasion and metastasis of colorectal cancer. Front. Cell Dev. Biol. 2024, 12, 1375543. [Google Scholar] [CrossRef]
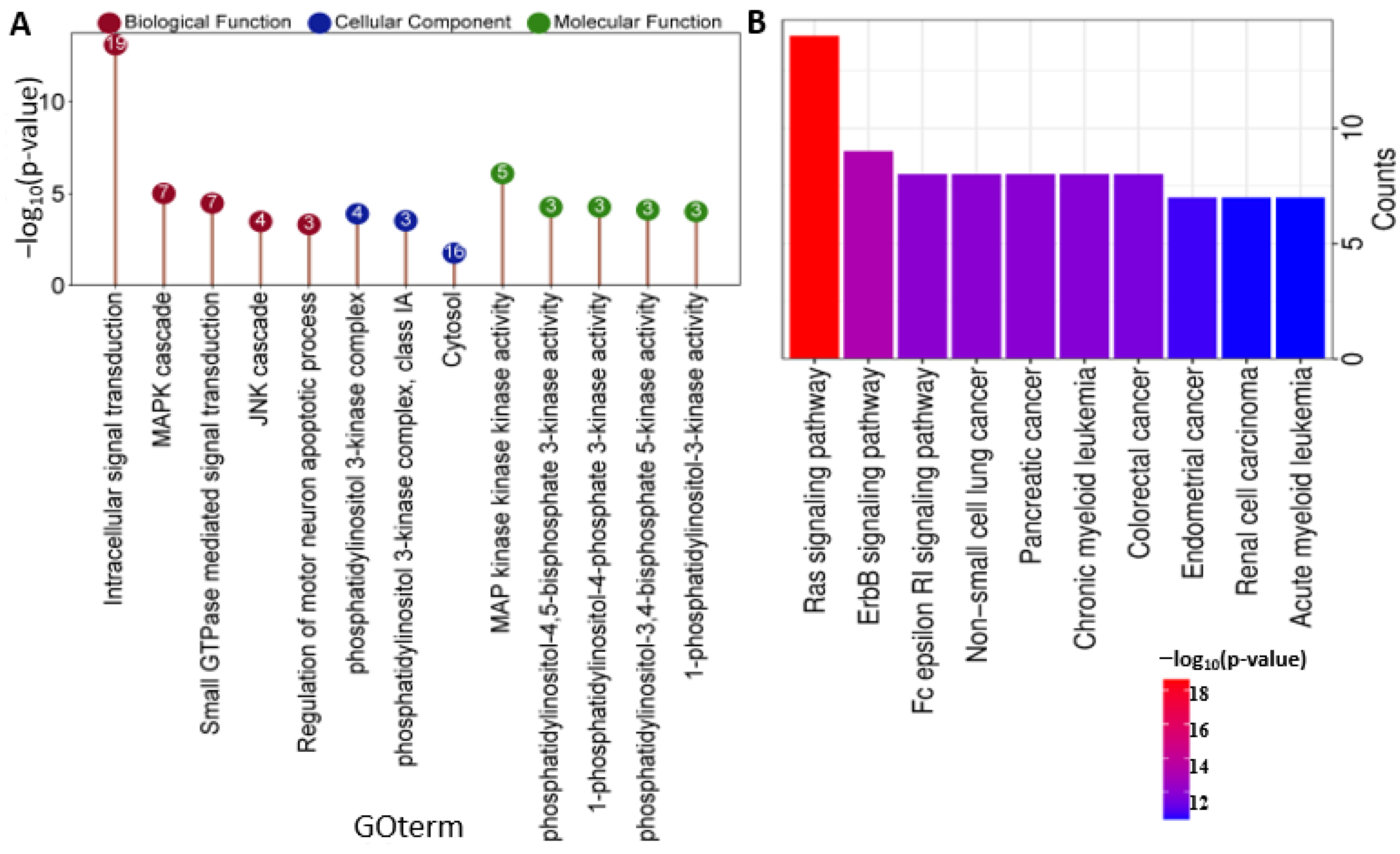



| Term | p-Value | Adj. p-Value | Genes |
|---|---|---|---|
| Sustaining proliferative signaling | 0.013518 | 0.015019988 | BRAF/MAP2K1/PIK3CA |
| Genome instability | 0.1420924 | 0.14209238 | MAP2K1 |
| Evading growth suppressors | 0.0105248 | 0.013155949 | BRAF/MAP2K1/PIK3CA |
| Evading immune destruction | 0.000124 | 0.000372266 | BRAF/MAP2K1/PIK3CA |
| Sustained angiogenesis | 0.0001489 | 0.000372266 | BRAF/MAP2K1/PIK3CA |
| Tissue invasion and metastasis | 0.0036863 | 0.005551356 | BRAF/MAP2K1/PIK3CA |
| Tumor-promoting inflammation | 0.0001342 | 0.000372266 | BRAF/MAP2K1/PIK3CA |
| Resisting cell death | 0.0021638 | 0.00432761 | BRAF/MAP2K1/PIK3CA |
| Reprogramming energy metabolism | 0.0001196 | 0.000372266 | BRAF/MAP2K1/PIK3CA |
| Replicative immortality | 0.0038859 | 0.005551356 | MAP2K1/PIK3CA |
| Formula and SMILE of Antrocin | Physicochemical Properties | Aqueous Solubility | Pharmacokinetics and Absorption | Drug-Likeness | Toxicity |
|---|---|---|---|---|---|
| Formula: C15H22O2 SMILE: CC1(C)CCCC23C(COC2=O)C(=C)CCC13 | Molecular weight: 234.33g/mol NHA: 2 NHD: 0 NRB: 0 Molar Refractivity: 68.17 Lipophilicity: 3.31 | Log S (Ali): −3.67 Log S (ESOL): −3.46 Log S (SILICOS-IT): −3.52 Class: Soluble | BBB permanent: Yes GI absorption: High TPSA: 26.30 Å2 (high crossing of biological barriers) Bioavailability score:0.55 CYP2C19: No CYP1A2: No CYP2C9:Yes | Lipinski: Yes Egan: Yes Veber: Yes Muegge: Yes Ghose: Yes | hERG blocking: (0–0.1): poor Rat acute toxicity (0.1–0.3): poor Genotoxicity: poor |
| Proteins and Gibbs Free Energy | Protein Interaction Site and Bonding Proximity | Bond Type |
|---|---|---|
| BRAF [ΔG = −8.5 kcal/mol] | CYS532 (3.53 Å), ALA481, VAL471, LEU514 | Alkyl |
| TRP531, PHE595, PHE583 | Pi–Alkyl | |
| MEK [ΔG = −7.3 kcal/mol] | SER212 (3.00 Å), VAL211 (3.12 Å) | Conventional hydrogen |
| LEU115, LEU215, ILE99, ILE111, ILE139 | Alkyl | |
| PI3K [ΔG = −6.9 kcal/mol] | LYS298 (3.77 Å), LEU211, ARG294 | Alkyl |
| HIS295, TRP201, TYR210 | Pi–Alkyl |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, J.-S.; Enwolo-Chibueze, C.G.; Chinyama, H.A.; Lai, C.-T.; Ezeala, I.C.; Huang, P.-Y.; Wu, A.T.H.; Huang, Y.-J. Integrated In Silico and Experimental Validation of Antrocin as a Plant-Derived Multi-Target Therapeutic for BRAF/MEK/PI3K-Driven Colorectal Cancer. Int. J. Mol. Sci. 2025, 26, 8780. https://doi.org/10.3390/ijms26188780
Chen J-S, Enwolo-Chibueze CG, Chinyama HA, Lai C-T, Ezeala IC, Huang P-Y, Wu ATH, Huang Y-J. Integrated In Silico and Experimental Validation of Antrocin as a Plant-Derived Multi-Target Therapeutic for BRAF/MEK/PI3K-Driven Colorectal Cancer. International Journal of Molecular Sciences. 2025; 26(18):8780. https://doi.org/10.3390/ijms26188780
Chicago/Turabian StyleChen, Jian-Syun, Chioma Grace Enwolo-Chibueze, Harold Arnold Chinyama, Cheng-Ta Lai, Ifeyinwa Chioma Ezeala, Po-Yang Huang, Alexander T. H. Wu, and Yan-Jiun Huang. 2025. "Integrated In Silico and Experimental Validation of Antrocin as a Plant-Derived Multi-Target Therapeutic for BRAF/MEK/PI3K-Driven Colorectal Cancer" International Journal of Molecular Sciences 26, no. 18: 8780. https://doi.org/10.3390/ijms26188780
APA StyleChen, J.-S., Enwolo-Chibueze, C. G., Chinyama, H. A., Lai, C.-T., Ezeala, I. C., Huang, P.-Y., Wu, A. T. H., & Huang, Y.-J. (2025). Integrated In Silico and Experimental Validation of Antrocin as a Plant-Derived Multi-Target Therapeutic for BRAF/MEK/PI3K-Driven Colorectal Cancer. International Journal of Molecular Sciences, 26(18), 8780. https://doi.org/10.3390/ijms26188780










