Deep Sequencing Reveals Novel Mutations in Androgen Receptor-Related Genes in Prostate Cancer
Abstract
1. Introduction
2. Results
2.1. Clinicopathologic Characteristics of Patients
2.2. Risk Factors Identified in Our Patient Cohorts
2.3. Sequencing Statistics
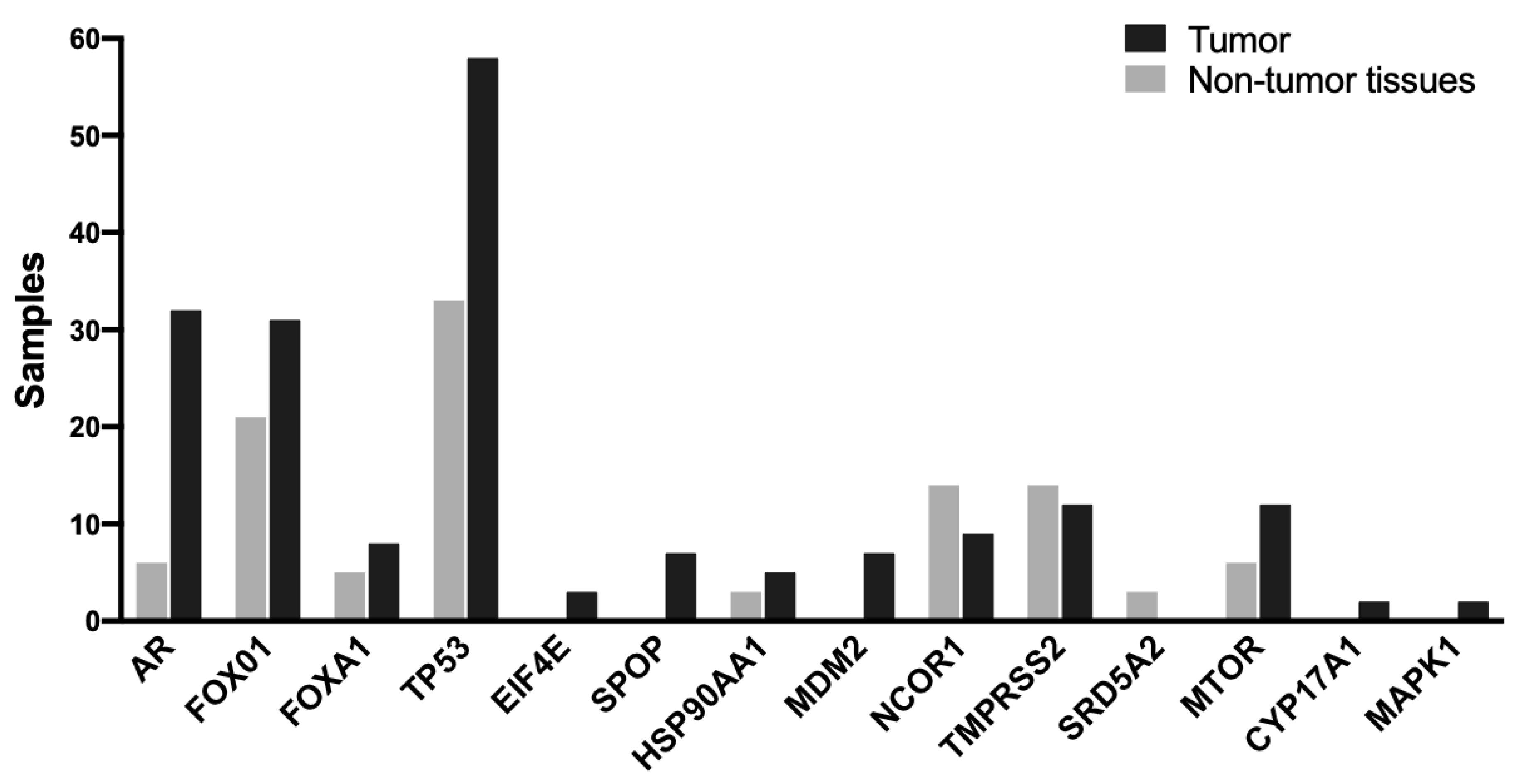
2.4. Genomic Alterations
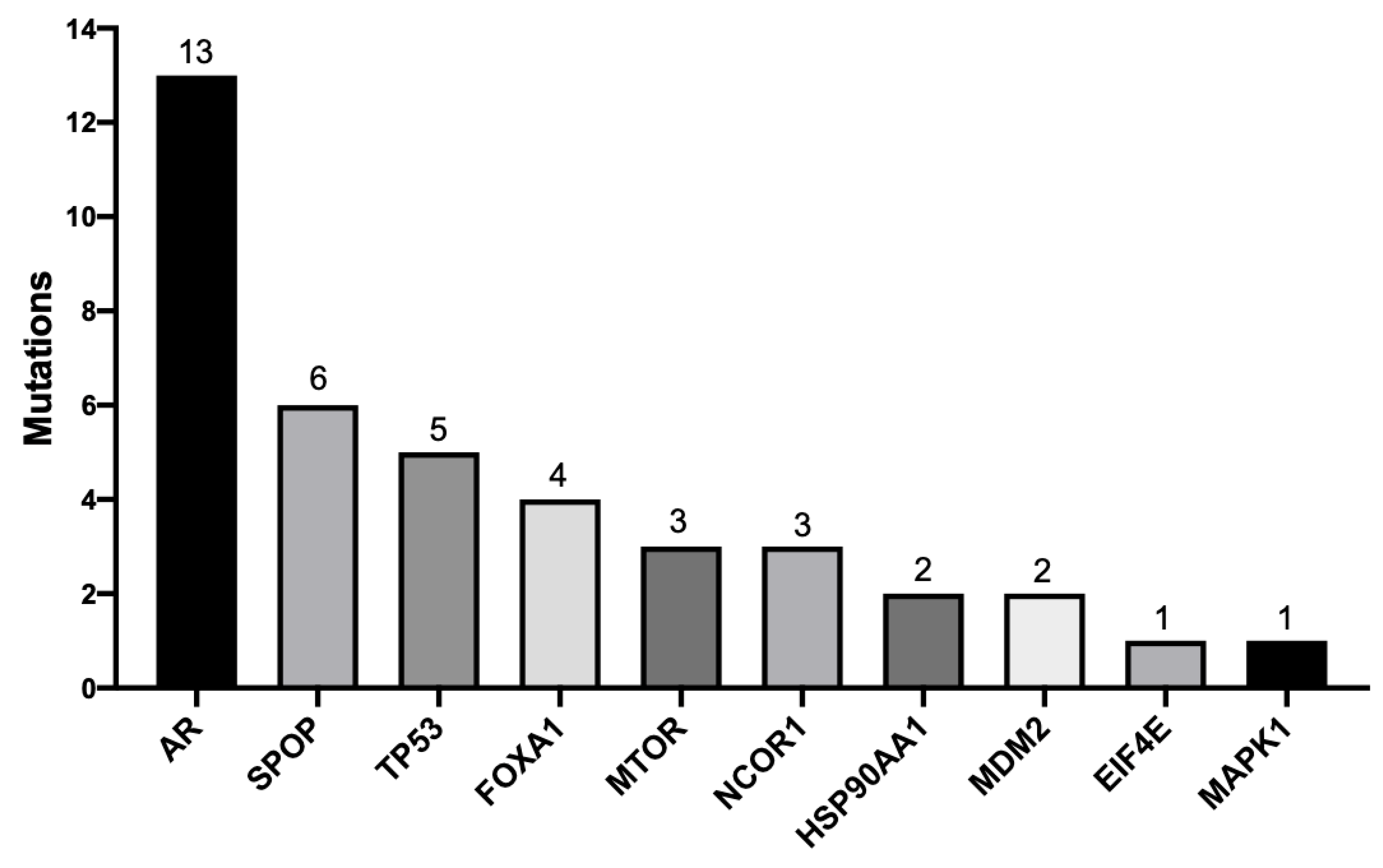
2.5. Pathogenic Mutations
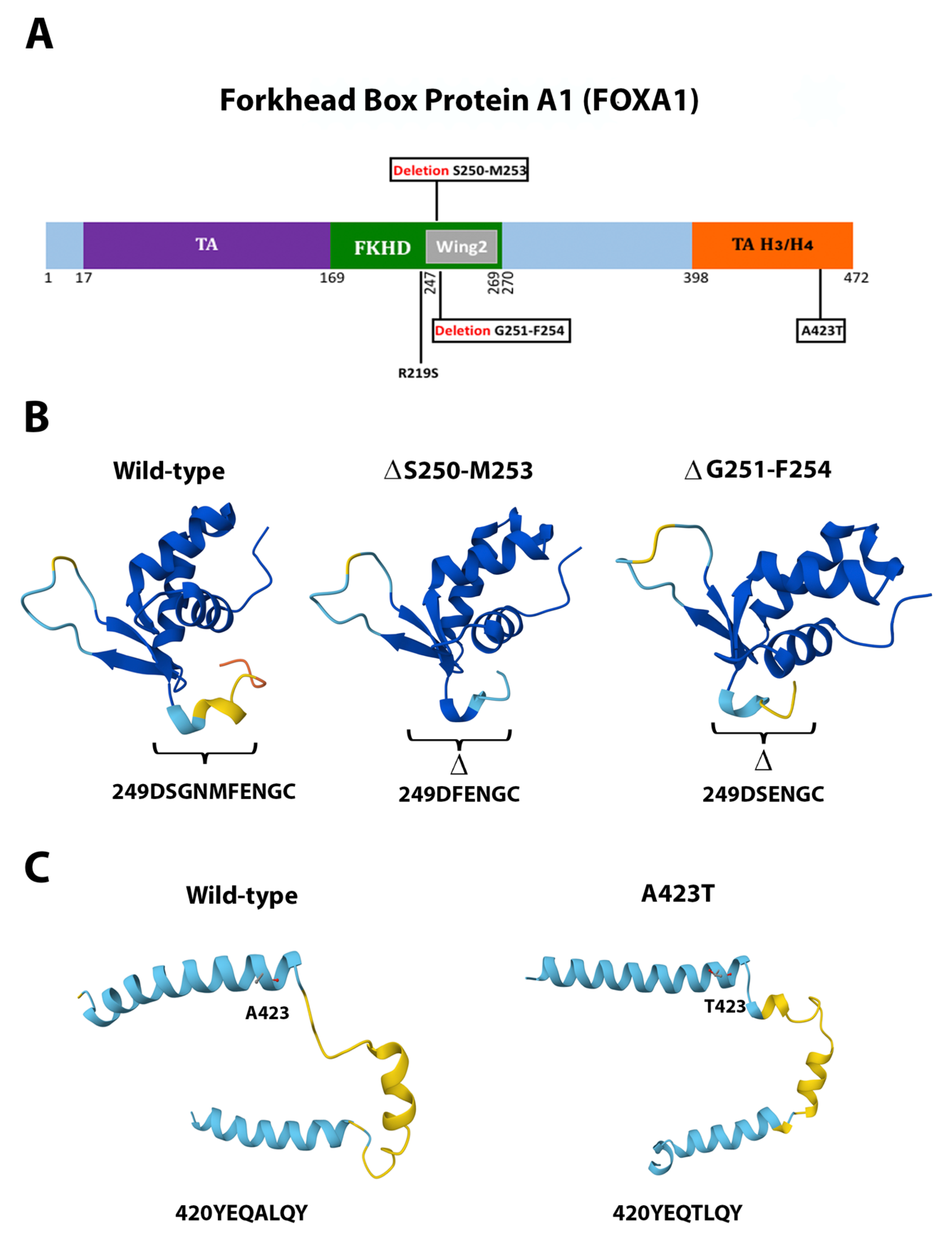
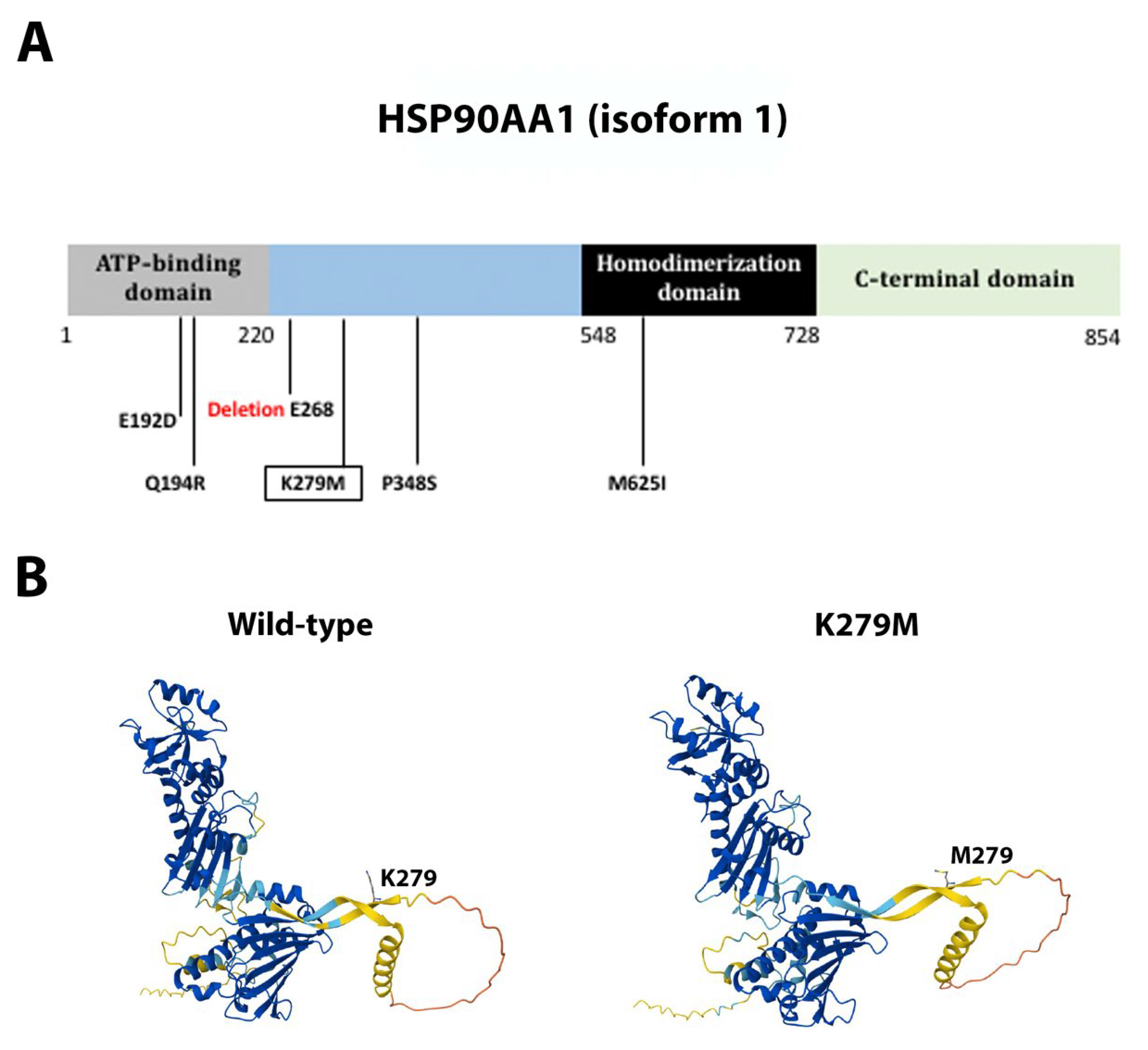
2.6. Novel Mutations
2.7. Association Between Genetic Mutations and Clinicopathologic Characteristics
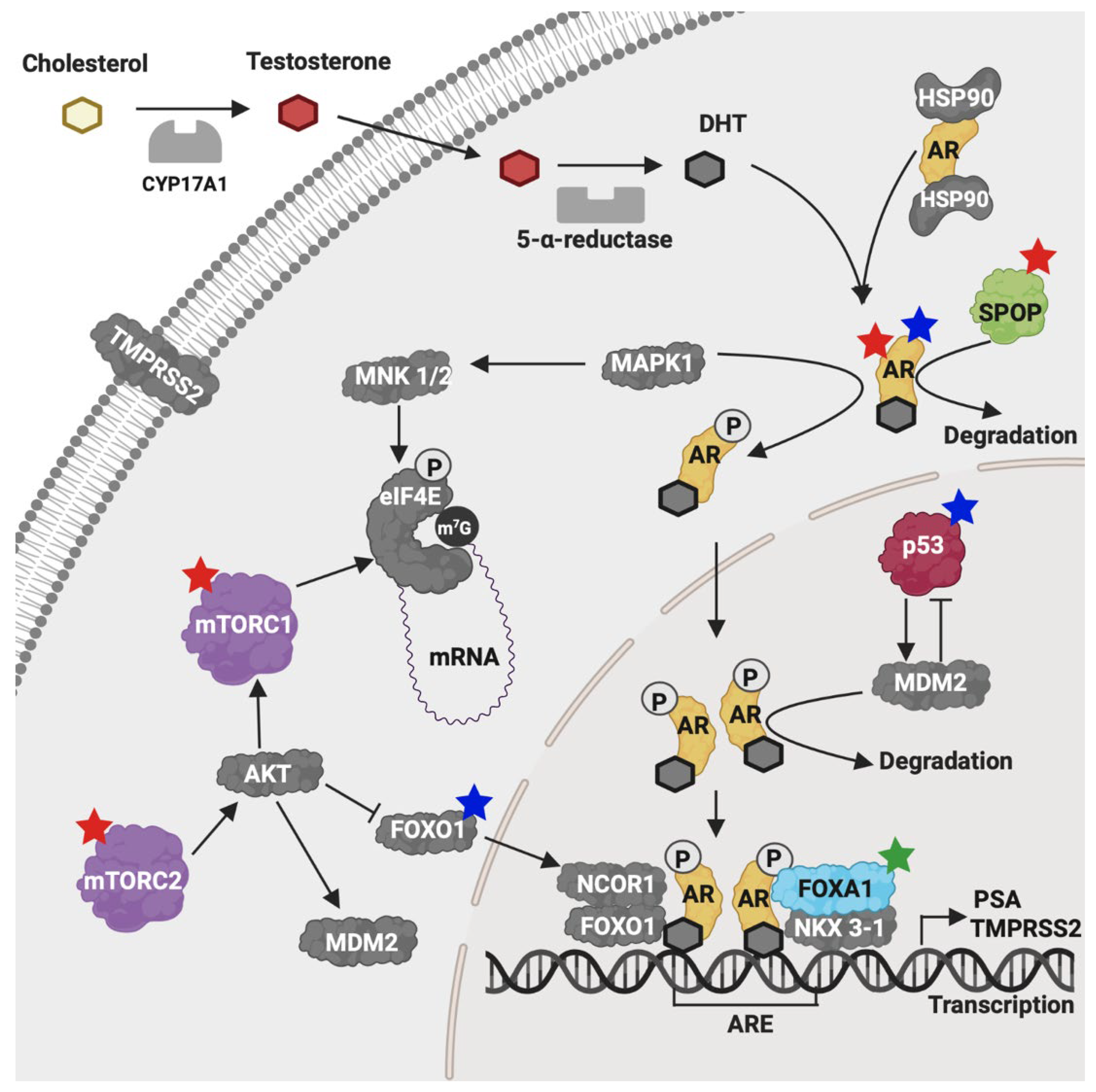
3. Discussion
3.1. Novel Mutations from a Novel Genetic Background
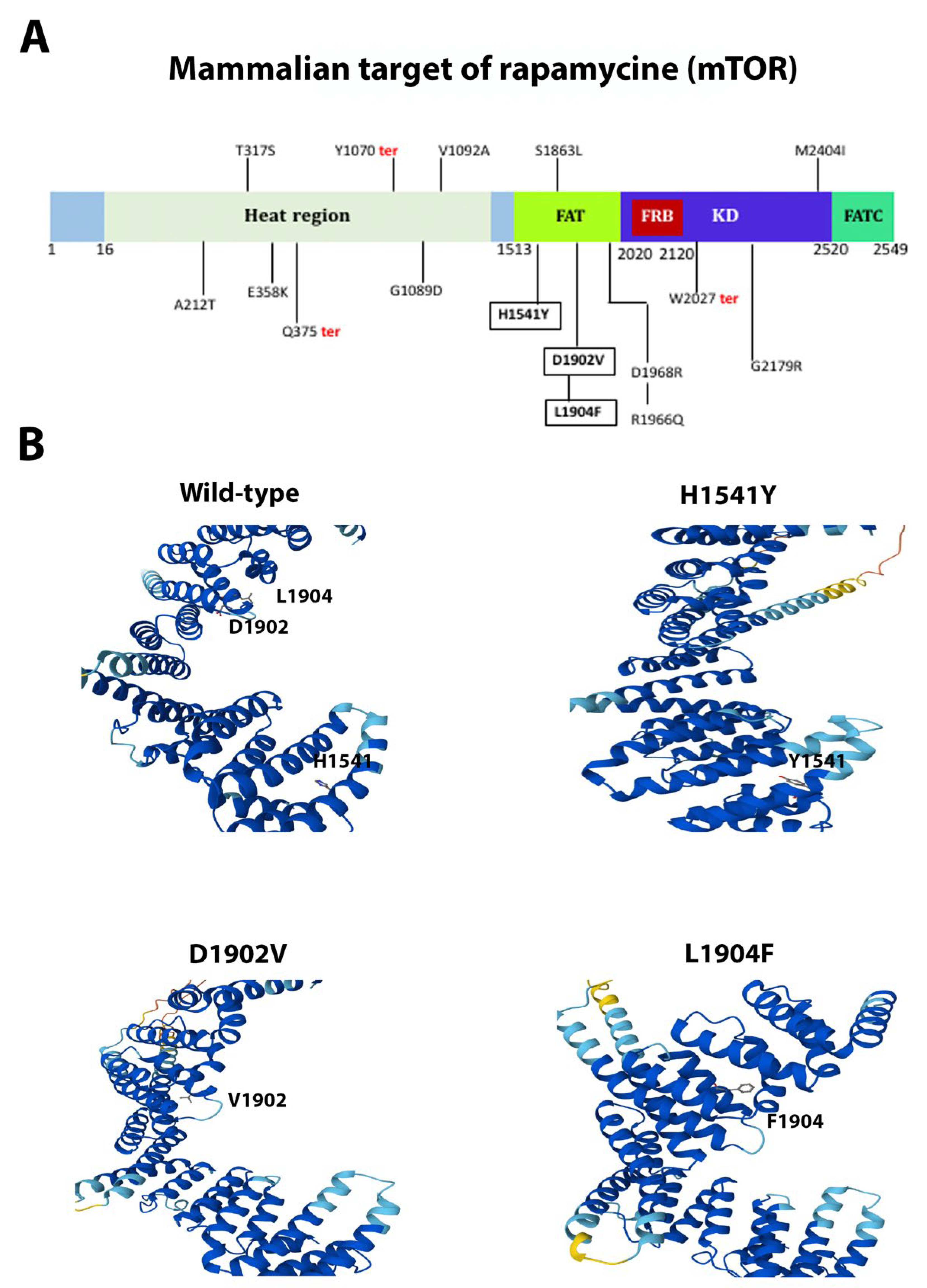
3.2. AR, FOXA1, MTOR, and MDM2 Carried the Highest Number of Novel and Pathogenic Mutations
3.3. Clinical Association of FOXA1 and Patient Survival
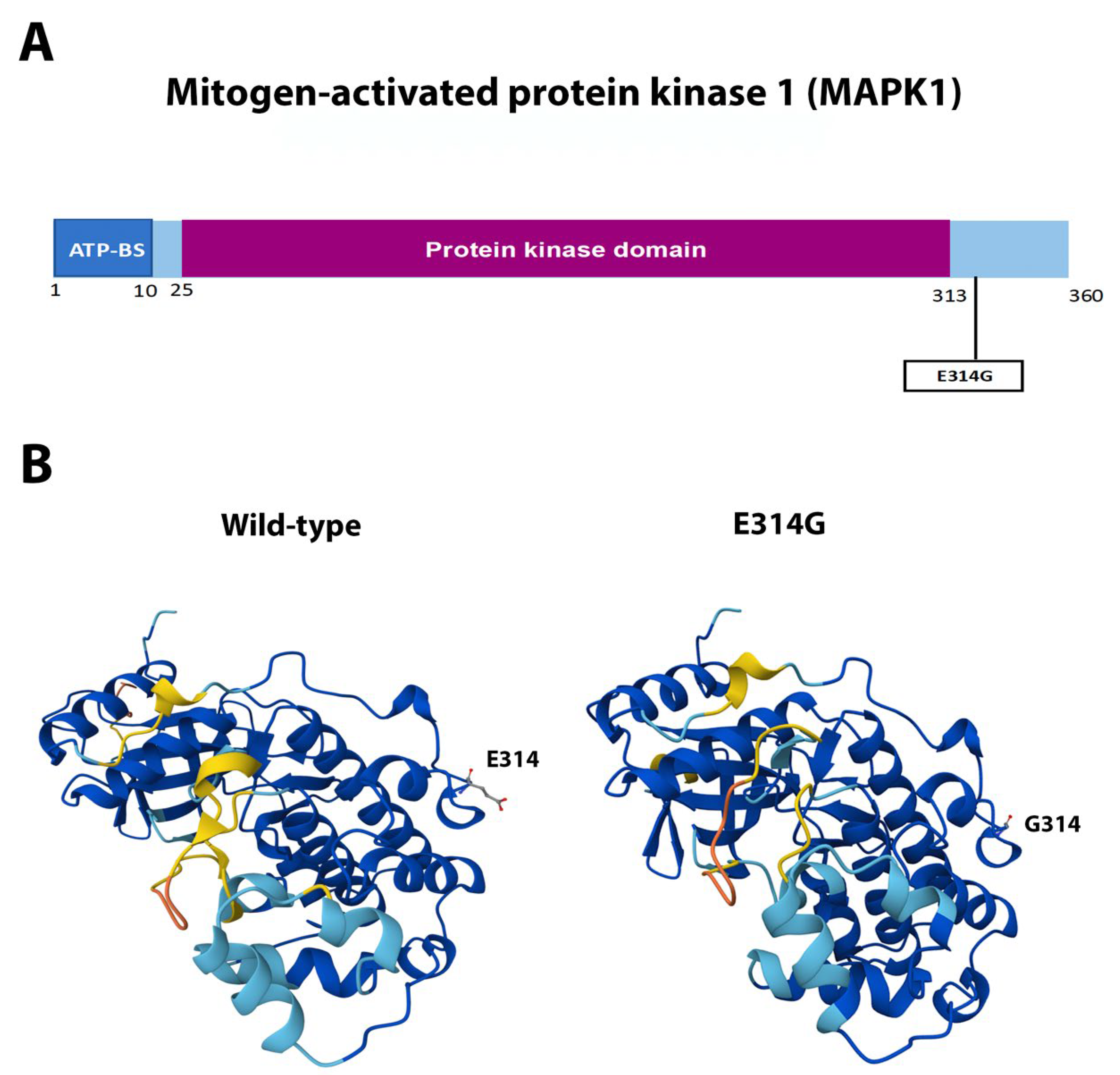
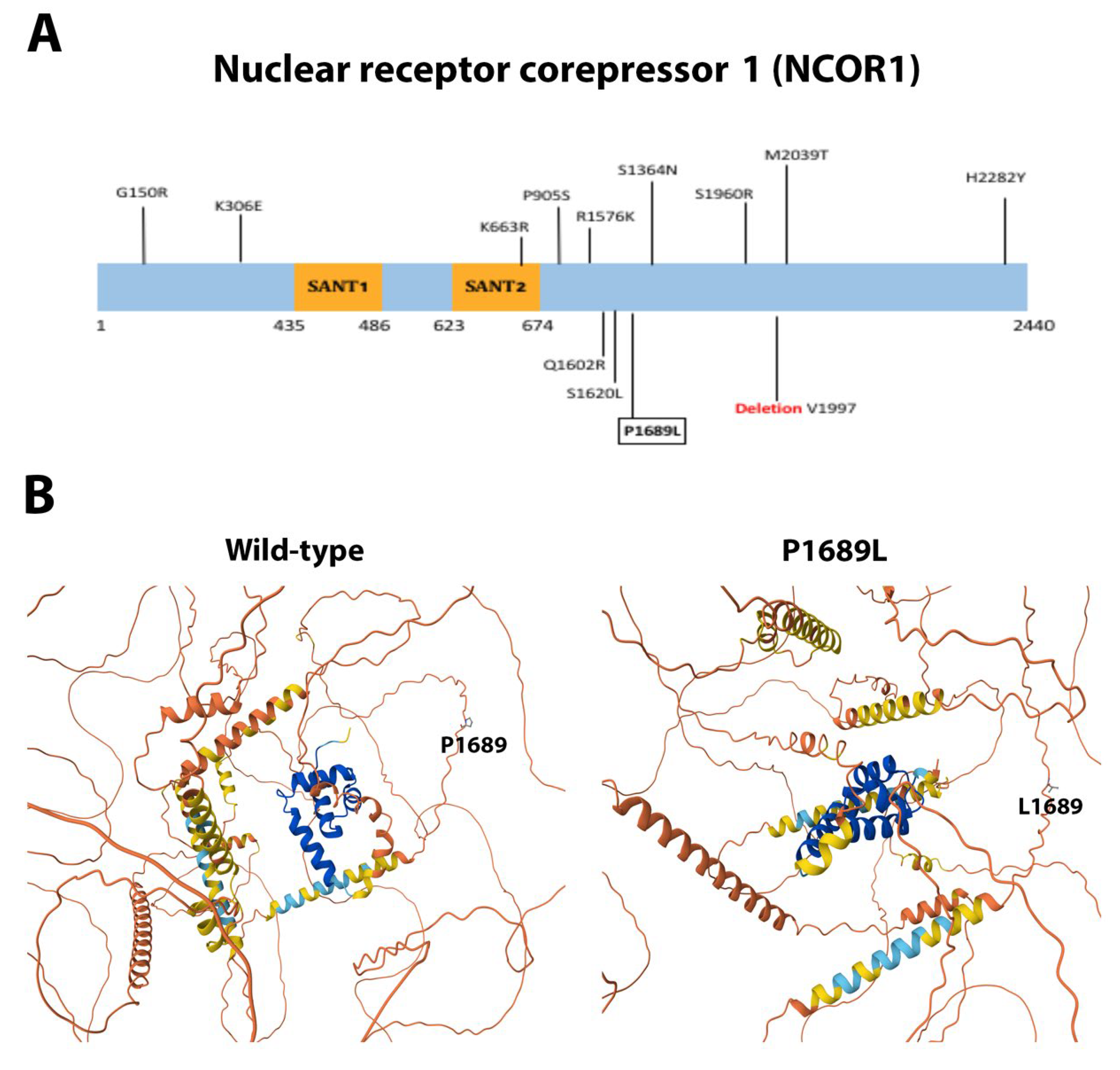
3.4. Single Novel Mutations in EIF4E, HSP90AA1, MAPK1, and NCOR1
4. Materials and Methods
4.1. Patients and Tissue Samples
4.2. Genomic DNA Extraction
4.3. Generation of Primer Pools for Target Genes
4.4. Amplicon Library Generation
4.5. Ion Torrent Sequencing
4.6. Data Analyses
4.7. Variant Classification
4.8. Three-Dimensional (3D) Structure Prediction
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wild, C.P.; Weiderpass, E.; Stewart, B.W. (Eds.) World Cancer Report. Cancer Research for Cancer Prevention; International Agency for Research on Cancer and World Health Organization: Lyon, France, 2020. [Google Scholar]
- Ferlay, J.; Ervik, M.; Laversanne, M.; Colombet, N.; Mery, L.; Piñeros, M.; Znaor, A.; Soerjomataram, I.; Bray, F. Global Cancer Observatory: Cancer Today; International Agency for Research on Cancer; World Health Organization: Geneva, Switzerland, 2024; Available online: https://gco.iarc.who.int/today (accessed on 15 April 2025).
- International Agency for Research on Cancer. Cancer Today. Data Version: Globocan 2022; World Health Organization: Geneva, Switzerland, 2022; Available online: https://gco.iarc.who.int (accessed on 15 April 2025).
- James, N.D.; Tannock, I.; N’Dow, J.; Feng, F.; Gillessen, S.; Ali, S.A.; Trujillo, B.; Al-Lazikanii, B.; Attard, G.; Bray, F.; et al. The Lancet Commission on prostate cancer: Planning for the surge in cases. Lancet 2024, 403, 1683–1722. [Google Scholar] [CrossRef] [PubMed]
- Packer, J.R.; Maitland, N.J. The molecular and cellular origin of human prostate cancer. Biochim. Biophys. Acta 2016, 1863, 1238–1260. [Google Scholar] [CrossRef] [PubMed]
- Humphrey, P.A. Histopathology of prostate cancer. Cold Spring Harb. Perspect. Med. 2017, 7, a030411. [Google Scholar] [CrossRef] [PubMed]
- Sekhoacha, M.; Riet, K.; Motloung, P.; Gumenku, L.; Adegoke, A.; Mashele, S. Prostate cancer review: Genetics, diagnosis, treatment options, and alternative approaches. Molecules 2022, 27, 5730. [Google Scholar] [CrossRef]
- Pernar, C.H.; Ebot, E.M.; Wilson, K.M.; Mucci, L.A. The epidemiology of prostate cancer. Cold Spring Harb. Perspect. Med. 2018, 8, a030361. [Google Scholar] [CrossRef]
- Porcaro, A.B.; Tafuri, A.; Inverardi, D.; Amigoni, N.; Sebben, M.; Pirozzi, M.; Processali, T.; Rizzetto, R.; Shakir, A.; Cerrato, C.; et al. Incidental prostate cancer after transurethral resection of the prostate: Analysis of incidence and risk factors in 458 patients. Minerva Urol. Nefrol. 2020. online ahead of print. [Google Scholar] [CrossRef]
- Rebbeck, T.R. Prostate cancer disparities by race and ethnicity: From nucleotide to Neighborhood. Cold Spring Harb. Perspect. Med. 2018, 8, a030387. [Google Scholar] [CrossRef]
- Jiang, S.; Narayan, V.; Warlick, C. Racial disparities and considerations for active surveillance of prostate cancer. Transl. Androl. Urol. 2018, 7, 214–220. [Google Scholar] [CrossRef]
- Tan, S.H.; Petrovics, G.; Srivastava, S. Prostate gancer genomics: Recent advances and the prevailing underrepresentation from racial and ethnic minorities. Int. J. Mol. Sci. 2018, 19, 1255. [Google Scholar] [CrossRef]
- Sarwar, S.; Adil, M.A.; Nyamath, P.; Ishaq, M. Biomarkers of prostatic cancer: An attempt to categorize patients into prostatic carcinoma, benign prostatic hyperplasia, or prostatitis based on serum prostate specific antigen, prostatic acid phosphatase, calcium, and phosphorus. Prostate Cancer 2017, 2017, 5687212. [Google Scholar] [CrossRef]
- Herget, K.A.; Patel, D.P.; Hanson, H.A.; Sweeney, C.; Lowrance, W.T. Recent decline in prostate cancer incidence in the United States, by age, stage, and Gleason score. Cancer Med. 2016, 5, 136–141. [Google Scholar] [CrossRef]
- Martin, R.M.; Donovan, J.L.; TRurner, E.L.; Metcalfe, C.; Young, G.J.; Walshh, E.I.; Lane, J.A.; Noble, S.; Oliver, S.E.; Evans, S.; et al. Effect of a low-intensity PSA-based ccreening intervention on prostate cancer mortality: The CAP randomized clinical trial. J. Am. Med. Asoc. 2018, 319, 883–895. [Google Scholar] [CrossRef] [PubMed]
- Jones, C.; Fam, M.M.; Davies, B. Expanded criteria for active surveillance in prostate cancer: A review of the current data. Transl. Androl. Urol. 2018, 7, 221–227. [Google Scholar] [CrossRef] [PubMed]
- McCrea, E.M.; Lee, D.K.; Sissung, T.M.; Figg, W.D. Precision medicine applications in prostate cancer. Ther. Adv. Med. Oncol. 2018, 10, 1758835918776920. [Google Scholar] [CrossRef]
- Li, Y.; Roberts, N.D.; Wala, J.A.; Shapira, O.; Schumacher, S.E.; Kumar, K.; Khurana, E.; Waszak, S.; Korbel, J.O.; Haber, J.E.; et al. Patterns of somatic structural variation in human cancer genomes. Nature 2020, 578, 112–121, Correction in Nature 2023, 614, E38. [Google Scholar] [CrossRef]
- Rubin, M.A.; Demichelis, F. The genomics of prostate cancer: A historic perspective. Cold Spring Harb. Perspect. Med. 2019, 31, a034942. [Google Scholar] [CrossRef]
- Khemlina, G.; Ikeda, S.; Kurzrock, R. Molecular landscape of prostate cancer: Implications for current clinical trials. Cancer Treat. Rev. 2015, 41, 761–766. [Google Scholar] [CrossRef]
- Taylor, B.S.; Schultz, N.; Hieronymus, H.; Gopalan, A.; Xiao, Y.; Carver, B.S.; Arora, V.K.; Kaushik, P.; Cerami, E.; Reva, B.; et al. Integrative genomic profiling of human prostate cancer. Cancer Cell 2010, 18, 11–22. [Google Scholar] [CrossRef]
- Grasso, C.S.; Wu, Y.M.; Robinson, D.R.; Cao, X.; Dhanasekaran, S.M.; Khan, A.P.; Quiste, M.J.; Jing, X.; Lonigro, R.J.; Brenner, J.C.; et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature 2012, 487, 239–243. [Google Scholar] [CrossRef]
- Baca, S.C.; Prandi, D.; Lawrence, M.S.; Mosquera, J.M.; Romanel, A.; Drier, Y.; Park, K.; Kitabayashi, N.; MacDonald, T.Y.; Ghandi, M.; et al. Punctuated evolution of prostate cancer genomes. Cell 2013, 153, 666–677. [Google Scholar] [CrossRef]
- Robinson, D.; Van Allen, E.M.; Wu, Y.M.; Schultz, N.; Lonigro, R.J.; Mosquera, J.M.; Montgomery, B.; Taplin, M.E.; Pritchard, C.C.; Attard, G.; et al. Integrative clinical genomics of advanced prostate cancer. Cell 2015, 161, 1215–1228. [Google Scholar] [CrossRef]
- Rheinbay, E.; Nielsen, M.M.; Abascal, F.; Wala, J.A.; Shapira, O.; Tiao, G.; Hornshøj, H.; Hess, J.M.; Juul, R.I.; Lin, Z.; et al. Analyses of non-coding somatic drivers in 2,658 cancer whole genomes. Nature 2020, 578, 102–111. [Google Scholar] [CrossRef] [PubMed]
- Blattner, M.; Lee, D.J.; O’Reilly, C.; Park, K.; MacDonald, T.Y.; Khani, F.; Turner, K.R.; Chiu, Y.L.; Wild, P.J.; Dolgalev, I.; et al. SPOP mutations in prostate cancer across demographically diverse patient cohorts. Neoplasia 2014, 16, 14–20. [Google Scholar] [CrossRef] [PubMed]
- Magi-Galluzzi, C.; Tsusuki, T.; Elson, P.; Simmerman, K.; LaFargue, C.; Esgueva, R.; Klein, E.; Rubin, M.A.; Zhou, M. TMPRSS2-ERG gene fusion prevalence and class are significantly different in prostate cancer of Caucasian, African-American and Japanese patients. Prostate 2011, 71, 489–497. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Mao, X.; Huang, X.; Zhao, J.; Wang, L.; Xu, J.; Zhang, H.; Lu, Y.; Yu, Y. TMPRSS2:ERG fusion gene occurs less frequently in Chinese patients with prostate cancer. Tumor Biol. 2016, 37, 12397–12402. [Google Scholar] [CrossRef]
- Zhou, C.K.; Young, D.; Yeboah, E.D.; Coburn, S.B.; Tettey, Y.; Biritwum, R.B.; Adjei, A.A.; Tay, E.; Niwa, S.; Truelove, A.; et al. TMPRSS2:ERG gene fusions in prostate cancer of west African men and a meta-analysis of racial differences. Am. J. Epidemiol. 2017, 186, 1352–1361. [Google Scholar] [CrossRef]
- Wong, M.; Bierman, Y.; Pettaway, C.; Kittles, R.; Mims, M.; Jones, J.; Ittmann, M. Comparative analysis of p16 expression among African American and European American prostate cancer patients. Prostate 2019, 79, 1274–1283. [Google Scholar] [CrossRef]
- Shina, M.; Hashimoto, Y.; Kato, T.; Yamamura, S.; Tanaka, Y.; Majid, S.; Saini, S.; Varahram, S.; Kulkarni, P.; Dasgupta, P. Differential expression of miR-34b and androgen receptor pathway regulate prostate cancer aggressiveness between African-Americans and Caucasians. Oncotarget 2017, 8, 8356–8368. [Google Scholar] [CrossRef]
- Li, J.; Xu, C.; Lee, H.J.; Ren, S.; Zi, X.; Zhang, Z.; Wang, H.; Yu, Y.; Yang, C.; Gao, X.; et al. A genomic and epigenomic atlas of prostate cancer in Asian populations. Nature 2020, 580, 93–99. [Google Scholar] [CrossRef]
- Crawford, E.D.; Schellhammer, P.F.; McLeod, D.G.; Moul, J.W.; Higano, C.S.; Shore, N.; Denis, L.; Iversen, P.; Eisenberger, M.A.; Labrie, F. Androgen receptor-targeted treatments for prostate cancer: 35 years’ Progress with antiandrogens. J. Urol. 2018, 200, 956–966. [Google Scholar] [CrossRef]
- Tan, M.H.E.; Li, J.; Xu, H.E.; Melcher, K.; Yong, E.L. Androgen receptor: Structure, role in prostate cancer and drug discovery. Acta Pharma. Sin. 2015, 36, 3–23. [Google Scholar] [CrossRef]
- Aurilio, G.; Cimadamore, A.; Mazzucchelli, R.; Lopez-Beltran, A.; Verri, E.; Scarpelli, M.; Massari, F.; Cheng, L.; Santoni, M.; Montironi, R. Androgen Receptor signaling pathway in prostate cancer: From genetics to clinical applications. Cells 2020, 9, 2653. [Google Scholar] [CrossRef] [PubMed]
- Centenera, M.M.; Selth, L.A.; Ebrahimiie, E.; Butler, L.M.; Tilley, W.D. New opportunitie for targeting the Androgen receptor in prostate cancer. Cold Spring Harb. Perspect. Med. 2019, 15, 235–253. [Google Scholar]
- Visakorpi, T.; Hyytinen, E.; Koivisto, P.; Tanner, M.; Keinänen, R.; Palmberg, C.; Palotie, A.; Tammela, T.; Isola, J.; Kallioniemi, O.P. In vivo amplification of the androgen receptor gene and progression of human prostate cancer. Nat. Genet. 1995, 9, 401–406. [Google Scholar] [CrossRef] [PubMed]
- Linja, M.J.; Savinainen, K.J.; Saramäki, O.R.; Tammela, T.L.; Vessella, R.L.; Visakorpi, T. Amplification and overexpression of androgen receptor gene in hormone-refractory prostate cancer. Cancer Res. 2001, 61, 3550–3555. [Google Scholar]
- Gottlieb, B.; Beitel, L.K.; Nadarajah, A.; Paliouras, M.; Trifiro, M. The androgen receptor gene mutations database: 2012 Update. Hum. Mut. 2012, 33, 887–894. [Google Scholar] [CrossRef]
- Culig, Z.; Santer, F. Androgen receptor signaling in prostate cancer. Cancer Metastasis Rev. 2014, 33, 413–427. [Google Scholar] [CrossRef]
- Zaman, N.; Giannopoulos, P.N.; Chowdhury, S.; Bonneil, E.; Thibault, P.; Wang, E.; Trifiro, M.; Paliouras, M. Proteomic-coupled-network analysis of T877A Androgen Receptor interactomes can predict clinical prostate cancer outcomes between White (non-Hispanic) and African-American groups. PLoS ONE 2014, 9, e113190. [Google Scholar] [CrossRef]
- Sun, J.H.; Lee, S.A. Association between CAG repeat polymorphisms and the risk of prostate cancer: A meta-analysis by race, study design and the number of (CAG) n repeat polymorphisms. Int. J. Mol. Med. 2013, 32, 1195–1203. [Google Scholar] [CrossRef][Green Version]
- Qin, Z.; Li, X.; Han, P.; Zheng, Y.; Liu, H.; Tang, J.; Yang, C.; Zhang, J.; Wang, K.; Qi, X.; et al. Association between polymorphic CAG repeat lengths in the androgen receptor gene and susceptibility to prostate cancer: A systematic review and meta-analysis. Medicine 2017, 96, e7258. [Google Scholar] [CrossRef]
- Weng, H.; Li, S.; Huang, J.Y.; He, Z.Q.; Meng, X.Y.; Cao, Y.; Fang, C.; Zeng, X.T. Androgen receptor gene polymorphisms and risk of prostate cancer: A meta-analysis. Sci. Rep. 2017, 7, 40554. [Google Scholar] [CrossRef]
- Gu, M.; Dong, X.; Zhang, X.; Niu, W. The CAG repeat polymorphism of androgen receptor gene and prostate cancer: A meta-analysis. Mol. Biol. Rep. 2012, 39, 2615–2624. [Google Scholar] [CrossRef] [PubMed]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Weikum, E.R.; Liu, X.; Ortlund, E.A. The nuclear receptor super-family: A structural perspective. Protein Sci. 2018, 27, 1876–1892. [Google Scholar] [CrossRef] [PubMed]
- Callewaert, L.; Christiaens, V.; Haelens, A.; Verrijdt, G.; Verhoeven, G.; Claessens, F. Implications of a polyglutamine tract in the function of the human androgen receptor. Biochem. Biophys. Res. Commun. 2003, 306, 46–52. [Google Scholar] [CrossRef]
- Beilin, J.; Ball, E.M.; Favaloro, J.M.; Zajac, J.D. Effect of the androgen receptor CAG repeat polymorphism on transcriptional activity: Specificity in prostate and non-prostate cell lines. J. Mol. Endocrinol. 2000, 25, 85–96. [Google Scholar] [CrossRef]
- Parolia, A.; Cieslik, M.; Chu, S.-C.; Xiao, L.; Ouchi, T.; Zhang, Y.; Wang, X.; Vats, P.; Cao, X.; Pitchiaya, S.; et al. Distinct structural classes of activating FOXA1 alterations in advanced prostate cancer. Nature 2019, 571, 413–418. [Google Scholar] [CrossRef]
- Teng, M.; Zhou, S.; Cai, C.; Lupien, M.; He, H.H. Pioneer of prostate cancer: Past, present and the future of FOXA1. Prot. Cell 2021, 12, 29–38. [Google Scholar] [CrossRef]
- Adams, E.J.; Karthaus, W.R.; Hoover, E.; Liu, D.; Gruet, A.; Zhang, Z.; Cho, H.; DiLoreto, R.; Chhangawala, S.; Liu, Y.; et al. FOXA1 mutations alter pioneering activity, differentiation and prostate cancer phenotypes. Nature 2019, 571, 408–412, Correction in Nature 2020, 585, E20. [Google Scholar] [CrossRef]
- Zhu, J.; Wei, Y.; Deng, F.; Zhou, Y.; Yang, Z.; Ma, Y. The role of FOXA1 in human normal development and its functions in sex hormone-related cancers. Front. Biosc. 2024, 29, 225. [Google Scholar] [CrossRef]
- Marcotrigiano, J.; Gingras, A.C.; Sonenberg, N.; Burley, S.K. Cocrystal structure of the messenger RNA 5′ cap-binding protein (eIF4E) bound to 7-methyl-GDP. Cell 1997, 89, 951–961. [Google Scholar] [CrossRef]
- Tomoo, K.; Shen, J.; Okabe, K.; Nozoe, Y.; Fukuhara, S.; Morino, S.; Ishida, T.; Taniguchi, T.; Hasegawa, H.; Terashima, A.; et al. Crystal structures of 7-methylguanosine 5′-triphosphate (m7GTP)- and P1-7-methylguanosine-P3-adenosine-5′, 5′-triphosphate (m7GpppA)-bound human full-lenght eukaryotic initiation factor 4E: Biological importance of the C-terminal flexible region. Biochem. J. 2002, 362, 539–544. [Google Scholar] [CrossRef] [PubMed]
- Zuehlke, A.D.; Beebe, K.; Neckers, L.; Prince, T. Regulation and function of the human HSP90AA1 gene. Gene 2015, 570, 8–16. [Google Scholar] [CrossRef] [PubMed]
- Prodromou, C.; Roe, S.M.; O’Brien, R.; Ladbury, J.E.; Piiper, P.W.; Pearl, L.H. Identification and structural characterization of the ATP/ADP-binding site in the Hsp90 molecular chaperone. Cell 1997, 90, 65–75. [Google Scholar] [CrossRef] [PubMed]
- Petrosino, M.; Novak, L.; Pasquo, A.; Turina, P.; Capriotti, E.; Minicozzi, V.; Consalvi, V.; Chiaraluce, R. The complex impact of cancer-related missense mutations on the stability and on the biophysical and biochemical properties of MAPK1 and MAPK3 somatic variants. BMC Hum. Genet. 2023, 17, 95. [Google Scholar] [CrossRef]
- Sulaimani, M.N.; Ahmed, S.; Anjum, F.; Mohammad, T.; Shamsii, A.; Dohare, R.; Hassan, M.I. Structure-guided identification of mitogenactivated protein kinase-1 inhibitors towards anticancer therapeutics. PLoS ONE 2025, 20, e0311954. [Google Scholar] [CrossRef]
- Yang, H.; Rudge, D.G.; Koos, J.D.; Vaidialingam, B.; Yang, H.J.; Pavletich, N.P. mTOR kinase structure, mechanism and regulation. Nature 2013, 497, 217–223. [Google Scholar] [CrossRef]
- Marafie, S.K.; Al-Mulla, F.; Abubaker, J. mTOR: Its critical role in metabolic diseases, cancer, and the aging process. Int. J. Mol. Sci. 2024, 25, 6141. [Google Scholar] [CrossRef]
- Yu, J.; Li, Y.; Ishizuka, T.; Guenther, M.G.; Lazar, M.A. A SANT motif in the SMRT corepressor interprets the histone code and promotes histone deacetylation. EMBO J. 2003, 22, 3403–3410. [Google Scholar] [CrossRef]
- Zhang, Q.; Xiao, H.; Chai, S.C.; Hoang, Q.Q.; Lu, H. Hydrophilic residues are crucial for ribosomal protein L11 (RPL11) interaction with zinc finger domain of MDM2 and p53 protein activation. J. Biol. Chem. 2011, 286, 38264–38274. [Google Scholar] [CrossRef]
- Rebbeck, T.R. Prostate cancer. Challenges for prevention, detection and treatment. In World Cancer Report; Wild, C.P., Weiderpass, E., Stewart, B.W., Eds.; International Agency for Research in Cancer; World Health Organization: Lyon, France, 2020; pp. 421–429. [Google Scholar]
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.; Gastier-Foster, J.; Grody, W.W.; Hedge, M.; Lyon, E.; Spector, E.; et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015, 17, 405–424. [Google Scholar] [CrossRef]
- Landrum, M.J.; Lee, J.M.; Benson, M.; Brown, G.R.; Chao, C.; Chitipiralla, S.; Gu, B.; Hart, J.; Hoffman, D.; Jang, W.; et al. ClinVar: Improving access to variant interpretations and supporting evidence. Nucleic Acid Res. 2018, 46, 1062–1067. [Google Scholar] [CrossRef] [PubMed]
- Bustamante, C.D.; Burchard, E.G.; De la Vega, F.M. Genomics for the world. Nature 2011, 475, 163–165. [Google Scholar] [CrossRef] [PubMed]
- Goldstein, D.B.; Cavalleri, G.L. Understanding human diversity. Nature 2005, 437, 1241–1242. [Google Scholar] [CrossRef]
- Sirugo, G.; Williams, S.M.; Tishkoff, S.A. The missing diversity in human genetic studies. Cell 2019, 177, 26–31. [Google Scholar] [CrossRef]
- Park, S.L.; Cheng, I.; Haiman, C.A. Genome-wide association studies of cancer in diverse populations. Cancer Epidemiol. Biomark. Prev. 2018, 27, 405–417. [Google Scholar] [CrossRef]
- Paliouras, M.; Zaman, N.; Lumbroso, R.; Kapogeorgakis, L.; Beitel, L.K.; Wang, E.; Trifiro, M. Dynamic rewiring of the androgen receptor protein interaction network correlates with prostate cancer clinical outcomes. Integr. Biol. 2011, 3, 1020–1032. [Google Scholar] [CrossRef]
- Gaddipati, J.P.; McLeod, D.G.; Heidenberg, H.B.; Sesterhenn, I.A.; Finger, M.J.; Moul, J.W.; Srivastava, S. Frequent detection of codon 877 mutation in the androgen receptor gene in advanced prostate cancers. Cancer Res. 1994, 54, 2861–2864. [Google Scholar]
- Thompson, J.; Hyytinen, E.R.; Haapala, K.; Rantala, I.; Helin, H.J.; Jänne, O.A.; Palvimo, J.J.; Koivisto, P.A. Androgen receptor mutations in high-grade prostate cancer before hormonal therapy. Lab. Investig. 2003, 83, 1709–1713. [Google Scholar] [CrossRef][Green Version]
- Tilley, W.D.; Buchanan, G.; Hickey, T.E.; Bentel, J.M. Mutations in the androgen receptor gene are associated with progression of human prostate cancer to androgen independence. Clin. Cancer Res. 1996, 2, 277–285. [Google Scholar][Green Version]
- Yeh, S.H.; Chiu, C.M.; Chen, C.L.; Lu, S.F.; Hsu, H.C.; Che, D.S.; Chen, P.J. Somatic mutations at the trinucleotide repeats of androgen receptor gene in male epatocellular carcinoma. Int. J. Cancer 2007, 120, 1610–1617. [Google Scholar] [CrossRef]
- Pugh, T.J.; Weeraratne, S.D.; Archer, T.C.; Kummel, D.A.P.; Auclair, D.; Bochicchio, J.; Carneiro, M.O.; Carter, S.L.; Cibulskis, K.; Erlich, R.L.; et al. Medulloblastoma exome sequencing uncovers subtype-specific somatic mutations. Nature 2012, 488, 106–110. [Google Scholar] [CrossRef] [PubMed]
- Lefebvre, C.; Bachelot, T.; Filleron, T.; Pedrero, M.; Campone, M.; Soria, J.C.; Massard, C.; Lévy, C.; Arnedos, M.; Lacroix-Triki, M.; et al. Mutational profile of metastatic breast cancers: A retrospective analysis. PLoS Med. 2016, 13, e1002201. [Google Scholar] [CrossRef] [PubMed]
- Giannakis, M.; Mu, X.J.; Shukla, S.A.; Qian, Z.R.; Cohen, O.; Nishihara, R.; Bahl, S.; Cao, Y.; Amin-Mansour, A.; Yamauchi, M.; et al. Genomic correlates of immune-cell infiltrates in colorectal carcinoma. Cell Rep. 2016, 15, 857–865. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-González, G.; Cabrera, S.; Ramírez-Moreno, R.; Bilbao, C.; Díaz-Chico, J.C.; Serra, L.; Chesa, N.; Cabrera, J.J.; Díaz-Chico, B.N. Short alleles of both GGN and CAG repeats at the exon-1 of the androgen receptor gene are associated to increased PSA staining and a higher Gleason score in human prostatic cancer. J. Steroid Biochem. Mol. Biol. 2009, 113, 85–91. [Google Scholar] [CrossRef]
- Sahu, B.; Laakso, M.; Ovaska, K.; Mirtti, T.; Lundin, J.; Rannikko, A.; Sankila, A.; Turunen, J.P.; Lundin, M.; Konsti, J.; et al. Dual role of FoxA1 in androgen receptor binding to chromatin, androgen signalling and prostate cancer. EMBO J. 2011, 30, 3962–3976. [Google Scholar] [CrossRef]
- Bernardo, G.M.; Keri, R.A. FOXA1: A transcription factor with parallel functions in development and cancer. Biosci. Rep. 2012, 32, 113–130. [Google Scholar] [CrossRef]
- Gerhardt, J.; Montani, M.; Wild, P.; Beer, M.; Huber, F.; Hermanns, T.; Müntener, M.; Kristiansen, G. FOXA1 promotes tumor progression in prostate cancer and represents a novel hallmark of castration-resistant prostate cancer. Am. J. Pathol. 2012, 180, 848–861. [Google Scholar] [CrossRef]
- Jin, H.J.; Zhao, J.C.; Ogden, I.; Bergan, R.C.; Yu, J. Androgen receptor-independent function of FoxA1 in prostate cancer metastasis. Cancer Res. 2013, 73, 3725–3736. [Google Scholar] [CrossRef]
- Barbieri, C.E.; Baca, S.C.; Lawrence, M.S.; Demichelis, F.; Blattner, M.; Theurillat, J.P.; White, T.A.; Stojanov, P.; Allen, E.V.; Stransky, N.; et al. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat. Genet. 2012, 44, 685–689. [Google Scholar] [CrossRef]
- Cancer Genome Atlas Research Network. The molecular taxonomy of primary prostate cancer. Cell 2015, 163, 1011–1025. [Google Scholar] [CrossRef]
- Espiritu, S.M.G.; Liu, L.Y.; Rubanova, Y.; Bhandari, V.; Espiritu, S.M.G.; Holgersen, E.M.; Szyca, L.M.; Espiritu, S.M.G.; Fox, N.S.; Chua, M.L.; et al. The evolutionary landscape of localized prostate cancers drives clinical aggression. Cell 2018, 173, 1003–1013.e15. [Google Scholar] [CrossRef] [PubMed]
- Huang, F.W.; Mosquera, J.M.; Garofalo, A.; Oh, C.; Baco, M.; Amin-Mansour, A.; Rabasha, B.; Bahl, S.; Mullane, S.A.; Robinson, B.D.; et al. Exome sequencing of African-American prostate cancer reveals loss-of-function ERF mutations. Cancer Discov. 2017, 7, 973–983. [Google Scholar] [CrossRef] [PubMed]
- Hernández-Llodrà, S.; Segalés, L.; Safont, A.; Juanpere, N.; Lorenzo, M.; Fumadó, L.; Rodríguez-Vida, A.; Cecchini, L.; Bellmunt, J.; Lloreta-Trull, J. SPOP and FOXA1 mutations are associated with PSA recurrence in ERG wt tumors, and SPOP downregulation with ERG-rearranged. Prostate 2019, 79, 1156–1165. [Google Scholar] [CrossRef] [PubMed]
- Xu, B.; Song, B.; Lu, X.; Kim, J.; Hu, M.; Zhao, J.C.; Yu, J. Altered chromatin recruitment by FOXA1 mutations promotes androgen independence and prostate cancer progression. Cell Res. 2019, 29, 773–775. [Google Scholar] [CrossRef]
- Shorning, B.Y.; Dass, M.S.; Smalley, M.J.; Pearson, H.B. The PI3K-AKT-mTOR pathway and prostate cancer: At the crossroads of AR, MAPK, and WNT signaling. Int. J. Mol. Sci. 2020, 21, 4507. [Google Scholar] [CrossRef]
- Proud, C.G. Phosphorylation and signal transduction pathways in translational control. Cold Spring Harb. Perspect. Biol. 2019, 11, a033050. [Google Scholar] [CrossRef]
- Makwana, V.; Rudrawar, S.; Anoopkumar-Dukie, S. Signalling transduction of O-GlcNAcylation and PI3K/AKT/mTOR-axis in prostate cancer. Biochim. Biophys. Acta Mol. Basis Dis. 2021, 1867, 166129. [Google Scholar] [CrossRef]
- Dufor, C.R.; Scholtes, C.; Yan, M.; Chen, Y.; Han, L.; Li, T.; Xia, H.; Deng, Q.; Vernier, M.; Giguere, V. The mTOR chromatin-bound interactome in prostate cancer. Cell Rep. 2022, 38, 110534. [Google Scholar] [CrossRef]
- Chen, Y.; Han, L.; Dufor, C.R.; Alfonso, A.; Giguere, V. Canonical and nuclear mTOR specify distinct transcriptional programs in androgen-dependent prostate cancer cells. Mol. Cancer Res. 2024, 22, 113–124. [Google Scholar] [CrossRef]
- Audet-Walsh, E.; Dufor, C.R.; Yee, T.; Zouanat, F.Z.; Yang, M.; Kalloghlian, G.; Vernier, M.; Caron, M.; Bourque, G.; Scarlata, E.; et al. Nuclear mTOR acts as a transcriptional integrator of the androgen signaling pathway in prostate cancer. Genes Dev. 2017, 31, 1228–1242. [Google Scholar] [CrossRef]
- Giguere, V. DNA-PK, nuclear mTOR, and the androgen pathway in prostate cancer. Trends Cancer 2020, 6, 337–347. [Google Scholar] [CrossRef] [PubMed]
- Tan, B.X.; Liew, H.P.; Chua, J.S.; Ghadessy, F.J.; Tan, Y.S.; Lane, D.P.; Coffill, C.R. Anatomy of Mdm2 and Mdm4 in evolution. J. Mol. Cell Biol. 2017, 9, 3–15. [Google Scholar] [CrossRef]
- Brown, D.R.; Thomas, C.A.; Deb, S.P. The human oncoprotein MDM2 arrests the cell cycle: Elimination of its cell-cycle-inhibitory function induces tumorigenesis. EMBO J. 1998, 17, 2513–2525. [Google Scholar] [CrossRef] [PubMed]
- Manfredi, J.J. The Mdm2-p53 relationship evolves: Mdm2 swings both ways as an oncogene and a tumor suppressor. Genes Dev. 2010, 24, 1580–1589. [Google Scholar] [CrossRef] [PubMed]
- Alt, J.R.; Bouska, A.; Fernandez, M.R.; Cerny, L.; Xiao, H.; Eischen, C.M. Mdm2 binds to Nbs1 at sites of DNA damage and regulates double strand break repair. J. Biol. Chem. 2005, 280, 18771–18781. [Google Scholar] [CrossRef]
- Gu, L.; Zhang, H.; He, J.; Li, J.; Huang, M.; Zhou, M. MDM2 regulates MYCN mRNA stabilization and translation in human neuroblastoma cells. Oncogene 2012, 31, 1342–1353. [Google Scholar] [CrossRef]
- Jung, C.-H.; Kim, J.; Park, J.K.; Hwang, S.-G.; Moon, S.-K.; Kim, W.-J.; Um, H.-D. Mdm2 increases cellular invasiveness by binding to and stabilizing the Slug mRNA. Cancer Lett. 2013, 335, 270–277. [Google Scholar] [CrossRef]
- Wang, W.; Albadari, N.; Du, Y.; Fowler, J.F.; Sang, H.T.; Xian, W.; McKeon, F.; Li, W.; Zhou, J.; Zhang, R. MDM2 Inhibitors for cancer therapy: The past, present, and future. Pharmacol. Rev. 2024, 76, 414–453. [Google Scholar] [CrossRef]
- Xue, L.; Han, X.; Liu, R.; Wang, Z.; Li, H.; Chen, Q.; Zhang, P.; Wang, Z.; Chong, T. MDM2 and P53 polymorphisms contribute together to the risk and survival of prostate cancer. Oncotarget 2016, 7, 31825–31831. [Google Scholar] [CrossRef][Green Version]
- Khatiwada, P.; Rimal, U.; Han, Z.; Shemshedini, L. MDM2 regulates the stability of AR, AR-V7, and TM4SF3 proteins in prostate cancer. Endocr. Oncol. 2024, 4, e230017. [Google Scholar] [CrossRef]
- Vummidi Giridhar, P.; Williams, K.; VonHandorf, A.P.; Deford, P.L.; Kasper, S. Constant degradation of the androgen receptor by MDM2 conserves prostate cancer stem cell integrity. Cancer Res. 2019, 79, 1124–1137. [Google Scholar] [CrossRef] [PubMed]
- Hernández, G.; Vazquez-Pianzola, P. eIF4E as a molecular wildcard in metazoans RNA metabolism. Biol. Rev. Cambr. Phil. Soc. 2023, 98, 2284–2306. [Google Scholar] [CrossRef] [PubMed]
- Siddiqui, N.; Sonenberg, N. Signalling to eIF4E in cancer. Biochem. Soc. Trans. 2015, 43, 763–772. [Google Scholar] [CrossRef] [PubMed]
- Kremer, C.L.; Klein, R.R.; Mendelson, J.; Browne, W.; Samadzedeh, L.K.; Vanpatten, K.; Highstrom, L.; Pestano, G.A.; Nagle, R.B. Expression of mTOR signaling pathway markers in prostate cancer progression. Prostate 2006, 66, 1203–1212. [Google Scholar] [CrossRef]
- Mamane, Y.; Petroulakis, E.; Martineau, Y.; Sato, T.A.; Larsson, O.; Rajasekhar, V.; Sonenberg, N. Epigenetic activation of a subset of mRNAs by eIF4E explains its effects on cell proliferation. PLoS ONE 2007, 2, e242. [Google Scholar] [CrossRef]
- Yanagiya, A.; Suyama, E.; Adachi, H.; Svitkin, Y.V.; Aza-Blanc, P.; Imataka, H.; Mikami, S.; Martineau, Y.; Ronai, Z.A.; Sonenberg, N. Translational homeostasis via the mRNA Cap-binding protein, eIF4E. Mol. Cell 2012, 46, 847–858. [Google Scholar] [CrossRef]
- Truitt, M.L.; Conn, C.S.; Shi, Z.; Pang, X.; Tokuyasu, T.; Coady, A.M.; Seo, Y.; Barna, M.; Ruggero, D. Differential requirements for eIF4E dose in normal development and cancer. Cell 2015, 162, 59–71. [Google Scholar] [CrossRef]
- Trepel, J.; Mollapour, M.; Giaccone, G.; Neckers, L. Targeting the dynamic HSP90 complex in cancer. Nat. Rev. Cancer 2010, 10, 537–549. [Google Scholar] [CrossRef]
- Bohonowych, J.E.; Hance, M.W.; Nolan, K.D.; Defee, M.; Parsons, C.H.; Isaacs, J.S. Extracellular Hsp90 mediates an NF-kappaB dependent inflammatory stromal program: Implications for the prostate tumor microenvironment. Prostate 2014, 74, 395–407. [Google Scholar] [CrossRef]
- Lavoie, H.; Gagnon, J.; Therrien, M. ERK signalling: A master regulator of cell behaviour, life and fate. Nat. Rev. Mol. Cell Biol. 2020, 21, 607–632. [Google Scholar] [CrossRef]
- Guo, Y.-J.; OPan, W.W.; Liu, S.B.; Shen, Z.F.; Xu, Y.; Hu, L.L. ERK/MAPK signalling pathway and tumorigenesis. Exp. Ther. Med. 2020, 19, 1997–2007. [Google Scholar] [CrossRef] [PubMed]
- Roskoski, R.J. Targeting ERK1/2 protein-serine/threonine kinases in human cancers. Pharmacol. Res. 2019, 142, 151–168, Erratum in Pharmacol. Res. 2019, 143, 206. [Google Scholar] [CrossRef] [PubMed]
- Hernández, G.; Ramírez, J.L.; Pedroza-Torres, A.; Herrera, L.A.; Jiménez-Ríos, M.A. The secret life of translation initiation in prostate cancer. Front. Genet. 2019, 10, 14. [Google Scholar] [CrossRef] [PubMed]
- Paluvai, H.; Shanmukha, K.D.; Tyedmers, J.; Backs, J. Insights into the function of HDAC3 and NCoR1/NCoR2 co-repressor complex in metabolic diseases. Front. Mol. Biosci. 2023, 10, 1190094. [Google Scholar] [CrossRef]
- TI, L.; Valentim, R.R.; Araújo, H.N.; Oliveira, A.G.; Favero, B.C.; Menezes, E.S.; Araújo, R.; Silveira, L.R. Role of NCoR1 in mitochondrial function and energy metabolism. Cell Biol. Int. 2018, 42, 734–741. [Google Scholar]
- St-Jean, S.; De Castro, A.C.; Lecours, M.; Jones, C.; Rivard, N.; Rodier, F.; Perreault, N.; Boudreau, F. NCOR1 sustains colorectal cancer cell growth and protects against cellular senescence. Cancers 2021, 13, 4414. [Google Scholar] [CrossRef]
- Lopez, S.M.; Agoulnik, A.I.; Zhang, M.; Peterson, L.E.; Suarez, E.; Gandarillas, G.A.; Frolov, A.; Li, R.; Rajapakshe, K.; Coarfa, C.; et al. Nuclear receptor corepressor 1 expression and output declines with prostate cancer progression. Clin. Cancer Res. 2016, 22, 3937–3949. [Google Scholar] [CrossRef]
- Aylon, Y.; Furth, N.; Mallel, G.; Friedlander, G.; Nataraj, N.B.; Dong, M.; Hassin, O.; Zoabi, R.; Cohen, B.; Drendel, V.; et al. Breast cancer plasticity is restricted by a LATS1-NCOR1 repressive axis. Nat. Commun. 2022, 13, 7199, Correction in Nat. Comm. 2023, 14, 133. [Google Scholar] [CrossRef]
- Bertucci, F.; Ng, C.K.Y.; Patsouris, A.; Droiin, N.; Piscouglio, S.; Carbuccia, N.; Soria, J.C.; Dien, A.T.; Adnani, Y.; Kamal, M.; et al. Genomic characterization of metastatic breast cancers. Nature 2019, 569, 560–564, Correction in Nature 2019, 572, E7. [Google Scholar] [CrossRef]
- Shu, S.; Li, Z.; Liu, L.; Ying, X.; Zhang, Y.; Wang, T.; Zhou, X.; Jiang, P.; Lv, W. HPV16 E6-activated OCT4 promotes cervical cancer progression by suppressing p53 expression via co-repressor NCOR1. Front. Oncol. 2022, 12, 900856. [Google Scholar] [CrossRef]
- Lin, A.; Qiu, Z.; Zhang, J.; Luo, P. Effect of NCOR1 mutations on immune microenvironment and efficacy of immune checkpoint inhibitors in patient with bladder cancer. Front. Immunol. 2021, 12, 630773. [Google Scholar] [CrossRef] [PubMed]
- Tang, L.; Zhang, L.; Liu, L.; Dong, L.; Dong, Y.; Zhu, W.; Wang, H. NCOR1 may be a potential biomarker of a novel molecular subtype of prostate cancer. FEBS Open Bio 2020, 10, 2678–2686. [Google Scholar] [CrossRef] [PubMed]
- Tate, J.G.; Bamford, S.; Jubb, H.C.; Sondka, Z.; Beare, D.M.; Bindal, N.; Boutselakis, H.; Cole, C.G.; Creatore, C.; Dawson, E.; et al. COSMIC: The catalogue of somatic mutations in cancer. Nucleic Acids Res. 2019, 47, D941–D947. [Google Scholar] [CrossRef] [PubMed]





| Characteristic | Tumor Samples n = 64 (66.6%) | Non-Tumor Samples n = 32 (33.4%) |
|---|---|---|
| Age | ≤50: 4 (6.25%) | ≤50: 2 (6.25%) |
| >50: 60 (93.75%) | >50: 30 (93.75%) | |
| Median = 68-years-old | Median = 6-years-old | |
| PSA at diagnosis | ≤10 ng/mL: 11 (17.18) | ≤10 ng/mL: 18 (56.25%) |
| 10.1–20 ng/mL: 11 (17.18%) | 10.1–20 ng/mL: 11 (34.37%) | |
| >20 ng/mL: 42 (65.6%) | >20 ng/mL: 3 (9.37%) | |
| Median = 305 ng/mL | Median = 10.5 ng/mL | |
| Body mass index (BMI) | <18.5: 2 (3.12%) | <18.5: 5 (15.62%) |
| 18.5–24.9: 10 (15.62%) | 18.5–24.9: 10 (31.25%) | |
| 25–29.9: 26 (40.65%) | 25–29.9: 6 (18.75%) | |
| ≥30: 11 (17.18%) | ≥30: 6 (18.75%) | |
| Without data: 15 (23.43%) | Without data: 5 (15.62%) | |
| Median = 27 | Median = 25.9 | |
| Addictions | Smoking: 4 (6.25%) Alcohol: 11 (17.18%) Smoking and alcohol: 22 (34.37%) Without data: 27 (42.18%) | Without data |
| Gleason score | <7: 6 (9.37%) =7: 25 (39.06%) >7: 31 (50%) Without data: 2 (3.12%) | NA |
| Perineural invasion | Yes: 41 (64.06%) No: 15 (23.43%) Without data: 2 (3.12%) | NA |
| Metastasis | Yes: 19 (29.68%) No: 31 (48.43%) Without data: 14 (21.87%) | NA |
| Nationality | 100% Mexicans with all four Mexican grandparents | 100% Mexicans with all four Mexican grandparents |
| Gen | Mutation Type | Change in DNA | Change in Protein | Status a | Functional Prediction b |
|---|---|---|---|---|---|
| AR | Frameshift Insertion | c.230_231insA | p.Gln78fs | Novel | Probably pathogenic |
| AR | Frameshift Insertion | c.203_204insA | p.Gln69fs | Novel | Probably pathogenic |
| AR | Frameshift Insertion | c.206_207insA | p.Gln70fs | Novel | Probably pathogenic |
| AR | Missense | c.61G>A | p.Gly21Arg | Novel | Pathogenic |
| AR | Frameshift Insertion | c.227_228insA | p.Gln77fs | Novel | Probably pathogenic |
| AR | Missense | c.206A>G | p.Gln69Arg | Novel | Probably pathogenic |
| AR | Frameshift Insertion | c.1981_1982insAAAA | p.Thr661fs | Novel | Probably pathogenic |
| AR | Missense | c.1115C>T | p.Ala372Val | Previously reported | Probably pathogenic |
| AR | Missense | c.170T>A | p.Leu57Gln | Previously reported | Probably benign |
| AR | Missense | c.92G>A | p.Arg31His | Previously reported | Pathogenic |
| AR | Missense | c.215A>T | p.Gln72Leu | Previously reported | Probably benign |
| EIF4E | Frameshift Deletion | c.144_145delTA | p.Phe48fs | Novel | Probably pathogenic |
| FOXA1 | Missense | c.655C>A | p.Arg219Ser | Previously reported | Pathogenic |
| FOXA1 | Missense | c.1267G>A | p.Ala423Thr | Novel | Pathogenic |
| FOXA1 | Non-frameshift Deletion | c.752_763delGCAACATGTTCG | p.Gly251_Phe254del | Novel | Probably pathogenic |
| FOXA1 | Non-frameshift Deletion | c.749_760delCCGGCAACATGT | p.Ser250_Met253del | Novel | Probably pathogenic |
| HSP90AA1 | Missense | c.836A>T | p.Lys279Met | Novel | Pathogenic |
| HSP90AA1 | Missense | c.581A>G | p.Gln194Arg | Previously reported | Pathogenic |
| MAPK1 | Missense | c.941A>G | p.Glu314Gly | Novel | Pathogenic |
| MDM2 | Missense | c.845A>G | p.Tyr282Cys | Novel | Pathogenic |
| MDM2 | Missense | c.848A>G | p.Gln283Arg | Novel | Probably benign |
| MTOR | Missense | c.5710C>T | p.Leu1904Phe | Novel | Pathogenic |
| MTOR | Missense | c.4621C>T | p.His1541Tyr | Novel | Pathogenic |
| MTOR | Missense | c.5705A>T | p.Asp1902Val | Novel | Pathogenic |
| NCOR1 | Missense | c.5066C>T | p.Pro1689Leu | Novel | Pathogenic |
| NCOR1 | Non-frameshift Deletion | c.5989_5991delGTT | p.Val1997del | Previously reported | Probably pathogenic |
| NCOR1 | Missense | c.4805A>G | p.Gln1602Arg | Previously reported | Probably pathogenic |
| SPOP | Missense | c.397T>G | p.Phe133Val | Previously reported | Probably pathogenic |
| SPOP | Missense | c.469C>T | p.Leu157Phe | Previously reported | VUS |
| SPOP | Missense | c.398T>C | p.Phe133Ser | Previously reported | Probably pathogenic |
| SPOP | Missense | c.398T>G | p.Phe133Cys | Previously reported | Probably pathogenic |
| SPOP | Missense | c.305T>G | p.Phe102Cys | Previously reported | Probably pathogenic |
| SPOP | Missense | c.304T>G | p.Phe102Val | Previously reported | Probably pathogenic |
| TP53 | Missense | c.524G>A | p.Arg175His | Previously reported | Pathogenic |
| TP53 | Missense | c.578A>C | p.His193Pro | Previously reported | Pathogenic |
| TP53 | Missense | c.1096T>G | p.Ser366Ala | Previously reported | VUS |
| TP53 | Nonsense | c.916C>T | p.Arg306Ter | Previously reported | Pathogenic |
| TP53 | Missense | c.808T>A | p.Phe270Ile | Previously reported | Pathogenic |
| Gene Name | Chromosome | Chr. Start | Chr. End | Num. Amplicons | Total Bases | Covered Bases | Missed Bases | Coverage (%) |
|---|---|---|---|---|---|---|---|---|
| MTOR | 1 | 11107479 | 11259414 | 70 | 8220 | 8183 | 37 | 99.5 |
| CYP17A1 | 10 | 102830696 | 102837366 | 14 | 1607 | 1607 | 0 | 100 |
| MDM2 | 12 | 68808472 | 68839854 | 17 | 1604 | 1604 | 0 | 100 |
| FOXO1 | 13 | 40559517 | 40666217 | 10 | 1988 | 1500 | 488 | 75.5 |
| HSP90AA1 | 14 | 102081706 | 102139409 | 20 | 2685 | 2685 | 0 | 100 |
| FOXA1 | 14 | 37591359 | 37594977 | 9 | 1439 | 1296 | 143 | 90.1 |
| TP53 | 17 | 7669603 | 7676599 | 14 | 1383 | 1307 | 76 | 94.5 |
| SPOP | 17 | 49600372 | 49622815 | 9 | 1215 | 1215 | 0 | 100 |
| NCOR1 | 17 | 16032290 | 16194574 | 73 | 7829 | 7829 | 0 | 100 |
| SRD5A2 | 2 | 31526190 | 31580905 | 8 | 815 | 804 | 11 | 98.7 |
| TMPRSS2 | 21 | 41466136 | 41508009 | 16 | 1730 | 1693 | 37 | 97.9 |
| MAPK1 | 22 | 21769198 | 21867445 | 11 | 1163 | 1160 | 3 | 99.7 |
| EIF4E | 4 | 98881022 | 98929117 | 10 | 915 | 915 | 0 | 100 |
| NKX3-1 | 8 | 23681215 | 23682894 | 5 | 725 | 700 | 25 | 96.6 |
| AR | X | 67545141 | 67723846 | 19 | 2873 | 2873 | 0 | 100 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pedroza-Torres, A.; Baranda-Avila, N.; Ramírez, J.L.; González, M.; González, P.A.; Torres, B.L.; Jiménez-Ríos, M.A.; Méndez-Tenorio, A.; Álvarez-Gómez, R.M.; Hernández, G. Deep Sequencing Reveals Novel Mutations in Androgen Receptor-Related Genes in Prostate Cancer. Int. J. Mol. Sci. 2025, 26, 8758. https://doi.org/10.3390/ijms26188758
Pedroza-Torres A, Baranda-Avila N, Ramírez JL, González M, González PA, Torres BL, Jiménez-Ríos MA, Méndez-Tenorio A, Álvarez-Gómez RM, Hernández G. Deep Sequencing Reveals Novel Mutations in Androgen Receptor-Related Genes in Prostate Cancer. International Journal of Molecular Sciences. 2025; 26(18):8758. https://doi.org/10.3390/ijms26188758
Chicago/Turabian StylePedroza-Torres, Abraham, Noemí Baranda-Avila, Jorge L. Ramírez, Maricruz González, Pamela A. González, Blanca L. Torres, Miguel A. Jiménez-Ríos, Alfonso Méndez-Tenorio, Rosa María Álvarez-Gómez, and Greco Hernández. 2025. "Deep Sequencing Reveals Novel Mutations in Androgen Receptor-Related Genes in Prostate Cancer" International Journal of Molecular Sciences 26, no. 18: 8758. https://doi.org/10.3390/ijms26188758
APA StylePedroza-Torres, A., Baranda-Avila, N., Ramírez, J. L., González, M., González, P. A., Torres, B. L., Jiménez-Ríos, M. A., Méndez-Tenorio, A., Álvarez-Gómez, R. M., & Hernández, G. (2025). Deep Sequencing Reveals Novel Mutations in Androgen Receptor-Related Genes in Prostate Cancer. International Journal of Molecular Sciences, 26(18), 8758. https://doi.org/10.3390/ijms26188758







