Long Noncoding RNAs as Emerging Regulators of Seed Development, Germination, and Senescence
Abstract
1. Introduction
2. Long Noncoding RNAs
3. LncRNA Biogenesis
4. LncRNA Classification
4.1. Natural Antisense Transcripts (NATs)
4.2. Sense Long Noncoding RNAs
4.3. Bidirectional Long Noncoding RNAs
4.4. Intergenic Long Noncoding RNAs (lincRNAs)
4.5. Intronic lncRNAs (incRNAs)
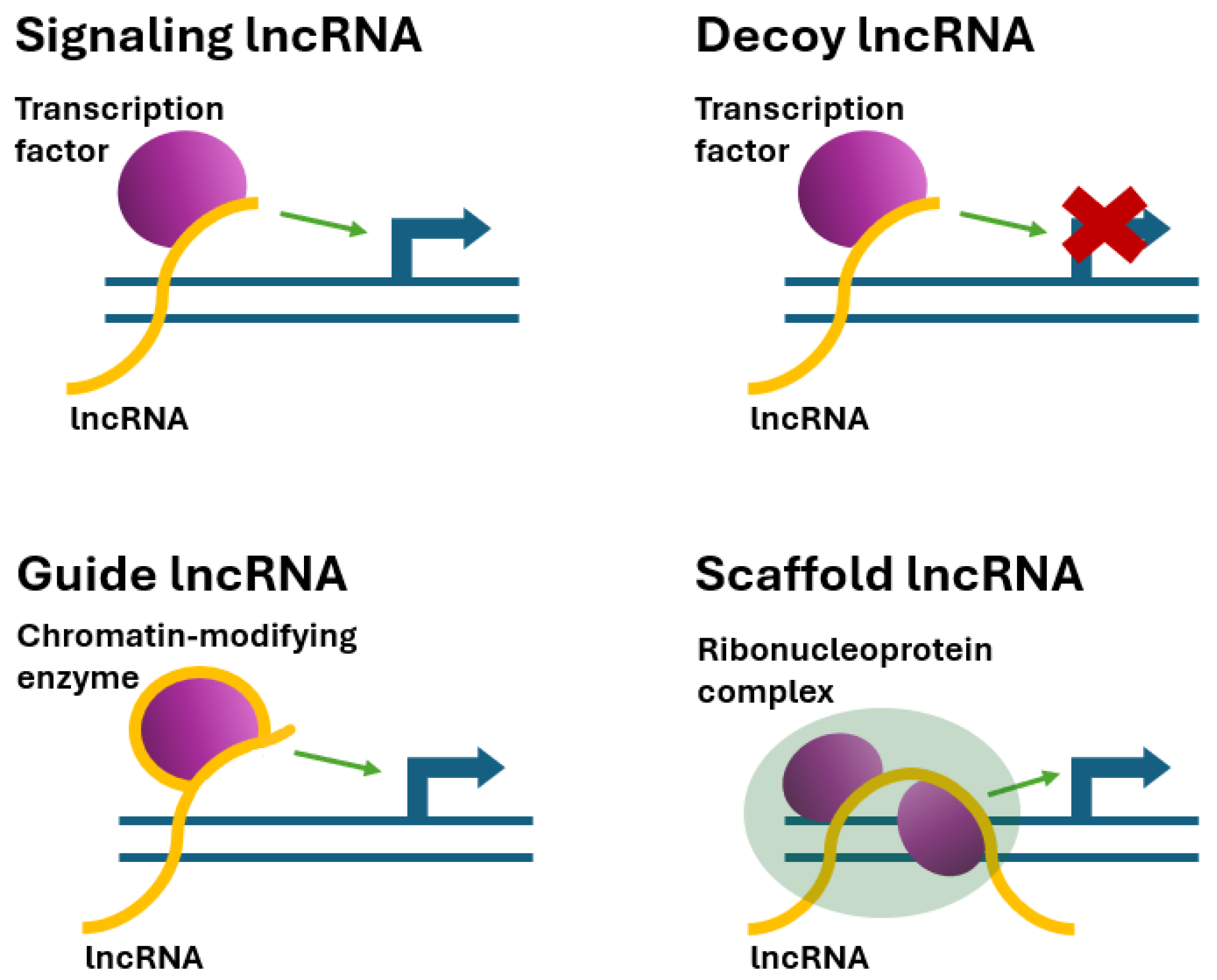
5. Mechanism of Action of Long Noncoding RNAs
5.1. Signaling lncRNAs
5.2. Decoy (Sponge) lncRNAs
5.3. Guide lncRNAs
5.4. Scaffold lncRNAs
6. LncRNA in Seeds
6.1. Role of lncRNAs in Seed Development
6.1.1. Imprinted LncRNA Sculpt Endosperm Morphogenesis
6.1.2. Cell Type-Specific lncRNA Networks Drive Endosperm Differentiation
6.1.3. Testa lncRNAs Link Differentiation to Protective Metabolism
6.1.4. lncRNA Regulators Fine-Tune Lipid Deposition During Maturation
6.2. Role of lncRNAs in Seed Dormancy
6.2.1. Cis-Antisense lncRNAs Fine-Tune Master Dormancy Genes
6.2.2. Dormancy-Enforcing lncRNAs Integrate ABA Signaling with Chromatin Repression
6.2.3. Trans-Acting lncRNAs Rebalance ABA/GA Metabolism to Modulate Dormancy Release
6.2.4. LncRNAs Act as Competitive Endogenous RNAs (ceRNAs) Within the ABA Core Circuitry
6.2.5. Epitranscriptomic Marks Stabilize Dormancy-Associated lncRNAs
6.3. Role of lncRNAs in Seed Germination
6.3.1. Light-Responsive lncRNA Gating of the phyB–ABA/GA Module in Seed Germination
6.3.2. Auxin-Linked lncRNA Control of Seed Vigor
6.3.3. Thermoresponsive lncRNA Networks Recapitulate Dormancy Under Heat
6.3.4. Single-Cell Atlases Reveal Cell Type-Specific lncRNA Bursts
6.3.5. Stress-Adaptive lncRNAs Modulate Reactive-Oxygen Homeostasis
6.4. Role of lncRNAs in Seed Senescence
6.4.1. Global Attrition of the lncRNA Transcriptome During Aging
6.4.2. A Small Cohort of Longevity-Associated lncRNAs Resists Decay
6.4.3. Alternative Splicing Intensifies Under Aging Stress
6.4.4. LncRNA-Centered ceRNA Networks Balance Antioxidant Defense and PCD
6.4.5. Domestication Reshaped lncRNA Loci Governing Storability
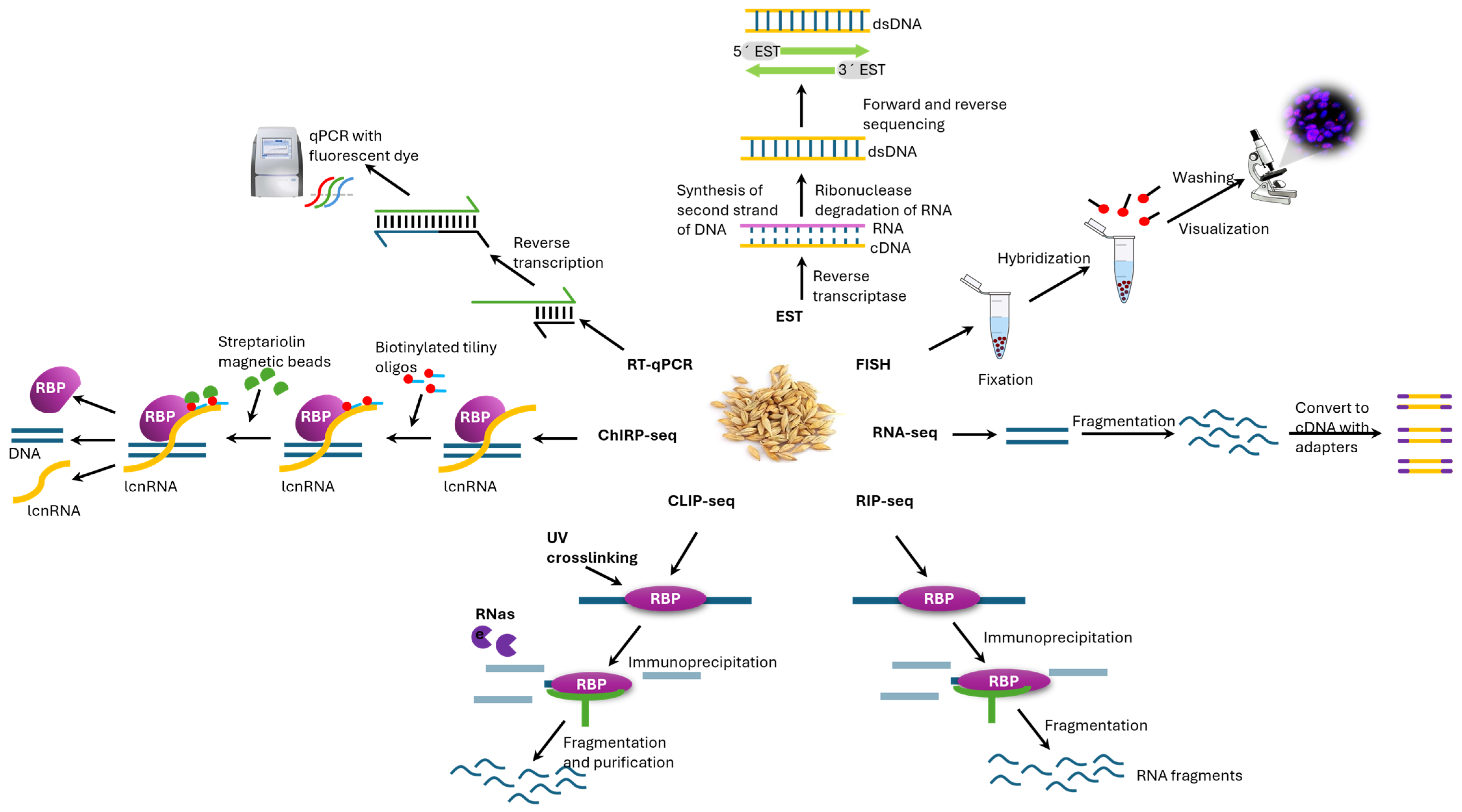
7. Methods for Identifying Long Noncoding RNAs
7.1. Hybridization and Sequence-Tag Approaches
7.2. Short-Read RNA-Seq
7.3. Long-Read and Direct RNA Sequencing
7.4. Expression and Localization Validation
7.5. RNA–Protein Interactome Mapping
7.6. RNA–Chromatin Target Mapping
7.7. Key Caveats and Best Practices
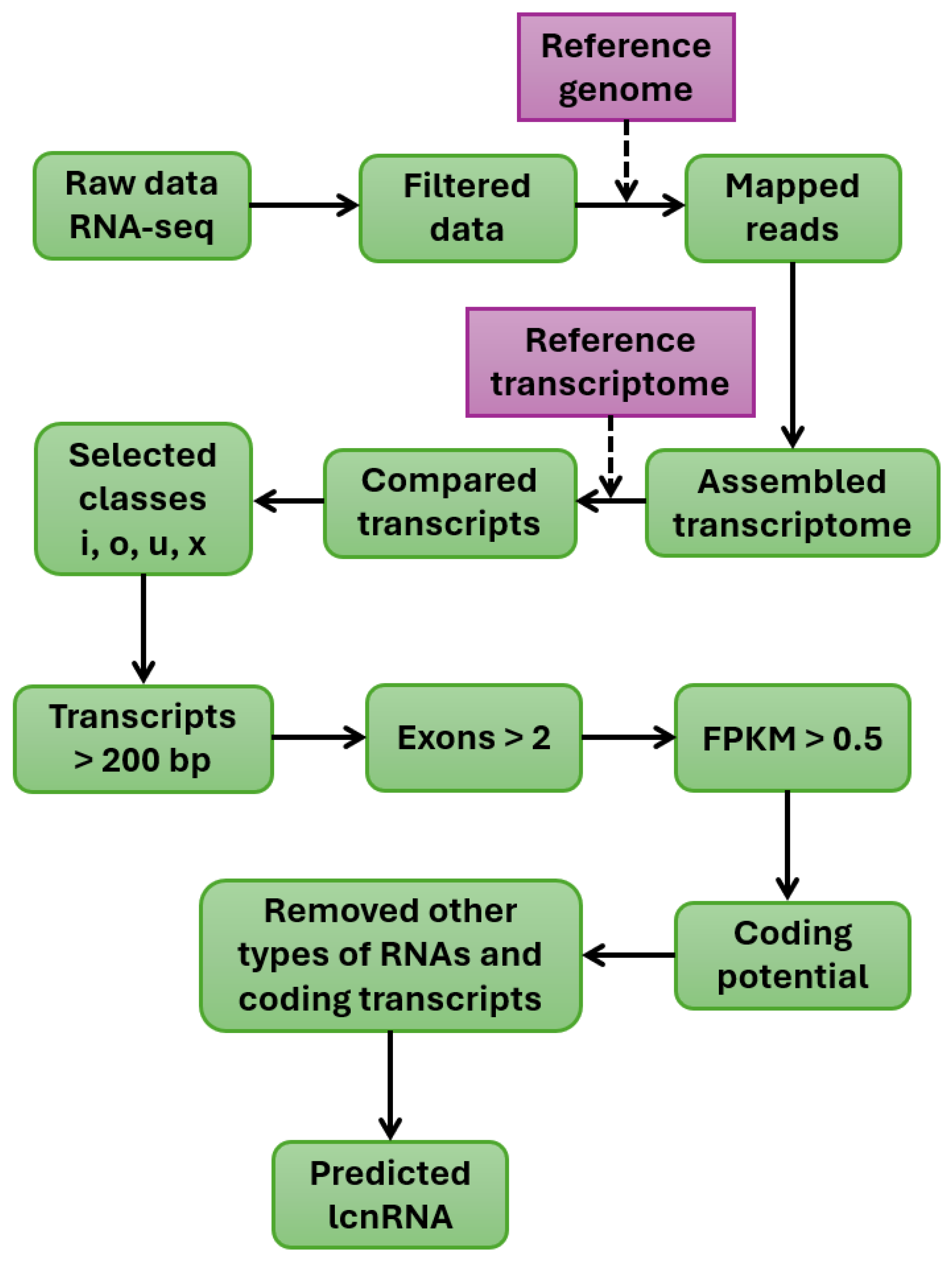
8. Bioinformatics Analysis of Long Noncoding RNAs
9. Prospects
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Li, X.; Liu, Q.; Liu, J. Long Non-Coding RNAs: Discoveries, Mechanisms, and Research Strategies in Seeds. Genes 2023, 14, 2214. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Fan, Y.; Fan, D.; Zhou, X.; Jiao, Y.; Deng, X.W.; Zhu, D. The noncoding RNA HIDDEN TREASURE 1 promotes phytochrome B-dependent seed germination by repressing abscisic acid biosynthesis. Plant Cell 2023, 35, 700–716. [Google Scholar] [CrossRef]
- Garcia-Gomez, M.L.; Castillo-Jimenez, A.; Martinez-Garcia, J.C.; Alvarez-Buylla, E.R. Multi-level gene regulatory network models to understand complex mechanisms underlying plant development. Curr. Opin. Plant Biol. 2020, 57, 171–179. [Google Scholar] [CrossRef]
- Ariel, F.; Romero-Barrios, N.; Jegu, T.; Benhamed, M.; Crespi, M. Battles and hijacks: Noncoding transcription in plants. Trends Plant Sci. 2015, 20, 362–371. [Google Scholar] [CrossRef]
- Bai, Y.; Dai, X.; Harrison, A.P.; Chen, M. RNA regulatory networks in animals and plants: A long noncoding RNA perspective. Brief. Funct. Genom. 2015, 14, 91–101. [Google Scholar] [CrossRef] [PubMed]
- Palazzo, A.F.; Lee, E.S. Non-coding RNA: What is functional and what is junk? Front. Genet. 2015, 6, 2. [Google Scholar] [CrossRef]
- Jarroux, J.; Morillon, A.; Pinskaya, M. History, discovery, and classification of lncRNAs. In Long Non Coding RNA Biology; Springer: Singapore, 2017; pp. 1–46. [Google Scholar]
- Crespi, M.; Jurkevitch, E.; Poiret, M.; d’Aubenton-Carafa, Y.; Petrovics, G.; Kondorosi, E.; Kondorosi, A. enod40, a gene expressed during nodule organogenesis, codes for a non-translatable RNA involved in plant growth. EMBO J. 1994, 13, 5099–5112. [Google Scholar] [CrossRef]
- Saha, C.; Saha, S.; Bhattacharyya, N.P. LncRNAOmics: A Comprehensive Review of Long Non-Coding RNAs in Plants. Genes 2025, 16, 765. [Google Scholar] [CrossRef]
- Ma, L.; Bajic, V.B.; Zhang, Z. On the classification of long non-coding RNAs. RNA Biol. 2013, 10, 924–933. [Google Scholar] [CrossRef]
- Palos, K.; Yu, L.; Railey, C.E.; Nelson Dittrich, A.C.; Nelson, A.D. Linking discoveries, mechanisms, and technologies to develop a clearer perspective on plant long noncoding RNAs. Plant Cell 2023, 35, 1762–1786. [Google Scholar] [CrossRef] [PubMed]
- Ponting, C.P.; Oliver, P.L.; Reik, W. Evolution and functions of long noncoding RNAs. Cell 2009, 136, 629–641. [Google Scholar] [CrossRef] [PubMed]
- Statello, L.; Guo, C.-J.; Chen, L.-L.; Huarte, M. Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 2021, 22, 96–118, Erratum in Nat. Rev. Mol. Cell Biol. 2021, 22, 159. [Google Scholar] [CrossRef]
- Chekanova, J.A. Long non-coding RNAs and their functions in plants. Curr. Opin. Plant Biol. 2015, 27, 207–216. [Google Scholar] [CrossRef] [PubMed]
- Tseng, K.C.; Wu, N.Y.; Chow, C.N.; Zheng, H.Q.; Chou, C.Y.; Yang, C.W.; Wang, M.J.; Chang, S.B.; Chang, W.C. JustRNA: A database of plant long noncoding RNA expression profiles and functional network. J. Exp. Bot. 2023, 74, 4949–4958. [Google Scholar] [CrossRef]
- Kapusta, A.; Feschotte, C. Volatile evolution of long noncoding RNA repertoires: Mechanisms and biological implications. Trends Genet. 2014, 30, 439–452. [Google Scholar] [CrossRef]
- Deng, P.; Liu, S.; Nie, X.; Weining, S.; Wu, L. Conservation analysis of long non-coding RNAs in plants. Sci. China Life Sci. 2018, 61, 190–198. [Google Scholar] [CrossRef]
- Shafiq, S.; Li, J.; Sun, Q. Functions of plants long non-coding RNAs. Biochim. Biophys. Acta 2016, 1859, 155–162. [Google Scholar] [CrossRef]
- Wang, H.V.; Chekanova, J.A. Long Noncoding RNAs in Plants. Adv. Exp. Med. Biol. 2017, 1008, 133–154. [Google Scholar] [CrossRef]
- Chen, L.; Zhu, Q.-H.; Kaufmann, K. Long non-coding RNAs in plants: Emerging modulators of gene activity in development and stress responses. Planta 2020, 252, 92. [Google Scholar] [CrossRef]
- Kościelniak, P.; Walas, Ł.; Konecka, A.; Buraczyk, W.; Klupczyńska, E.A. Plant long noncoding RNAs: Why do we not know more? Biol. Res. 2025, 58, 37. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Zhou, W.; Zhang, X.; Li, Y. Roles of long non-coding RNAs in plant immunity. PLoS Path. 2023, 19, e1011340. [Google Scholar] [CrossRef]
- Budak, H.; Kaya, S.B.; Cagirici, H.B. Long Non-coding RNA in Plants in the Era of Reference Sequences. Front. Plant Sci. 2020, 11, 276. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Wu, H.J.; Fang, J.; Chu, C.; Wang, X.J. A long noncoding RNA involved in rice reproductive development by negatively regulating osa-miR160. Sci. Bull. 2017, 62, 470–475. [Google Scholar] [CrossRef]
- Bhat, S.A.; Najar, M.A.; Wani, A.A.; Qadir, S.; John, R. The Long-noncoding RNAs: Effective players in plant development and stress responses. J. Plant Biochem. Biotechnol. 2025, 34, 35–61. [Google Scholar] [CrossRef]
- Quinn, J.J.; Chang, H.Y. Unique features of long non-coding RNA biogenesis and function. Nat. Rev. Genet. 2016, 17, 47–62. [Google Scholar] [CrossRef]
- Dominguez-Rosas, E.; Hernandez-Onate, M.A.; Fernandez-Valverde, S.L.; Tiznado-Hernandez, M.E. Plant long non-coding RNAs: Identification and analysis to unveil their physiological functions. Front. Plant Sci. 2023, 14, 1275399. [Google Scholar] [CrossRef]
- Liang, W.; Dong, H.; Guo, X.; Rodríguez, V.; Cheng, M.; Li, M.; Benech-Arnold, R.; Pu, Z.; Wang, J. Identification of long-lived and stable mRNAs in the aged seeds of wheat. Seed Biol. 2023, 2, 14. [Google Scholar] [CrossRef]
- Tuncel, O.; Kara, M.; Yaylak, B.; Erdogan, I.; Akgul, B. Noncoding RNAs in apoptosis: Identification and function. Turk. J. Biol. 2022, 46, 1–40. [Google Scholar] [CrossRef]
- Wang, H.; Chung, P.J.; Liu, J.; Jang, I.-C.; Kean, M.J.; Xu, J.; Chua, N.-H. Genome-wide identification of long noncoding natural antisense transcripts and their responses to light in Arabidopsis. Genome Res. 2014, 24, 444–453. [Google Scholar] [CrossRef]
- Meena, S.K.; Quevedo, M.; Nardeli, S.M.; Verez, C.; Bhat, S.S.; Zacharaki, V.; Kindgren, P. Antisense transcription from stress-responsive transcription factors fine-tunes the cold response in Arabidopsis. Plant Cell 2024, 36, 3467–3482. [Google Scholar] [CrossRef] [PubMed]
- Britto-Kido, S.d.A.; Ferreira Neto, J.R.C.; Pandolfi, V.; Marcelino-Guimarães, F.C.; Nepomuceno, A.L.; Vilela Abdelnoor, R.; Benko-Iseppon, A.M.; Kido, E.A. Natural antisense transcripts in plants: A review and identification in soybean infected with Phakopsora pachyrhizi SuperSAGE Library. Sci. World J. 2013, 2013, 219798. [Google Scholar] [CrossRef]
- Lapidot, M.; Pilpel, Y. Genome-wide natural antisense transcription: Coupling its regulation to its different regulatory mechanisms. EMBO Rep. 2006, 7, 1216–1222. [Google Scholar] [CrossRef]
- Zhao, X.; Li, J.; Lian, B.; Gu, H.; Li, Y.; Qi, Y. Global identification of Arabidopsis lncRNAs reveals the regulation of MAF4 by a natural antisense RNA. Nat. Commun. 2018, 9, 5056. [Google Scholar] [CrossRef]
- Xu, Y.-C.; Zhang, J.; Zhang, D.-Y.; Nan, Y.-H.; Ge, S.; Guo, Y.-L. Identification of long noncoding natural antisense transcripts (lncNATs) correlated with drought stress response in wild rice (Oryza nivara). BMC Genom. 2021, 22, 424. [Google Scholar] [CrossRef]
- Santini, L.; Yoshida, L.; de Oliveira, K.D.; Lembke, C.G.; Diniz, A.L.; Cantelli, G.C.; Nishiyama-Junior, M.Y.; Souza, G.M. Antisense transcription in plants: A systematic review and an update on cis-NATs of sugarcane. Int. J. Mol. Sci. 2022, 23, 11603. [Google Scholar] [CrossRef]
- Liu, J.; Wang, H.; Chua, N.H. Long noncoding RNA transcriptome of plants. Plant Biotechnol. J. 2015, 13, 319–328. [Google Scholar] [CrossRef]
- Zhou, H.; Ren, F.; Wang, X.; Qiu, K.; Sheng, Y.; Xie, Q.; Shi, P.; Zhang, J.; Pan, H. Genome-wide identification and characterization of long noncoding RNAs during peach (Prunus persica) fruit development and ripening. Sci. Rep. 2022, 12, 11044. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Wang, W.; Zhu, W.; Dong, J.; Cheng, Y.; Yin, Z.; Shen, F. Mechanisms and functions of long non-coding RNAs at multiple regulatory levels. Int. J. Mol. Sci. 2019, 20, 5573. [Google Scholar] [CrossRef]
- Dutta, T.K. Technical Advancements in Functional Mutagenesis of Plant Long Noncoding RNAs Using CRISPR/Cas Technology. In Genome and Epigenome Editing for Stress—Tolerant Crops; John Wiley & Sons Ltd.: Hoboken, NJ, USA, 2025; pp. 219–237. [Google Scholar]
- Tremblay, B.J.; Santini, C.P.; Cheng, Y.; Zhang, X.; Rosa, S.; Qüesta, J.I. Interplay between coding and non-coding regulation drives the Arabidopsis seed-to-seedling transition. Nat. Commun. 2024, 15, 1724. [Google Scholar] [CrossRef] [PubMed]
- He, Z.; Lan, Y.; Zhou, X.; Yu, B.; Zhu, T.; Yang, F.; Fu, L.-Y.; Chao, H.; Wang, J.; Feng, R.-X. Single-cell transcriptome analysis dissects lncRNA-associated gene networks in Arabidopsis. Plant Commun. 2024, 5, 100717. [Google Scholar] [CrossRef] [PubMed]
- Zhang, A.; Pi, W.; Wang, Y.; Li, Y.; Wang, J.; Liu, S.; Cui, X.; Liu, H.; Yao, D.; Zhao, R. Update on functional analysis of long non-coding RNAs in common crops. Front. Plant Sci. 2024, 15, 1389154. [Google Scholar] [CrossRef] [PubMed]
- Waseem, M.; Liu, Y.; Xia, R. Long non-coding RNAs, the dark matter: An emerging regulatory component in plants. Int. J. Mol. Sci. 2020, 22, 86. [Google Scholar] [CrossRef] [PubMed]
- Szcześniak, M.W.; Wanowska, E. CANTATAdb 3.0: An Updated Repository of Plant Long Non-Coding RNAs. Plant Cell Physiol. 2024, 65, 1486–1493. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Zang, S.; Zou, W.; Pan, Y.-B.; Yao, W.; You, C.; Que, Y. Long non-coding RNAs: New players in plants. Int. J. Mol. Sci. 2022, 23, 9301. [Google Scholar] [CrossRef]
- Corona-Gomez, J.A.; Coss-Navarrete, E.L.; Garcia-Lopez, I.J.; Klapproth, C.; Pérez-Patiño, J.A.; Fernandez-Valverde, S.L. Transcriptome-guided annotation and functional classification of long non-coding RNAs in Arabidopsis thaliana. Sci. Rep. 2022, 12, 14063. [Google Scholar] [CrossRef]
- Clark, M.B.; Johnston, R.L.; Inostroza-Ponta, M.; Fox, A.H.; Fortini, E.; Moscato, P.; Dinger, M.E.; Mattick, J.S. Genome-wide analysis of long noncoding RNA stability. Genome Res. 2012, 22, 885–898. [Google Scholar] [CrossRef]
- Xia, S.-W.; Song, C.-J.; Zheng, X.-F.; Zhang, Y.; Anwar, W.; Dissanayaka, D.D.; Zhu, P.-Z.; Liu, J.-L.; Lai, Y.-S. Identification and characterization of long non-coding RNA (lncRNA) in wild and semi-wild cucumbers. BMC Plant Biol. 2025, 25, 748. [Google Scholar] [CrossRef]
- Staněk, D. Long non-coding RNAs and splicing. Essays Biochem. 2021, 65, 723–729. [Google Scholar] [CrossRef]
- Basu, K.; Dey, A.; Kiran, M. Inefficient splicing of long non-coding RNAs is associated with higher transcript complexity in human and mouse. RNA Biol. 2023, 20, 563–572. [Google Scholar] [CrossRef]
- Krchňáková, Z.; Thakur, P.K.; Krausová, M.; Bieberstein, N.; Haberman, N.; Müller-McNicoll, M.; Staněk, D. Splicing of long non-coding RNAs primarily depends on polypyrimidine tract and 5′ splice-site sequences due to weak interactions with SR proteins. Nucleic Acids Res. 2019, 47, 911–928. [Google Scholar] [CrossRef]
- Lucero, L.; Fonouni-Farde, C.; Crespi, M.; Ariel, F. Long noncoding RNAs shape transcription in plants. Transcription 2020, 11, 160–171. [Google Scholar] [CrossRef]
- Gonzales, L.R.; Blom, S.; Henriques, R.; Bachem, C.W.; Immink, R.G. LncRNAs: The art of being influential without protein. Trends Plant Sci. 2024, 29, 770–785. [Google Scholar] [CrossRef] [PubMed]
- Chorostecki, U.; Bologna, N.G.; Ariel, F. The plant noncoding transcriptome: A versatile environmental sensor. EMBO J. 2023, 42, e114400. [Google Scholar] [CrossRef]
- Ferrer, J.; Dimitrova, N. Transcription regulation by long non-coding RNAs: Mechanisms and disease relevance. Nat. Rev. Mol. Cell Biol. 2024, 25, 396–415. [Google Scholar] [CrossRef]
- Jin, X.; Wang, Z.; Li, X.; Ai, Q.; Wong, D.C.J.; Zhang, F.; Yang, J.; Zhang, N.; Si, H. Current perspectives of lncRNAs in abiotic and biotic stress tolerance in plants. Front. Plant Sci. 2024, 14, 1334620. [Google Scholar] [CrossRef]
- Yadav, V.K.; Jalmi, S.K.; Tiwari, S.; Kerkar, S. Deciphering shared attributes of plant long non-coding RNAs through a comparative computational approach. Sci. Rep. 2023, 13, 15101. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Dong, Q.; Wang, Z.; Liu, Q.; Yu, H.; Sun, W.; Cheema, J.; You, Q.; Ding, L.; Cao, X. A fine-scale Arabidopsis chromatin landscape reveals chromatin conformation-associated transcriptional dynamics. Nat. Commun. 2024, 15, 3253. [Google Scholar] [CrossRef]
- Liang, Q.; Zhang, J.; Lam, H.-M.; Chan, T.-F. Nanopore direct RNA sequencing reveals N 6-methyladenosine and polyadenylation landscapes on long non-coding RNAs in Arabidopsis thaliana. BMC Plant Biol. 2024, 24, 1126. [Google Scholar] [CrossRef]
- Jeon, M.; Jeong, G.; Yang, Y.; Luo, X.; Jeong, D.; Kyung, J.; Hyun, Y.; He, Y.; Lee, I. Vernalization-triggered expression of the antisense transcript COOLAIR is mediated by CBF genes. eLife 2023, 12, e84594. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.; Li, Q.; Feng, X.; Yang, B.; Zhong, X.; Zhou, Y.; Wang, Q.; Mao, Y.; Xie, W.; Liu, T. Identification and functional analysis of drought-responsive long noncoding RNAs in maize roots. Int. J. Mol. Sci. 2023, 24, 15039. [Google Scholar] [CrossRef]
- Thieffry, A.; Vigh, M.L.; Bornholdt, J.; Ivanov, M.; Brodersen, P.; Sandelin, A. Characterization of Arabidopsis thaliana Promoter Bidirectionality and Antisense RNAs by Inactivation of Nuclear RNA Decay Pathways. Plant Cell 2020, 32, 1845–1867. [Google Scholar] [CrossRef]
- Jin, Y.; Ivanov, M.; Dittrich, A.N.; Nelson, A.D.; Marquardt, S. LncRNA FLAIL affects alternative splicing and represses flowering in Arabidopsis. EMBO J. 2023, 42, e110921. [Google Scholar] [CrossRef]
- Qi, J.; Li, S.; Su, J.; Lu, Y.; Yu, W.; Li, C. The role of m6A in plant development, stress response, and agricultural practices. Hortic. Plant J. 2025, in press. [Google Scholar] [CrossRef]
- Zhu, Y.; Chen, L.; Hong, X.; Shi, H.; Li, X. Revealing the novel complexity of plant long non-coding RNA by strand-specific and whole transcriptome sequencing for evolutionarily representative plant species. BMC Genom. 2022, 23, 381. [Google Scholar] [CrossRef] [PubMed]
- Golicz, A.A.; Singh, M.B.; Bhalla, P.L. The long intergenic noncoding RNA (LincRNA) landscape of the soybean genome. Plant Physiol. 2018, 176, 2133–2147. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zhu, Q.-F.; Li, W.-Y.; Chen, P.; Xue, J.; Yu, Y.; Feng, Y.-Z. The pivotal role of noncoding RNAs in flowering time regulation. Genes 2023, 14, 2114. [Google Scholar] [CrossRef] [PubMed]
- Traubenik, S.; Crespi, M. Spotlight: Antisense regulation of miRNA action during phosphate starvation. Mol. Plant 2023, 16, 1249–1251. [Google Scholar] [CrossRef]
- Chiang, C.P.; Li, J.L.; Chiou, T.J. Dose-dependent long-distance movement of microRNA399 duplex regulates phosphate homeostasis in Arabidopsis. New Phytol. 2023, 240, 802–814. [Google Scholar] [CrossRef]
- Seo, J.S.; Sun, H.-X.; Park, B.S.; Huang, C.-H.; Yeh, S.-D.; Jung, C.; Chua, N.-H. ELF18-INDUCED LONG-NONCODING RNA associates with mediator to enhance expression of innate immune response genes in Arabidopsis. Plant Cell 2017, 29, 1024–1038. [Google Scholar] [CrossRef]
- Nielsen, M.; Menon, G.; Zhao, Y.; Mateo-Bonmati, E.; Wolff, P.; Zhou, S.; Howard, M.; Dean, C. COOLAIR and PRC2 function in parallel to silence FLC during vernalization. Proc. Natl. Acad. Sci. USA 2024, 121, e2311474121. [Google Scholar] [CrossRef]
- Csorba, T. APOLO lncRNA, a self-calibrating switch of root development. Mol. Plant 2021, 14, 867–869. [Google Scholar] [CrossRef]
- Mammarella, M.F.; Lucero, L.; Hussain, N.; Muñoz-Lopez, A.; Huang, Y.; Ferrero, L.; Fernandez-Milmanda, G.L.; Manavella, P.; Benhamed, M.; Crespi, M. Long noncoding RNA-mediated epigenetic regulation of auxin-related genes controls shade avoidance syndrome in Arabidopsis. EMBO J. 2023, 42, e113941. [Google Scholar] [CrossRef] [PubMed]
- Ariel, F.; Lucero, L.; Christ, A.; Mammarella, M.F.; Jegu, T.; Veluchamy, A.; Mariappan, K.; Latrasse, D.; Blein, T.; Liu, C. R-loop mediated trans action of the APOLO long noncoding RNA. Mol. Cell 2020, 77, 1055–1065.e4. [Google Scholar] [CrossRef]
- Zheng, Q.; Rowley, M.J.; Böhmdorfer, G.; Sandhu, D.; Gregory, B.D.; Wierzbicki, A.T. RNA polymerase V targets transcriptional silencing components to promoters of protein-coding genes. Plant J. 2013, 73, 179–189. [Google Scholar] [CrossRef] [PubMed]
- Sigman, M.J.; Panda, K.; Kirchner, R.; McLain, L.L.; Payne, H.; Peasari, J.R.; Husbands, A.Y.; Slotkin, R.K.; McCue, A.D. An siRNA-guided ARGONAUTE protein directs RNA polymerase V to initiate DNA methylation. Nat. Plants 2021, 7, 1461–1474. [Google Scholar] [CrossRef] [PubMed]
- Lahmy, S.; Pontier, D.; Bies-Etheve, N.; Laudié, M.; Feng, S.; Jobet, E.; Hale, C.J.; Cooke, R.; Hakimi, M.-A.; Angelov, D. Evidence for ARGONAUTE4–DNA interactions in RNA-directed DNA methylation in plants. Genes Dev. 2016, 30, 2565–2570. [Google Scholar] [CrossRef]
- Emenecker, R.J.; Holehouse, A.S.; Strader, L.C. Emerging roles for phase separation in plants. Dev. Cell 2020, 55, 69–83. [Google Scholar] [CrossRef]
- Mattick, J.S.; Amaral, P.P.; Carninci, P.; Carpenter, S.; Chang, H.Y.; Chen, L.-L.; Chen, R.; Dean, C.; Dinger, M.E.; Fitzgerald, K.A. Long non-coding RNAs: Definitions, functions, challenges and recommendations. Nat. Rev. Mol. Cell Biol. 2023, 24, 430–447. [Google Scholar] [CrossRef]
- Wang, Y.; Luo, X.; Sun, F.; Hu, J.; Zha, X.; Su, W.; Yang, J. Overexpressing lncRNA LAIR increases grain yield and regulates neighbouring gene cluster expression in rice. Nat. Commun. 2018, 9, 3516. [Google Scholar] [CrossRef]
- Lei, M.-Q.; He, R.-R.; Zhou, Y.-F.; Yang, L.; Zhang, Z.-F.; Yuan, C.; Zhao, W.-L.; Cheng, Y.; Lian, J.-P.; Zhang, Y.-C. The long noncoding RNA ALEX1 confers a functional phase state of ARF3 to enhance rice resistance to bacterial pathogens. Mol. Plant 2025, 18, 114–129. [Google Scholar] [CrossRef]
- Fu, D.; Jiang, B. Liquid-liquid phase separation regulates gene expression in plants. Agric. Commun. 2025, 3, 100084. [Google Scholar] [CrossRef]
- Böhmdorfer, G.; Sethuraman, S.; Rowley, M.J.; Krzyszton, M.; Rothi, M.H.; Bouzit, L.; Wierzbicki, A.T. Long non-coding RNA produced by RNA polymerase V determines boundaries of heterochromatin. eLlife 2016, 5, e19092. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.; Zhang, Z.; Li, J.; Zhu, Y.; Yin, Y.; Zhang, X.; Dai, Y.; Zhang, A.; Li, C.; Zhu, Y. Genome-wide identification and characterization of lncRNAs in sunflower endosperm. BMC Plant Biol. 2022, 22, 494. [Google Scholar] [CrossRef]
- Yang, L.; Cheng, Y.; Yuan, C.; Zhou, Y.-F.; Huang, Q.-J.; Zhao, W.-L.; He, R.-R.; Jiang, J.; Qin, Y.-C.; Chen, Z.-T. The long noncoding RNA VIVIpary promotes seed dormancy release and pre-harvest sprouting through chromatin remodeling in rice. Mol. Plant 2025, 18, 978–994. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhang, R.; Meng, F.; Chen, Y.; Wang, W.; Yang, K.; Gao, Y.; Xin, M.; Du, J.; Hu, Z.; et al. A comprehensive atlas of long non-coding RNAs provides insight into grain development in wheat. Seed Biol. 2023, 2, 12. [Google Scholar] [CrossRef]
- Li, L.; Luo, H.; Lim, D.-H.; Han, L.; Li, Y.; Fu, X.-D.; Qi, Y. Global profiling of RNA–chromatin interactions reveals co-regulatory gene expression networks in Arabidopsis. Nat. Plants 2021, 7, 1364–1378. [Google Scholar] [CrossRef]
- Sacharowski, S.P.; Krzyszton, M.; Brzezniak, L.; Rogowski, K.J.; Montez, M.; Rosol, K.; Wrona, M.; Yatusevich, R.; Manjunath, V.H.; Szewc, L. Chromatin retained MUSHER lncRNA integrates ABA and DOG1 signalling pathways to enhance Arabidopsis seeds dormancy. Nat. Commun. 2025, 16, 7545. [Google Scholar] [CrossRef]
- Yuan, H.; Liu, S.; Yan, R.; Liu, Z.; Xu, K.; Huang, D.; Zhang, N.; Wu, Y.; Lan, X.; Yukawa, Y. AtR8 lncRNA integrates WRKY46 into ABA signaling to regulate seed and seeding growth in Arabidopsis. Plant Physiol. Biochem. 2025, 223, 109732. [Google Scholar] [CrossRef]
- Kuczynski, C.; McCorkle, S.; Keereetaweep, J.; Shanklin, J.; Schwender, J. An expanded role for the transcription factor WRINKLED1 in the biosynthesis of triacylglycerols during seed development. Front. Plant Sci. 2022, 13, 955589. [Google Scholar] [CrossRef]
- Song, P.; Wei, L.; Chen, Z.; Cai, Z.; Lu, Q.; Wang, C.; Tian, E.; Jia, G. m6A readers ECT2/ECT3/ECT4 enhance mRNA stability through direct recruitment of the poly (A) binding proteins in Arabidopsis. Genome Biol. 2023, 24, 103. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Fan, X.; Lin, F.; He, G.; Terzaghi, W.; Zhu, D.; Deng, X.W. Arabidopsis noncoding RNA mediates control of photomorphogenesis by red light. Proc. Natl. Acad. Sci. USA 2014, 111, 10359–10364. [Google Scholar] [CrossRef]
- Zicola, J. Spotlight on a long noncoding RNA: HID1 regulates seed germination after red light exposure. Plant Cell 2023, 35, 630–631. [Google Scholar] [CrossRef]
- Guo, G.; Liu, X.; Sun, F.; Cao, J.; Huo, N.; Wuda, B.; Xin, M.; Hu, Z.; Du, J.; Xia, R.; et al. Wheat miR9678 Affects Seed Germination by Generating Phased siRNAs and Modulating Abscisic Acid/Gibberellin Signaling. Plant Cell 2018, 30, 796–814. [Google Scholar] [CrossRef] [PubMed]
- Heo, J.B.; Sung, S. Vernalization-mediated epigenetic silencing by a long intronic noncoding RNA. Science 2011, 331, 76–79. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Fan, F.; Zhang, Q.; Luo, Y.; Liu, Q.; Gao, J.; Liu, J.; Chen, G.; Zhang, H. Identification and Functional Analysis of Long Non-Coding RNA (lncRNA) in Response to Seed Aging in Rice. Plants 2022, 11, 3223. [Google Scholar] [CrossRef]
- Sherazi, S.A.M.; Abbasi, A.; Jamil, A.; Uzair, M.; Ikram, A.; Qamar, S.; Olamide, A.A.; Arshad, M.; Fried, P.J.; Ljubisavljevic, M. Molecular hallmarks of long non-coding RNAs in aging and its significant effect on aging-associated diseases. Neural Regen. Res. 2023, 18, 959–968. [Google Scholar] [PubMed]
- Liew, L.C.; You, Y.; Auroux, L.; Oliva, M.; Peirats-Llobet, M.; Ng, S.; Tamiru-Oli, M.; Berkowitz, O.; Hong, U.V.T.; Haslem, A. Establishment of single-cell transcriptional states during seed germination. Nat. Plants 2024, 10, 1418–1434. [Google Scholar] [CrossRef]
- Zhang, M.; Zhao, H.; Xie, S.; Chen, J.; Xu, Y.; Wang, K.; Zhao, H.; Guan, H.; Hu, X.; Jiao, Y. Extensive, clustered parental imprinting of protein-coding and noncoding RNAs in developing maize endosperm. Proc. Natl. Acad. Sci. USA 2011, 108, 20042–20047. [Google Scholar] [CrossRef]
- Wolff, P.; Weinhofer, I.; Seguin, J.; Roszak, P.; Beisel, C.; Donoghue, M.T.; Spillane, C.; Nordborg, M.; Rehmsmeier, M.; Köhler, C. High-resolution analysis of parent-of-origin allelic expression in the Arabidopsis endosperm. PLoS Genet. 2011, 7, e1002126. [Google Scholar] [CrossRef]
- Fu, Y.; Xiao, W.; Tian, L.; Guo, L.; Ma, G.; Ji, C.; Huang, Y.; Wang, H.; Wu, X.; Yang, T. Spatial transcriptomics uncover sucrose post-phloem transport during maize kernel development. Nat. Commun. 2023, 14, 7129, Erratum in Nat. Commun. 2024, 15, 279. [Google Scholar] [CrossRef]
- Yuan, Y.; Huo, Q.; Zhang, Z.; Wang, Q.; Wang, J.; Chang, S.; Cai, P.; Song, K.M.; Galbraith, D.W.; Zhang, W. Decoding the gene regulatory network of endosperm differentiation in maize. Nat. Commun. 2024, 15, 34. [Google Scholar] [CrossRef]
- Qu, J.; Ma, C.; Feng, J.; Xu, S.; Wang, L.; Li, F.; Li, Y.; Zhang, R.; Zhang, X.; Xue, J. Transcriptome dynamics during maize endosperm development. PLoS ONE 2016, 11, e0163814. [Google Scholar] [CrossRef]
- Kim, E.D.; Xiong, Y.; Pyo, Y.; Kim, D.H.; Kang, B.H.; Sung, S. Spatio-temporal analysis of coding and long noncoding transcripts during maize endosperm development. Sci. Rep. 2017, 7, 3838. [Google Scholar] [CrossRef] [PubMed]
- Doll, N.M.; Just, J.; Brunaud, V.; Caïus, J.; Grimault, A.; Depège-Fargeix, N.; Esteban, E.; Pasha, A.; Provart, N.J.; Ingram, G.C. Transcriptomics at maize embryo/endosperm interfaces identifies a transcriptionally distinct endosperm subdomain adjacent to the embryo scutellum. Plant Cell 2020, 32, 833–852. [Google Scholar] [CrossRef]
- Zheng, K.; Wu, X.; Xue, X.; Li, W.; Wang, Z.; Chen, J.; Zhang, Y.; Qiao, F.; Zhao, H.; Zhang, F. Transcriptome screening of long noncoding RNAs and their target protein-coding genes unmasks a dynamic portrait of seed coat coloration associated with anthocyanins in Tibetan Hulless Barley. Int. J. Mol. Sci. 2023, 24, 10587. [Google Scholar] [CrossRef]
- Vaughan, S.P.; Baker, J.M.; Primavesi, L.F.; Patil, A.; King, R.; Hassani-Pak, K.; Kulasekaran, S.; Coghill, J.; Ward, J.L.; Huttly, A.K. Proanthocyanidin biosynthesis in the developing wheat seed coat investigated by chemical and RNA-Seq analysis. Plant Direct 2022, 6, e453. [Google Scholar] [CrossRef]
- Yao, X.; Yao, Y.; An, L.; Li, X.; Bai, Y.; Cui, Y.; Wu, K. Accumulation and regulation of anthocyanins in white and purple Tibetan Hulless Barley (Hordeum vulgare L. var. nudum Hook. f.) revealed by combined de novo transcriptomics and metabolomics. BMC Plant Biol. 2022, 22, 391. [Google Scholar] [CrossRef]
- Cappellini, F.; Marinelli, A.; Toccaceli, M.; Tonelli, C.; Petroni, K. Anthocyanins: From mechanisms of regulation in plants to health benefits in foods. Front. Plant Sci. 2021, 12, 748049. [Google Scholar] [CrossRef] [PubMed]
- Shen, E.; Zhu, X.; Hua, S.; Chen, H.; Ye, C.; Zhou, L.; Liu, Q.; Zhu, Q.H.; Fan, L.; Chen, X. Genome-wide identification of oil biosynthesis-related long non-coding RNAs in allopolyploid Brassica napus. BMC Genom. 2018, 19, 745. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Tan, Z.; Zeng, C.; Xiao, M.; Lin, S.; Yao, W.; Li, Q.; Guo, L.; Lu, S. Regulation of seed oil accumulation by lncRNAs in Brassica napus. Biotechnol. Biofuels Bioprod. 2023, 16, 22. [Google Scholar] [CrossRef]
- Chen, J.; Wu, Z.; Li, R.; Huang, D.; Zhai, W.; Chen, C. New insight into LncRNA-mRNA regulatory network associated with lipid biosynthesis using Hi-C data in seeds of tung tree (Vernicia fordii Hemsl.). Ind. Crops Prod. 2021, 164, 113321. [Google Scholar] [CrossRef]
- Yuan, Y.; Sun, H.; Wang, X.; Gao, L.; Liu, C.; Gai, S.; Zhang, Y. Transcriptome of Paeonia ostii developing seeds revealed transcriptional factors, mRNAs and LncRNAs related to fatty acid synthesis and accumulation. Ind. Crops Prod. 2022, 188, 115670. [Google Scholar] [CrossRef]
- Yatusevich, R.; Fedak, H.; Ciesielski, A.; Krzyczmonik, K.; Kulik, A.; Dobrowolska, G.; Swiezewski, S. Antisense transcription represses Arabidopsis seed dormancy QTL DOG 1 to regulate drought tolerance. EMBO Rep. 2017, 18, 2186–2196. [Google Scholar] [CrossRef]
- Krzyczmonik, K.; Wroblewska-Swiniarska, A.; Swiezewski, S. Developmental transitions in Arabidopsis are regulated by antisense RNAs resulting from bidirectionally transcribed genes. RNA Biol. 2017, 14, 838–842. [Google Scholar] [CrossRef] [PubMed]
- Cao, D. Paving the way to secondary dormancy: Mind the DOG’s tail. Plant Physiol. 2025, 197, kiaf008. [Google Scholar] [CrossRef] [PubMed]
- Kowalczyk, J.; Palusinska, M.; Wroblewska-Swiniarska, A.; Pietras, Z.; Szewc, L.; Dolata, J.; Jarmolowski, A.; Swiezewski, S. Alternative Polyadenylation of the Sense Transcript Controls Antisense Transcription of DELAY OF GERMINATION 1 in Arabidopsis. Mol. Plant 2017, 10, 1349–1352. [Google Scholar] [CrossRef]
- Madhawan, A.; Bhunia, R.K.; Kumar, P.; Sharma, V.; Sinha, K.; Fandade, V.; Rahim, M.S.; Parveen, A.; Mishra, A.; Roy, J. Interaction between long noncoding RNA (lnc663) and microRNA (miR1128) regulates PDAT-like gene activity in bread wheat (Triticum aestivum L.). Plant Physiol. Biochem. 2023, 203, 108040. [Google Scholar] [CrossRef]
- Wang, G.; Li, H.; Ye, C.; He, K.; Liu, S.; Jiang, B.; Ge, R.; Gao, B.; Wei, J.; Zhao, Y. Quantitative profiling of m6A at single base resolution across the life cycle of rice and Arabidopsis. Nat. Commun. 2024, 15, 4881. [Google Scholar] [CrossRef]
- Shen, L.; Liang, Z.; Gu, X.; Chen, Y.; Teo, Z.W.N.; Hou, X.; Cai, W.M.; Dedon, P.C.; Liu, L.; Yu, H. N6-methyladenosine RNA modification regulates shoot stem cell fate in Arabidopsis. Dev. Cell 2016, 38, 186–200. [Google Scholar] [CrossRef]
- Shen, L.; Ma, J.; Li, P.; Wu, Y.; Yu, H. Recent advances in the plant epitranscriptome. Genome Biol. 2023, 24, 43. [Google Scholar] [CrossRef] [PubMed]
- Gao, Q.; Liu, J.; Weng, H.; Yuan, X.; Xiao, W.; Wang, H. A long noncoding RNA derived from lncRNA–mRNA networks modulates seed vigor. Int. J. Mol. Sci. 2022, 23, 9472. [Google Scholar] [CrossRef]
- Jiang, H.; Gao, W.; Jiang, B.L.; Liu, X.; Jiang, Y.T.; Zhang, L.T.; Zhang, Y.; Yan, S.N.; Cao, J.J.; Lu, J.; et al. Identification and validation of coding and non-coding RNAs involved in high-temperature-mediated seed dormancy in common wheat. Front. Plant Sci. 2023, 14, 1107277. [Google Scholar] [CrossRef]
- Zhou, Y.F.; Zhang, Y.C.; Sun, Y.M.; Yu, Y.; Lei, M.Q.; Yang, Y.W.; Lian, J.P.; Feng, Y.Z.; Zhang, Z.; Yang, L.; et al. The parent-of-origin lncRNA MISSEN regulates rice endosperm development. Nat. Commun. 2021, 12, 6525. [Google Scholar] [CrossRef] [PubMed]
- Urquiaga, M.C.d.O.; Thiebaut, F.; Hemerly, A.S.; Ferreira, P.C.G. From trash to luxury: The potential role of plant LncRNA in DNA methylation during abiotic stress. Front. Plant Sci. 2021, 11, 603246. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Le, J.; Zhang, Y.; Wang, R.; Li, Q.; Lu, X.; Liu, J.; Deng, Z. Identification and Functional Analysis of LncRNAs in Response to Seed Aging in Metasequoia glyptostroboides by Third Generation Sequencing Technology. Forests 2022, 13, 1579. [Google Scholar] [CrossRef]
- Mirdar Mansuri, R.; Azizi, A.-H.; Sadri, A.-H.; Shobbar, Z.-S. Long non-coding RNAs as the regulatory hubs in rice response to salt stress. Sci. Rep. 2022, 12, 21696. [Google Scholar] [CrossRef]
- Liu, L.; Fang, F. Long noncoding RNA mediated regulation in human embryogenesis, pluripotency, and reproduction. Stem Cells Int. 2022, 2022, 8051717. [Google Scholar] [CrossRef]
- Kim, J.Y.; Lee, J.; Kang, M.H.; Trang, T.T.M.; Lee, J.; Lee, H.; Jeong, H.; Lim, P.O. Dynamic landscape of long noncoding RNAs during leaf aging in Arabidopsis. Front. Plant Sci. 2022, 13, 1068163. [Google Scholar] [CrossRef]
- Luo, Y.; Zhang, Y.; Le, J.; Li, Q.; Mou, J.; Deng, S.; Li, J.; Wang, R.; Deng, Z.; Liu, J. Full-length transcriptome sequencing reveals the molecular mechanism of Metasequoia glyptostroboides seed responding to aging. Antioxidants 2023, 12, 1353. [Google Scholar] [CrossRef]
- Zhao, B.; Zhang, H.; Chen, T.; Ding, L.; Zhang, L.; Ding, X.; Zhang, J.; Qian, Q.; Xiang, Y. Sdr4 dominates pre-harvest sprouting and facilitates adaptation to local climatic condition in Asian cultivated rice. J. Integr. Plant Biol. 2022, 64, 1246–1263. [Google Scholar] [CrossRef]
- Zheng, X.; Chen, J.; Pang, H.; Liu, S.; Gao, Q.; Wang, J.; Qiao, W.; Wang, H.; Liu, J.; Olsen, K. Genome-wide analyses reveal the role of noncoding variation in complex traits during rice domestication. Sci. Adv. 2019, 5, eaax3619. [Google Scholar] [CrossRef]
- He, H.; Zhou, Y.-F.; Yang, Y.-W.; Zhang, Z.; Lei, M.-Q.; Feng, Y.-Z.; Zhang, Y.-C.; Chen, Y.-Q.; Lian, J.-P.; Yu, Y. Genome-wide analysis identified a set of conserved lncRNAs associated with domestication-related traits in rice. Int. J. Mol. Sci. 2021, 22, 4742. [Google Scholar] [CrossRef]
- Yang, G.; Deng, P.; Guo, Q.; Shi, T.; Pan, W.; Cui, L.; Liu, X.; Nie, X. Population transcriptomic analysis identifies the comprehensive lncRNAs landscape of spike in wheat (Triticum aestivum L.). BMC Plant Biol. 2022, 22, 450. [Google Scholar] [CrossRef]
- Tianshun, Z.; Dong, Y.; Liubing, W.; Yusheng, X.; Meijuan, D.; Dingyang, Y. Seed storability in rice: Physiological foundations, molecular mechanisms, and applications in breeding. Rice Sci. 2024, 31, 401–416. [Google Scholar] [CrossRef]
- Zhu, Q.H.; Wang, M.B. Molecular Functions of Long Non-Coding RNAs in Plants. Genes 2012, 3, 176–190. [Google Scholar] [CrossRef]
- Mattick, J.S. The genetic signatures of noncoding RNAs. PLoS Genet. 2009, 5, e1000459. [Google Scholar] [CrossRef]
- Wang, X.; Fan, H.; Wang, B.; Yuan, F. Research progress on the roles of lncRNAs in plant development and stress responses. Front. Plant Sci. 2023, 14, 1138901. [Google Scholar] [CrossRef]
- Salehi, S.; Taheri, M.N.; Azarpira, N.; Zare, A.; Behzad-Behbahani, A. State of the art technologies to explore long non-coding RNAs in cancer. J. Cell Mol. Med. 2017, 21, 3120–3140. [Google Scholar] [CrossRef] [PubMed]
- Puchta-Jasinska, M.; Bolc, P.; Pietrusinska-Radzio, A.; Motor, A.; Boczkowska, M. Small Interfering RNAs as Critical Regulators of Plant Life Process: New Perspectives on Regulating the Transcriptomic Machinery. Int. J. Mol. Sci. 2025, 26, 1624. [Google Scholar] [CrossRef] [PubMed]
- Chekanova, J.A.; Gregory, B.D.; Reverdatto, S.V.; Chen, H.; Kumar, R.; Hooker, T.; Yazaki, J.; Li, P.; Skiba, N.; Peng, Q. Genome-wide high-resolution mapping of exosome substrates reveals hidden features in the Arabidopsis transcriptome. Cell 2007, 131, 1340–1353. [Google Scholar] [CrossRef] [PubMed]
- Laubinger, S.; Zeller, G.; Henz, S.R.; Sachsenberg, T.; Widmer, C.K.; Naouar, N.; Vuylsteke, M.; Schölkopf, B.; Rätsch, G.; Weigel, D. At-TAX: A whole genome tiling array resource for developmental expression analysis and transcript identification in Arabidopsis thaliana. Genome Biol. 2008, 9, R112. [Google Scholar] [CrossRef]
- Liu, J.; Jung, C.; Xu, J.; Wang, H.; Deng, S.; Bernad, L.; Arenas-Huertero, C.; Chua, N.H. Genome-wide analysis uncovers regulation of long intergenic noncoding RNAs in Arabidopsis. Plant Cell 2012, 24, 4333–4345. [Google Scholar] [CrossRef] [PubMed]
- Robinson, S.J.; Parkin, I.A. Differential SAGE analysis in Arabidopsis uncovers increased transcriptome complexity in response to low temperature. BMC Genom. 2008, 9, 434. [Google Scholar] [CrossRef]
- Meyers, B.C.; Vu, T.H.; Tej, S.S.; Ghazal, H.; Matvienko, M.; Agrawal, V.; Ning, J.; Haudenschild, C.D. Analysis of the transcriptional complexity of Arabidopsis thaliana by massively parallel signature sequencing. Nat. Biotechnol. 2004, 22, 1006–1011. [Google Scholar] [CrossRef]
- Stark, R.; Grzelak, M.; Hadfield, J. RNA sequencing: The teenage years. Nat. Rev. Genet. 2019, 20, 631–656. [Google Scholar] [CrossRef]
- Wang, L.; Park, H.J.; Dasari, S.; Wang, S.; Kocher, J.-P.; Li, W. CPAT: Coding-Potential Assessment Tool using an alignment-free logistic regression model. Nucleic Acids Res. 2013, 41, e74. [Google Scholar] [CrossRef]
- Kang, Y.-J.; Yang, D.-C.; Kong, L.; Hou, M.; Meng, Y.-Q.; Wei, L.; Gao, G. CPC2: A fast and accurate coding potential calculator based on sequence intrinsic features. Nucleic Acids Res. 2017, 45, W12–W16. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.; Lu, P.; Xu, Y.; Li, Z.; Yu, S.; Liu, J.; Wang, H.; Chua, N.-H.; Cao, P. PLncDB V2.0: A comprehensive encyclopedia of plant long noncoding RNAs. Nucleic Acids Res. 2021, 49, D1489–D1495. [Google Scholar] [CrossRef] [PubMed]
- Pardo-Palacios, F.J.; Wang, D.; Reese, F.; Diekhans, M.; Carbonell-Sala, S.; Williams, B.; Loveland, J.E.; De María, M.; Adams, M.S.; Balderrama-Gutierrez, G. Systematic assessment of long-read RNA-seq methods for transcript identification and quantification. Nat. Methods 2024, 21, 1349–1363. [Google Scholar] [CrossRef]
- Workman, R.E.; Tang, A.D.; Tang, P.S.; Jain, M.; Tyson, J.R.; Razaghi, R.; Zuzarte, P.C.; Gilpatrick, T.; Payne, A.; Quick, J. Nanopore native RNA sequencing of a human poly (A) transcriptome. Nat. Methods 2019, 16, 1297–1305, Erratum in Nat. Methods 2020, 17, 114. [Google Scholar] [CrossRef]
- Parker, M.T.; Knop, K.; Sherwood, A.V.; Schurch, N.J.; Mackinnon, K.; Gould, P.D.; Hall, A.J.; Barton, G.J.; Simpson, G.G. Nanopore direct RNA sequencing maps the complexity of Arabidopsis mRNA processing and m6A modification. eLife 2020, 9, e49658. [Google Scholar] [CrossRef]
- Zhang, R.; Kuo, R.; Coulter, M.; Calixto, C.P.; Entizne, J.C.; Guo, W.; Marquez, Y.; Milne, L.; Riegler, S.; Matsui, A. A high-resolution single-molecule sequencing-based Arabidopsis transcriptome using novel methods of Iso-seq analysis. Genome Biol. 2022, 23, 149. [Google Scholar] [CrossRef]
- Tang, X.; Zhang, N.; Si, H.; Calderón-Urrea, A. Selection and validation of reference genes for RT-qPCR analysis in potato under abiotic stress. Plant Methods 2017, 13, 85. [Google Scholar] [CrossRef]
- Rosa, S.; Duncan, S.; Dean, C. Mutually exclusive sense–antisense transcription at FLC facilitates environmentally induced gene repression. Nat. Commun. 2016, 7, 13031. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Fonseca, A.; Meschichi, A.; Sicard, A.; Rosa, S. Whole-mount smFISH allows combining RNA and protein quantification at cellular and subcellular resolution. Nat. Plants 2023, 9, 1094–1102. [Google Scholar] [CrossRef] [PubMed]
- Huang, T.; Guillotin, B.; Rahni, R.; Birnbaum, K.D.; Wagner, D. A rapid and sensitive, multiplex, whole mount RNA fluorescence in situ hybridization and immunohistochemistry protocol. Plant Methods 2023, 19, 131. [Google Scholar] [CrossRef] [PubMed]
- Duncan, S.; Olsson, T.S.; Hartley, M.; Dean, C.; Rosa, S. A method for detecting single mRNA molecules in Arabidopsis thaliana. Plant Methods 2016, 12, 13. [Google Scholar] [CrossRef]
- Duncan, S.; Olsson, T.S.; Hartley, M.; Dean, C.; Rosa, S. Single molecule RNA FISH in Arabidopsis root cells. Bio-Protocol 2017, 7, e2240. [Google Scholar] [CrossRef]
- Lin, X.; Lin, W.; Ku, Y.-S.; Wong, F.-L.; Li, M.-W.; Lam, H.-M.; Ngai, S.-M.; Chan, T.-F. Analysis of soybean long non-coding RNAs reveals a subset of small peptide-coding transcripts. Plant Physiol. 2020, 182, 1359–1374. [Google Scholar] [CrossRef]
- Xing, D.; Wang, Y.; Hamilton, M.; Ben-Hur, A.; Reddy, A.S. Transcriptome-wide identification of RNA targets of Arabidopsis SERINE/ARGININE-RICH45 uncovers the unexpected roles of this RNA binding protein in RNA processing. Plant Cell 2015, 27, 3294–3308, Erratum in Plant Cell. 2017, 29, 2304. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Gu, L.; Hou, Y.; Wang, L.; Deng, X.; Hang, R.; Chen, D.; Zhang, X.; Zhang, Y.; Liu, C. Integrative genome-wide analysis reveals HLP1, a novel RNA-binding protein, regulates plant flowering by targeting alternative polyadenylation. Cell Res. 2015, 25, 864–876. [Google Scholar] [CrossRef]
- Meyer, K.; Köster, T.; Nolte, C.; Weinholdt, C.; Lewinski, M.; Grosse, I.; Staiger, D. Adaptation of iCLIP to plants determines the binding landscape of the clock-regulated RNA-binding protein At GRP7. Genome Biol. 2017, 18, 204. [Google Scholar] [CrossRef]
- Lewinski, M.; Brüggemann, M.; Köster, T.; Reichel, M.; Bergelt, T.; Meyer, K.; König, J.; Zarnack, K.; Staiger, D. Mapping protein–RNA binding in plants with individual-nucleotide-resolution UV cross-linking and immunoprecipitation (plant iCLIP2). Nat. Protoc. 2024, 19, 1183–1234. [Google Scholar] [CrossRef]
- Arribas-Hernández, L.; Rennie, S.; Köster, T.; Porcelli, C.; Lewinski, M.; Staiger, D.; Andersson, R.; Brodersen, P. Principles of mRNA targeting via the Arabidopsis m6A-binding protein ECT2. eLife 2021, 10, e72375. [Google Scholar] [CrossRef]
- Lucero, L.; Ferrero, L.; Fonouni-Farde, C.; Ariel, F. Functional classification of plant long noncoding RNAs: A transcript is known by the company it keeps. New Phytol. 2021, 229, 1251–1260. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.-H.; Xi, Y.; Sung, S. Modular function of long noncoding RNA, COLDAIR, in the vernalization response. PLoS Genet. 2017, 13, e1006939. [Google Scholar] [CrossRef] [PubMed]
- Simon, M.D.; Wang, C.I.; Kharchenko, P.V.; West, J.A.; Chapman, B.A.; Alekseyenko, A.A.; Borowsky, M.L.; Kuroda, M.I.; Kingston, R.E. The genomic binding sites of a noncoding RNA. Proc. Natl. Acad. Sci. USA 2011, 108, 20497–20502. [Google Scholar] [CrossRef]
- Engreitz, J.M.; Pandya-Jones, A.; McDonel, P.; Shishkin, A.; Sirokman, K.; Surka, C.; Kadri, S.; Xing, J.; Goren, A.; Lander, E.S. The Xist lncRNA exploits three-dimensional genome architecture to spread across the X chromosome. Science 2013, 341, 1237973. [Google Scholar] [CrossRef] [PubMed]
- Chu, C.; Quinn, J.; Chang, H.Y. Chromatin isolation by RNA purification (ChIRP). J. Vis. Exp. JoVE 2012, 3912. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Weirather, J.L.; de Cesare, M.; Wang, Y.; Piazza, P.; Sebastiano, V.; Wang, X.-J.; Buck, D.; Au, K.F. Comprehensive comparison of Pacific Biosciences and Oxford Nanopore Technologies and their applications to transcriptome analysis. F1000Research 2017, 6, 100. [Google Scholar] [CrossRef]
- Liang, W.-W.; Müller, S.; Hart, S.K.; Wessels, H.-H.; Méndez-Mancilla, A.; Sookdeo, A.; Choi, O.; Caragine, C.M.; Corman, A.; Lu, L. Transcriptome-scale RNA-targeting CRISPR screens reveal essential lncRNAs in human cells. Cell 2024, 187, 7637–7654.e7629. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016, 11, 1650–1667. [Google Scholar] [CrossRef] [PubMed]
- Pertea, G.; Pertea, M. GFF Utilities: GffRead and GffCompare. F1000Research 2020, 9. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, H.; Li, Q.; Jin, J.; Chen, H.; Zou, Y.; Huang, X.; Ding, Y. Genome-wide identification of lncRNAs involved in fertility transition in the photo-thermosensitive genic male sterile rice line Wuxiang S. Front. Plant Sci. 2021, 11, 580050. [Google Scholar] [CrossRef]
- Jali, I.; Vanamamalai, V.K.; Garg, P.; Navarrete, P.; Gutiérrez-Adán, A.; Sharma, S. Identification and differential expression of long non-coding RNAs and their association with XIST gene during early embryonic developmental stages of Bos taurus. Int. J. Biol. Macromol. 2023, 229, 896–908. [Google Scholar] [CrossRef]
- Yan, X.; Ma, L.; Yang, M. Identification and characterization of long non-coding RNA (lncRNA) in the developing seeds of Jatropha curcas. Sci. Rep. 2020, 10, 10395. [Google Scholar] [CrossRef]
- Wagner, G.P.; Kin, K.; Lynch, V.J. Measurement of mRNA abundance using RNA-seq data: RPKM measure is inconsistent among samples. Theory Biosci. 2012, 131, 281–285. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef]
- Ontiveros-Palacios, N.; Cooke, E.; Nawrocki, E.P.; Triebel, S.; Marz, M.; Rivas, E.; Griffiths-Jones, S.; Petrov, A.I.; Bateman, A.; Sweeney, B. Rfam 15: RNA families database in 2025. Nucleic Acids Res. 2025, 53, D258–D267. [Google Scholar] [CrossRef] [PubMed]
- Hu, S.; Hu, Y.; Mei, H.; Li, J.; Xuan, W.; Jeyaraj, A.; Zhao, Z.; Zhao, Y.; Han, R.; Chen, X. Genome-wide analysis of long non-coding RNAs (lncRNAs) in tea plants (Camellia sinensis) lateral roots in response to nitrogen application. Front. Plant Sci. 2023, 14, 1080427. [Google Scholar] [CrossRef] [PubMed]

| lncRNA Name | Plant Species | Biological Function | References |
|---|---|---|---|
| MISSEN (XLOC_057324) | Oryza sativa | Imprinted lncRNA in the endosperm; acts as a “decoy” for HeFP helicase protein, disrupts microtubule polymerization, regulates nuclear division rate and grain size | [85] |
| asDOG1 | Arabidopsis thaliana | Antisense transcript of DOG1; recruits PRC2 and silences DOG1, regulating seed dormancy depth | [86,87] |
| MUSHER | Arabidopsis thaliana | Integrates ABA and DOG1 signaling; strengthens seed dormancy under stress conditions (drought, high temperature) | [88] |
| AtR8 | Arabidopsis thaliana | Pol III-dependent lncRNA; interacts with WRKY46, activates AtEM6 gene, increases ABA sensitivity and enhances dormancy | [89] |
| TraesLNC1D26001.1 | Triticum aestivum | Activates TaABI5; delays germination by reinforcing ABA signaling; potential ceRNA role | [90] |
| VIVIPARY | Oryza sativa | Forms a complex with OsMSI1 and OsHDAC1; suppresses ABA signaling, promotes germination and pre-harvest sprouting | [91] |
| HID1 (HIDDEN TREASURE 1) | Arabidopsis thaliana | Red-light induced; binds to NCED9 intron, suppresses ABA biosynthesis and promotes GA biosynthesis, facilitating germination | [2,92,93] |
| SVR (Seed Vigor-Related) | Oryza sativa | Endosperm-specific lncRNA; regulates seed vigor via neighboring SAUR genes involved in auxin metabolism | [94] |
| WSGAR | Triticum aestivum | lncRNA cleaved by miR9678; regulates ABA/GA balance, influences resistance to pre-harvest sprouting | [87] |
| lnc663 | Triticum aestivum | Acts as ceRNA for miR1128; derepresses PDAT-like gene, modulates embryo lipid metabolism | [95] |
| COOLAIR | Arabidopsis thaliana | Regulates the flowering locus C (FLC) gene during vernalization. It plays a key role in controlling flowering time | [36,96] |
| lncRNA_00185 | Metasequoia glyptostroboides | miR167 decoy, stabilizes RCD1; enhances oxidative stress tolerance and seed viability | [97,98] |
| LNC_037529 | Oryza sativa | Produces alternative splicing isoforms associated with seed aging resistance; interacts with repair proteins (e.g., OsXRCC1) | [99] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Motor, A.; Puchta-Jasińska, M.; Bolc, P.; Boczkowska, M. Long Noncoding RNAs as Emerging Regulators of Seed Development, Germination, and Senescence. Int. J. Mol. Sci. 2025, 26, 8702. https://doi.org/10.3390/ijms26178702
Motor A, Puchta-Jasińska M, Bolc P, Boczkowska M. Long Noncoding RNAs as Emerging Regulators of Seed Development, Germination, and Senescence. International Journal of Molecular Sciences. 2025; 26(17):8702. https://doi.org/10.3390/ijms26178702
Chicago/Turabian StyleMotor, Adrian, Marta Puchta-Jasińska, Paulina Bolc, and Maja Boczkowska. 2025. "Long Noncoding RNAs as Emerging Regulators of Seed Development, Germination, and Senescence" International Journal of Molecular Sciences 26, no. 17: 8702. https://doi.org/10.3390/ijms26178702
APA StyleMotor, A., Puchta-Jasińska, M., Bolc, P., & Boczkowska, M. (2025). Long Noncoding RNAs as Emerging Regulators of Seed Development, Germination, and Senescence. International Journal of Molecular Sciences, 26(17), 8702. https://doi.org/10.3390/ijms26178702