Pan-Plastome Analysis Reveals the Genetic Diversity and Genetic Divergence of Adenocaulon himalaicum (Asteraceae)
Abstract
1. Introduction
2. Results
2.1. Comparative Analysis of the Pan-Plastome in A. himalaicum
2.2. Codon Usage Bias and Repeat Sequence Analysis

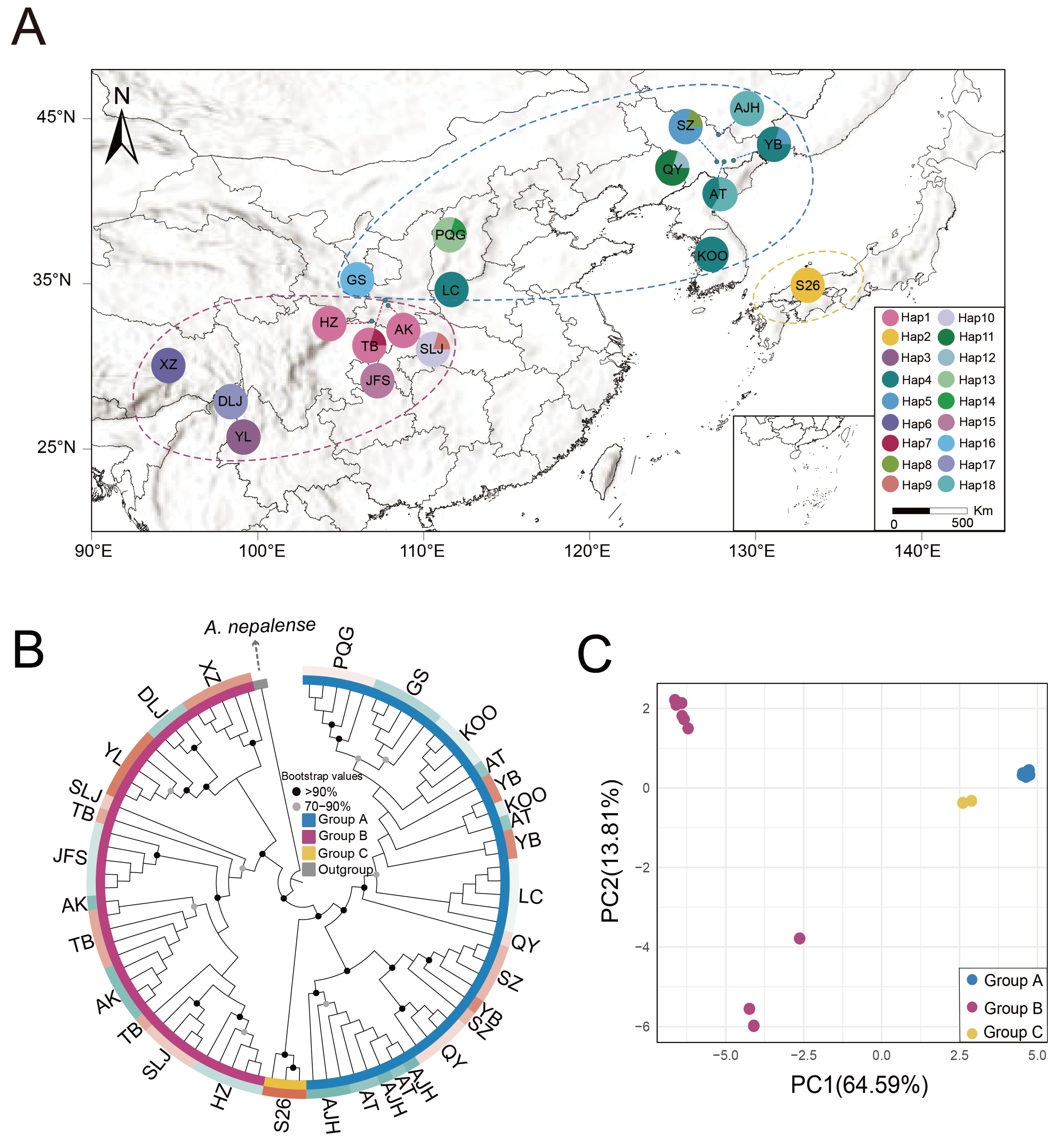
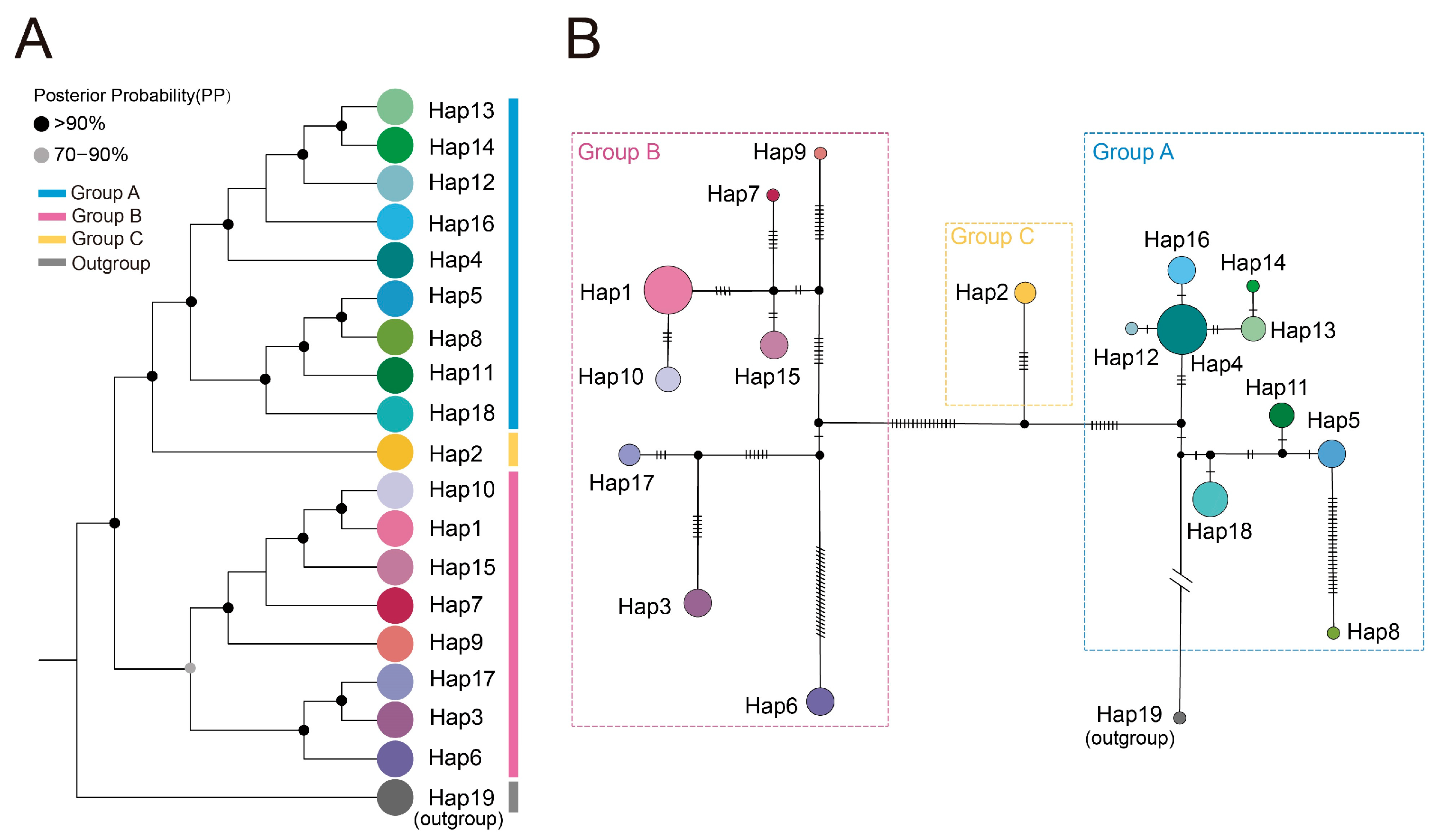
2.3. Population Structure and Haplotype Network Analysis
2.4. Analyses of Genetic Diversity and Genetic Differentiation
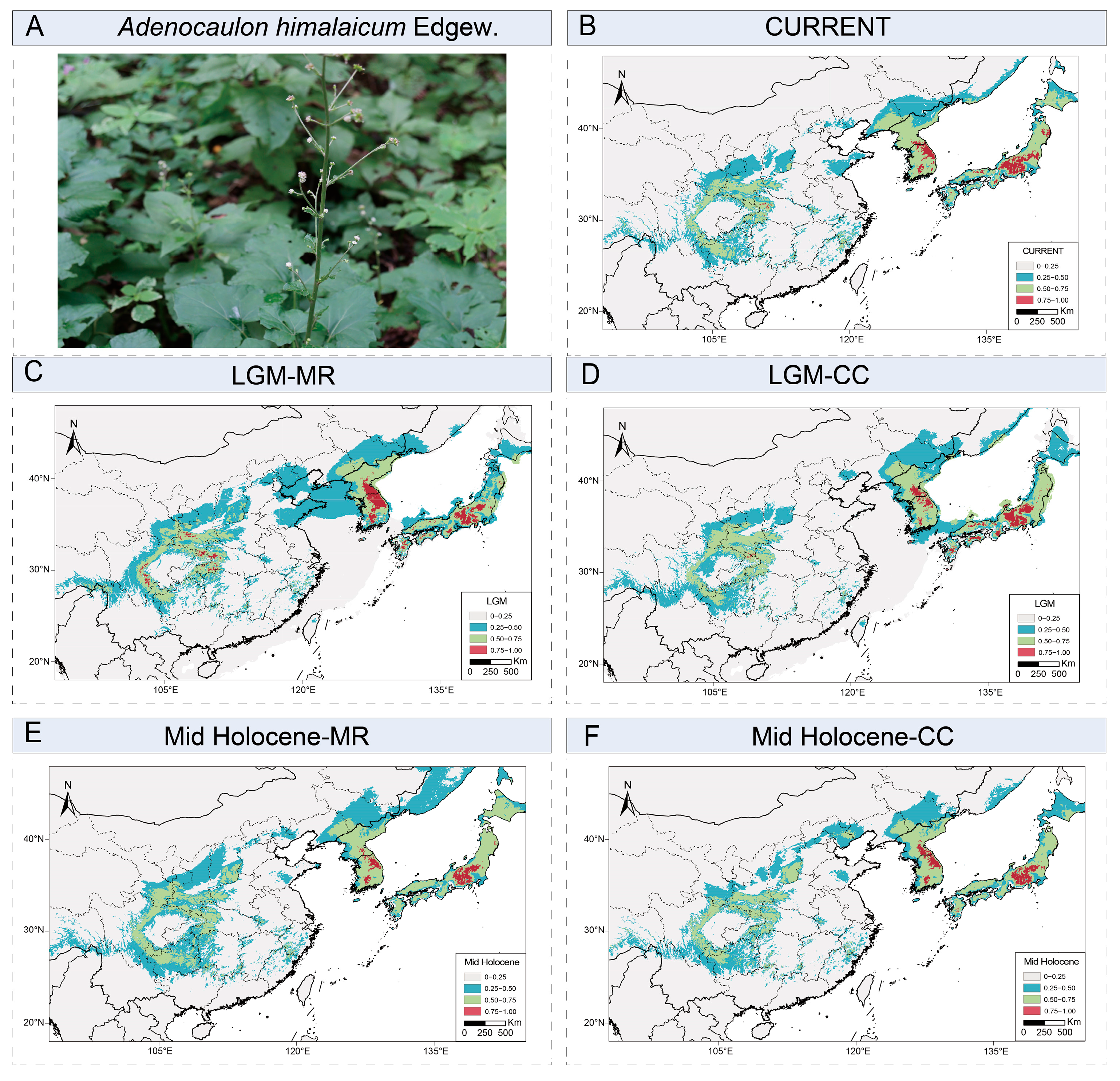
2.5. Ecological Niche Modeling

3. Discussion
3.1. Pan-Plastome Characteristics and Evolution in the Adenocaulon himalaicum
3.2. Genetic Diversity, Genetic Structure and Conservation Implications
4. Materials and Methods
4.1. Plant Materials and Plastome Assembly
4.2. Comparative Analysis of the Pan-Plastome
4.3. Population Structure and Phylogroup Analysis
4.4. Genetic Diversity and Genetic Differentiation
4.5. Species Distribution Modeling
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Deng, T.; Chen, Y.; Wang, H.; Zhang, X.; Volis, S.; Yusupov, Z.; Qian, H.; Sun, H. Molecular phylogeny and biogeography of Adenocaulon highlight the biogeographic links between New World and Old World. Front. Ecol. Evol. 2018, 5, 162. [Google Scholar] [CrossRef]
- Funk, V.A.; Hind, D.J. Typification of species names in Adenocaulon and Eriachaenium (Compositae/Asteraceae, Subfamily Mutisioideae, Tribe Mutisieae, Subtribe Adenocaulinae). PhytoKeys 2016, 69, 121–128. [Google Scholar] [CrossRef]
- Li, J. Flora of China. Harv. Pap. Bot. 2007, 13, 301–302. [Google Scholar] [CrossRef]
- Feng, J.X.; Sun, Y.; Xia, G.Q.; Li, L.; Zang, H. Research progress on the phytochemistry and pharmacological activities of the genus Adenocaulon Hook. Ginseng Res. 2023, 35, 49–51. [Google Scholar]
- Lee, S.Y.; Yoon, N.; Uy, N.P.; Choi, C.H.; Lee, S. Phytochemical Profiling and Antioxidant Activity of True Leaves and Cotyledons of Adenocaulon himalaicum. ChemEngineering 2025, 9, 31. [Google Scholar] [CrossRef]
- Ahn, H.S.; Kim, H.J.; Na, C.; Jang, D.S.; Shin, Y.K.; Lee, S.H. The protective effect of Adenocaulon himalaicum Edgew. and its bioactive compound neochlorogenic acid against UVB-Induced skin damage in human dermal fibroblasts and epidermal keratinocytes. Plants 2021, 10, 1669. [Google Scholar] [CrossRef]
- Yun, J.H.; Lee, S.B.; Kang, K.; Lee, E.H.; Lee, H.J.; Jung, S.H.; Nho, C.W. Bifunctional Chemopreventive Effects of Adenocaulon himalaicum Through Induction of Detoxification Enzymes and Apoptosis. J. Med. Food 2013, 16, 701–710. [Google Scholar] [CrossRef]
- Lee, Y.M.; Son, E.; Kim, D.S.; Shim, K.S.; Yu, S.H. Evaluating the Anti-Inflammatory and Chondroprotective Effects of Adenocaulon himalaicum Extract Through Network Pharmacology and Experimental Validation. Int. J. Mol. Sci. 2025, 26, 877. [Google Scholar] [CrossRef]
- Yu, J.; Yan, Y.; Wang, G.; Zhang, Q. Habitat fragmentation reduced plant functional diversity in the agro-pastoral ecotone of Inner Mongolia. Ecol. Indic. 2024, 169, 112975. [Google Scholar] [CrossRef]
- Haddad, N.M.; Brudvig, L.A.; Clobert, J.; Davies, K.F.; Gonzalez, A.; Holt, R.D.; Lovejoy, T.E.; Sexton, J.O.; Austin, M.P.; Collins, C.D.; et al. Habitat fragmentation and its lasting impact on Earth’s ecosystems. Sci. Adv. 2015, 1, e1500052. [Google Scholar] [CrossRef]
- Suyal, R.; Jugran, A.K.; Rawal, R.S.; Bhatt, I.D. Morphological, phytochemical and genetic diversity of threatened Polygonatum verticillatum (L.) All. populations of different altitudes and habitat types in Himalayan region. Physiol. Mol. Biol. Plants 2021, 27, 1795–1809. [Google Scholar] [CrossRef]
- Pais, A.L.; Li, X.; Jenny Xiang, Q.Y. Discovering variation of secondary metabolite diversity and its relationship with disease resistance in Cornus florida L. Ecol. Evol. 2018, 8, 5619–5636. [Google Scholar] [CrossRef] [PubMed]
- Kim, G.B.; Lim, C.E.; Kim, J.S.; Kim, K.; Lee, J.H.; Yu, H.J.; Mun, J.H. Comparative chloroplast genome analysis of Artemisia (Asteraceae) in East Asia: Insights into evolutionary divergence and phylogenomic implications. BMC Genom. 2020, 21, 415. [Google Scholar] [CrossRef]
- Mahai, R.; Sheng, S.; Wang, X.; Yuan, J.; Mu, Z. Comparative analysis of complete chloroplast genomes of 14 Asteraceae species. Mol. Biol. Rep. 2024, 51, 1094. [Google Scholar] [CrossRef]
- Chen, C.; Luo, D.; Wang, Z.; Miao, Y.; Liu, Q.; Zhao, T.; Liu, D. Complete chloroplast genomes of eight Artemisia species: Comparative analysis, molecular identification, and phylogenetic analysis. Plant Biol. 2024, 26, 257–269. [Google Scholar] [CrossRef]
- Zhou, J.; Zhang, S.; Wang, J.; Shen, H.; Ai, B.; Gao, W.; Zhang, C.; Fei, Q.; Yuan, D.; Wu, Z.; et al. Chloroplast genomes in Populus (Salicaceae): Comparisons from an intensively sampled genus reveal dynamic patterns of evolution. Sci. Rep. 2021, 11, 9471. [Google Scholar] [CrossRef] [PubMed]
- Daniell, H.; Jin, S.; Zhu, X.G.; Gitzendanner, M.A.; Soltis, D.E.; Soltis, P.S. Green giant—A tiny chloroplast genome with mighty power to produce high-value proteins: History and phylogeny. Plant Biotechnol. J. 2021, 19, 430–447. [Google Scholar] [CrossRef]
- Wang, N.; Chen, S.; Xie, L.; Wang, L.; Feng, Y.; Lv, T.; Fang, Y.; Ding, H. The complete chloroplast genomes of three Hamamelidaceae species: Comparative and phylogenetic analyses. Ecol. Evol. 2022, 12, e8637. [Google Scholar] [CrossRef]
- Chang, H.; Zhang, L.; Xie, H.; Liu, J.; Xi, Z.; Xu, X. The conservation of chloroplast genome structure and improved resolution of infrafamilial relationships of Crassulaceae. Front. Plant Sci. 2021, 12, 631884. [Google Scholar] [CrossRef]
- Yang, L.; Abduraimov, O.; Tojibaev, K.; Shomurodov, K.; Zhang, Y.M.; Li, W.J. Analysis of complete chloroplast genome sequences and insight into the phylogenetic relationships of Ferula L. BMC Genom. 2022, 23, 643. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Zhang, X.; Landis, J.B.; Zhang, H.; Deng, T.; Sun, H.; Wang, H. Plastome evolution in Dolomiaea (Asteraceae, Cardueae) using phylogenomic and comparative analyses. Front. Plant Sci. 2020, 11, 376. [Google Scholar] [CrossRef]
- Cho, M.-S.; Yang, J.; Kim, S.-H.; Crawford, D.J.; Stuessy, T.F.; López-Sepúlveda, P.; Kim, S.-C. Plastid phylogenomics of Robinsonia (Senecioneae; Asteraceae), endemic to the Juan Fernández Islands: Insights into structural organization and molecular evolution. BMC Plant Biol. 2024, 24, 1016. [Google Scholar] [CrossRef]
- Jia, M.; Wang, J.; Cao, D.; Jiang, C.; Li, W.; Tembrock, L.R.; Xing, G.; Li, S.; Wu, Z. The pan-plastome of Hemerocallis citrina reveals new insights into the genetic diversity and cultivation history of an economically important food plant. BMC Plant Biol. 2024, 24, 44. [Google Scholar] [CrossRef] [PubMed]
- Chen, N.; Sha, L.N.; Wang, Y.L.; Yin, L.J.; Zhang, Y.; Wang, Y.; Wu, D.D.; Kang, H.Y.; Zhang, H.Q.; Zhou, Y.H. Variation in plastome sizes accompanied by evolutionary history in monogenomic Triticeae (Poaceae: Triticeae). Front. Plant Sci. 2021, 12, 741063. [Google Scholar] [CrossRef]
- Xia, L.; Wang, H.; Zhao, X.; Obel, H.O.; Yu, X.; Lou, Q.; Chen, J.; Cheng, C. Chloroplast pan-genomes and comparative transcriptomics reveal genetic variation and temperature adaptation in the cucumber. Int. J. Mol. Sci. 2023, 24, 8943. [Google Scholar] [CrossRef] [PubMed]
- Wicke, S.; Schneeweiss, G.M.; de Pamphilis, C.W.; Müller, K.F.; Quandt, D. The evolution of the plastid chromosome in land plants: Gene content, gene order, gene function. Plant Mol. Biol. 2011, 76, 273–297. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.; Aishan, S.; Zhu, J.; Qin, Y.; Liu, J.; Liu, H.; Tie, J.; Wang, J.; Qin, R. Chloroplast genomes and phylogenetic analysis of three Carthamus (Asteraceae) Species. Int. J. Mol. Sci. 2023, 24, 15634. [Google Scholar] [CrossRef]
- Pascual-Díaz, J.P.; Garcia, S.; Vitales, D. Plastome Diversity and Phylogenomic Relationships in Asteraceae. Plants 2021, 10, 2699. [Google Scholar] [CrossRef]
- Shen, X.; Guo, S.; Yin, Y.; Zhang, J.; Yin, X.; Liang, C.; Wang, Z.; Huang, B.; Liu, Y.; Xiao, S.; et al. Complete chloroplast genome sequence and phylogenetic analysis of Aster tataricus. Molecules 2018, 23, 2426. [Google Scholar] [CrossRef]
- Ping, J.; Hao, J.; Li, J.; Yang, Y.; Su, Y.; Wang, T. Loss of the IR region in conifer plastomes: Changes in the selection pressure and substitution rate of protein-coding genes. Ecol. Evol. 2022, 12, e8499. [Google Scholar] [CrossRef]
- Maréchal, A.; Brisson, N. Recombination and the maintenance of plant organelle genome stability. New Phytol. 2010, 186, 299–317. [Google Scholar] [CrossRef] [PubMed]
- Li, F.W.; Kuo, L.Y.; Pryer, K.M.; Rothfels, C.J. Genes translocated into the plastid inverted repeat show decelerated substitution rates and elevated GC content. Genome Biol. Evol. 2016, 8, 2452–2458. [Google Scholar] [CrossRef]
- Sun, Y.; Moore, M.J.; Yue, L.; Feng, T.; Chu, H.; Chen, S.; Ji, Y.; Wang, H.; Li, J. Chloroplast phylogeography of the East Asian Arcto-Tertiary relict Tetracentron sinense (Trochodendraceae). J. Biogeogr. 2014, 41, 1721–1732. [Google Scholar] [CrossRef]
- Chumley, T.W.; Palmer, J.D.; Mower, J.P.; Fourcade, H.M.; Calie, P.J.; Boore, J.L.; Jansen, R.K. The complete chloroplast genome sequence of Pelargonium x hortorum: Organization and evolution of the largest and most highly rearranged chloroplast genome of land plants. Mol. Biol. Evol. 2006, 23, 2175–2190. [Google Scholar] [CrossRef]
- Chen, L.Q.; Li, X.; Yao, X.; Li, D.Z.; Barrett, C.; dePamphilis, C.W.; Yu, W.B. Variations and reduction of plastome are associated with the evolution of parasitism in Convolvulaceae. Plant Mol. Biol. 2024, 114, 40, Erratum in Plant Mol. Biol. 2024, 114, 58. [Google Scholar] [CrossRef]
- Zhang, Y.; Ma, J.; Yang, B.; Li, R.; Zhu, W.; Sun, L.; Tian, J.; Zhang, L. The complete chloroplast genome sequence of Taxus chinensis var. mairei (Taxaceae): Loss of an inverted repeat region and comparative analysis with related species. Gene 2014, 540, 201–209. [Google Scholar] [CrossRef]
- Lin, C.P.; Huang, J.P.; Wu, C.S.; Hsu, C.Y.; Chaw, S.M. Comparative chloroplast genomics reveals the evolution of pinaceae genera and subfamilies. Genome Biol. Evol. 2010, 2, 504–517. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.S.; Chaw, S.M. Highly rearranged and size-variable chloroplast genomes in conifers II clade (cupressophytes): Evolution towards shorter intergenic spacers. Plant Biotechnol. J. 2014, 12, 344–353. [Google Scholar] [CrossRef]
- Kim, K.J.; Choi, K.S.; Jansen, R.K. Two chloroplast DNA inversions originated simultaneously during the early evolution of the sunflower family (Asteraceae). Mol. Biol. Evol. 2005, 22, 1783–1792. [Google Scholar] [CrossRef]
- Walker, J.F.; Zanis, M.J.; Emery, N.C. Correction to “Comparative analysis of complete chloroplast genome sequence and inversion variation in Lasthenia burkei (Madieae, Asteraceae)”. Am. J. Bot. 2015, 102, 1008. [Google Scholar] [CrossRef]
- Sablok, G.; Amiryousefi, A.; He, X.; Hyvönen, J.; Poczai, P. Sequencing the plastid genome of giant ragweed (Ambrosia trifida, Asteraceae) from a herbarium specimen. Front. Plant Sci. 2019, 10, 218. [Google Scholar] [CrossRef]
- Qu, T.; Li, T.; Chen, X.; Zheng, X.; Chen, H.; Pang, L.; Fu, Z. The complete chloroplast genome of Leibnitzia nepalensis (Kunze) Kitamura, 1983 (Asteraceae, Mutisieae) and its phylogenetic analysis. Mitochondrial DNA Part B 2025, 10, 212–217. [Google Scholar] [CrossRef]
- Ru, B.; Wang, T.; Liu, Y.; Zhao, X.; Lei, M. The complete chloroplast genome sequence of leibnitzia anandria (linnaeus) turczaninow. Mitochondrial DNA B Resour. 2024, 9, 578–582. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Chen, J.; Li, Y.; Liu, A.; Li, A.; Yin, M.; Shrestha, N.; Liu, J.; Ren, G. Extensive genomic rearrangements mediated by repetitive sequences in plastomes of Medicago and its relatives. BMC Plant Biol 2021, 21, 421. [Google Scholar] [CrossRef]
- Sawicki, J.; Bączkiewicz, A.; Buczkowska, K.; Górski, P.; Krawczyk, K.; Mizia, P.; Myszczyński, K.; Ślipiko, M.; Szczecińska, M. The increase of simple sequence repeats during diversification of Marchantiidae, an early land plant lineage, leads to the first known expansion of inverted repeats in the evolutionarily-stable structure of liverwort plastomes. Genes 2020, 11, 299. [Google Scholar] [CrossRef] [PubMed]
- Crow, T.; Ta, J.; Nojoomi, S.; Aguilar-Rangel, M.R.; Torres Rodríguez, J.V.; Gates, D.; Rellán-Álvarez, R.; Sawers, R.; Runcie, D. Gene regulatory effects of a large chromosomal inversion in highland maize. PLoS Genet. 2020, 16, e1009213. [Google Scholar] [CrossRef]
- Wu, C.-S.; Sudianto, E.; Chaw, S.-M. Tight association of genome rearrangements with gene expression in conifer plastomes. BMC Plant Biol. 2021, 21, 33. [Google Scholar] [CrossRef]
- Sasaki, Y.; Nagano, Y. Plant Acetyl-CoA Carboxylase: Structure, biosynthesis, regulation, and sene manipulation for plant breeding. Biosci. Biotechnol. Biochem. 2004, 68, 1175–1184. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Yang, J.B.; Wang, H.; Song, Y.; Corlett, R.T.; Yao, X.; Li, D.Z.; Yu, W.B. Plastid NDH pseudogenization and gene loss in a recently derived lineage from the largest hemiparasitic plant genus Pedicularis (Orobanchaceae). Plant Cell Physiol. 2021, 62, 971–984. [Google Scholar] [CrossRef]
- Yu, J.; Li, J.; Zuo, Y.; Qin, Q.; Zeng, S.; Rennenberg, H.; Deng, H. Plastome variations reveal the distinct evolutionary scenarios of plastomes in the subfamily Cereoideae (Cactaceae). BMC Plant Biol. 2023, 23, 132. [Google Scholar] [CrossRef]
- Rousseau-Gueutin, M.; Huang, X.; Higginson, E.; Ayliffe, M.; Day, A.; Timmis, J.N. Potential functional replacement of the plastidic Acetyl-CoA carboxylase subunit (accD) gene by recent transfers to the nucleus in some angiosperm lineages. Plant Physiol. 2013, 161, 1918–1929. [Google Scholar] [CrossRef]
- Sudianto, E.; Chaw, S.M. Two independent plastid accD transfers to the nuclear genome of gnetum and other insights on Acetyl-CoA carboxylase evolution in gymnosperms. Genome Biol. Evol. 2019, 11, 1691–1705. [Google Scholar] [CrossRef] [PubMed]
- Shrestha, B.; Gilbert, L.E.; Ruhlman, T.A.; Jansen, R.K. Rampant Nuclear Transfer and Substitutions of Plastid Genes in Passiflora. Genome Biol. Evol. 2020, 12, 1313–1329. [Google Scholar] [CrossRef]
- Lyko, P.; Wicke, S. Genomic reconfiguration in parasitic plants involves considerable gene losses alongside global genome size inflation and gene births. Plant Physiol 2021, 186, 1412–1423. [Google Scholar] [CrossRef] [PubMed]
- McNeal, J.R.; Kuehl, J.V.; Boore, J.L.; de Pamphilis, C.W. Complete plastid genome sequences suggest strong selection for retention of photosynthetic genes in the parasitic plant genus Cuscuta. BMC Plant Biol. 2007, 7, 57. [Google Scholar] [CrossRef]
- Cusimano, N.; Wicke, S. Massive intracellular gene transfer during plastid genome reduction in nongreen Orobanchaceae. New Phytol. 2016, 210, 680–693. [Google Scholar] [CrossRef]
- Slotte, T.; Foxe, J.P.; Hazzouri, K.M.; Wright, S.I. Genome-wide evidence for efficient positive and purifying selection in Capsella grandiflora, a plant species with a large effective population size. Mol. Biol. Evol. 2010, 27, 1813–1821. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Han, Z.; Jiang, H.; Tian, D.; Yang, S. Strong positive selection drives rapid diversification of R-genes in Arabidopsis relatives. J. Mol. Evol. 2010, 70, 137–148. [Google Scholar] [CrossRef]
- Piot, A.; Hackel, J.; Christin, P.A.; Besnard, G. One-third of the plastid genes evolved under positive selection in PACMAD grasses. Planta 2018, 247, 255–266. [Google Scholar] [CrossRef]
- Robbins, E.H.; Kelly, S. The evolutionary constraints on angiosperm chloroplast adaptation. Genome Biol. Evol. 2023, 15, evad101. [Google Scholar] [CrossRef]
- Camus, M.F.; Alexander-Lawrie, B.; Sharbrough, J.; Hurst, G.D.D. Inheritance through the cytoplasm. Heredity 2022, 129, 31–43. [Google Scholar] [CrossRef]
- Keeling, P.J. The endosymbiotic origin, diversification and fate of plastids. Philos. Trans. R. Soc. B Biol. Sci. 2010, 365, 729–748. [Google Scholar] [CrossRef] [PubMed]
- Han, B.; Zhang, M.-J.; Xian, Y.; Xu, H.; Cui, C.-C.; Liu, D.; Wang, L.; Li, D.-Z.; Li, W.-Q.; Xie, X.-M. Variations in genetic diversity in cultivated Pistacia chinensis. Front. Plant Sci. 2022, 13, 1030647. [Google Scholar] [CrossRef]
- Jordano, P. Pollen, seeds and genes: The movement ecology of plants. Heredity 2010, 105, 329–330. [Google Scholar] [CrossRef]
- Petit, R.J.; Duminil, J.; Fineschi, S.; Hampe, A.; Salvini, D.; Vendramin, G.G. Comparative organization of chloroplast, mitochondrial and nuclear diversity in plant populations. Mol. Ecol. 2005, 14, 689–701. [Google Scholar] [CrossRef] [PubMed]
- Bai, W.N.; Wang, W.T.; Zhang, D.Y. Phylogeographic breaks within Asian butternuts indicate the existence of a phytogeo-graphic divide in East Asia. New Phytol. 2016, 209, 1757–1772. [Google Scholar] [CrossRef]
- Guo, X.D.; Wang, H.F.; Bao, L.; Wang, T.M.; Bai, W.N.; Ye, J.W.; Ge, J.P. Evolutionary history of a widespread tree species Acer mono in East Asia. Ecol. Evol. 2014, 4, 4332–4345. [Google Scholar] [CrossRef]
- Ye, J.W.; Bai, W.N.; Bao, L.; Wang, T.M.; Wang, H.F.; Ge, J.P. Sharp genetic discontinuity in the aridity-sensitive Lindera obtusiloba (Lauraceae): Solid evidence supporting the Tertiary floral subdivision in East Asia. J. Biogeogr. 2017, 44, 2082–2095. [Google Scholar] [CrossRef]
- Qiu, Y.; Lu, Q.; Zhang, Y.; Cao, Y. Phylogeography of East Asia’s Tertiary relict plants: Current progress and future prospects. Biodivers. Sci. 2017, 25, 136. [Google Scholar] [CrossRef]
- Qiu, Y.X.; Sun, Y.; Zhang, X.P.; Lee, J.; Fu, C.X.; Comes, H.P. Molecular phylogeography of East Asian Kirengeshoma (Hydrangeaceae) in relation to Quaternary climate change and landbridge configurations. New Phytol. 2009, 183, 480–495. [Google Scholar] [CrossRef]
- Qi, X.S.; Yuan, N.; Comes, H.P.; Sakaguchi, S.; Qiu, Y.X. A strong ‘filter’ effect of the East China Sea land bridge for East Asia’s temperate plant species: Inferences from molecular phylogeography and ecological niche modelling of Platycrater arguta (Hydrangeaceae). BMC Evol. Biol. 2014, 14, 41. [Google Scholar] [CrossRef] [PubMed]
- Mao, K.S.; Wang, Y.; Liu, J.Q. Evolutionary origin of species diversity on the Qinghai–Tibet Plateau. J. Syst. Evol. 2021, 59, 1142–1158. [Google Scholar] [CrossRef]
- Shi, Y.; Zheng, B.; Li, S. Last glaciation and maximum glaciation in the Qinghai-Xizang (Tibet) Plateau: A controversy to M. Kuhle’s Ice Sheet hypothesis. Chin. Geogr. Sci. 1992, 2, 293–311. [Google Scholar] [CrossRef]
- Li, X.; Ruhsam, M.; Wang, Y.; Zhang, H.Y.; Fan, X.Y.; Zhang, L.; Wang, J.; Mao, K.S. Wind-dispersed seeds blur phylogeographic breaks: The complex evolutionary history of Populus lasiocarpa around the Sichuan Basin. Plant Divers 2023, 45, 156–168. [Google Scholar] [CrossRef]
- Kirchner, N.; Greve, R.; Stroeven, A.P.; Heyman, J. Paleoglaciological reconstructions for the Tibetan plateau during the last glacial cycle: Evaluating numerical ice sheet simulations driven by GCM-ensembles. Quat. Sci. Rev. 2011, 30, 248–267. [Google Scholar] [CrossRef]
- Xie, X.F.; Yan, H.F.; Wang, F.Y.; Ge, X.J.; Hu, C.M.; Hao, G. Chloroplast DNA phylogeography of Primula ovalifolia in central and adjacent southwestern China: Past gradual expansion and geographical isolation. J. Syst. Evol. 2012, 50, 284–294. [Google Scholar] [CrossRef]
- Guan, B.C.; Fu, C.X.; Qiu, Y.X.; Zhou, S.L.; Comes, H.P. Genetic structure and breeding system of a rare understory herb, Dysosma versipellis (Berberidaceae), from temperate deciduous forests in China. Am. J. Bot. 2010, 97, 111–122. [Google Scholar] [CrossRef] [PubMed]
- Luo, S.; He, Y.; Ning, G.; Zhang, J.; Ma, G.; Bao, M. Genetic diversity and genetic structure of different populations of the endangered species Davidia involucrata in China detected by inter-simple sequence repeat analysis. Trees 2011, 25, 1063–1071. [Google Scholar] [CrossRef]
- Li, Z.; Ji, Q.; Yang, Y.; Xu, M.; Guan, Y. Low genetic diversity and weak population structure of Albizia odoratissima on Hainan Island. BMC Plant Biol. 2025, 25, 395. [Google Scholar] [CrossRef]
- Navascués, M.; Vaxevanidou, Z.; González-Martínez, S.C.; Climent, J.; Gil, L.; Emerson, B.C. Chloroplast microsatellites reveal colonization and metapopulation dynamics in the Canary Island pine. Mol. Ecol. 2006, 15, 2691–2698. [Google Scholar] [CrossRef]
- Merklinger, F.; Zheng, Y.; Luebert, F.; Harpke, D.; Böhnert, T.; Stoll, A.; Koch, M.; Blattner, F.; Wiehe, T.; Quandt, D. Population genomics of Tillandsia landbeckii reveals unbalanced genetic diversity and founder effects in the Atacama Desert. Glob. Planet. Change 2020, 184, 103076. [Google Scholar] [CrossRef]
- Vinogradova, Y.K.; Galkina, M.A.; Mayorov, S.R.; Kartashova, A.S.; Shelepova, O.V. Biomorpholgy and taxonomic Status of Adenocaulon adhaerescens Maxim. (Asteraceae), an invasive species in the moscow region. Russ. J. Biol. Invasions 2022, 13, 439–453. [Google Scholar] [CrossRef]
- Cao, B.; Yang, X.; Li, B.; Lu, Y.; Wen, J. Diurnal variation in cloud and precipitation characteristics in summer over the Tibetan plateau and Sichuan basin. Remote Sens. 2022, 14, 2711. [Google Scholar] [CrossRef]
- Chung, M.Y.; Chung, M.G.; López-Pujol, J.; Ren, M.X.; Zhang, Z.Y.; Park, S.J. Were the main mountain ranges in the Korean Peninsula a glacial refugium for plants? Insights from the congeneric pair Lilium cernuum—Lilium amabile. Biochem. Syst. Ecol. 2014, 53, 36–45. [Google Scholar] [CrossRef]
- Ikeda, H.; Senni, K.; Fujii, N.; Setoguchi, H. High mountains of the Japanese archipelago as refugia for arctic–alpine plants: Phylogeography of Loiseleuria procumbens (L.) Desvaux (Ericaceae). Biol. J. Linn. Soc. 2009, 97, 403–412. [Google Scholar] [CrossRef]
- Ikeda, H.; Yakubov, V.; Barkalov, V.; Setoguchi, H. Molecular evidence for ancient relicts of arctic-alpine plants in East Asia. New Phytol. 2014, 203, 980–988. [Google Scholar] [CrossRef]
- Doyle, J.J.; Doyle, J.L. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 1987, 19, 11–15. [Google Scholar]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; Subgroup, G.P.D.P. The sequence alignment/map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed]
- Dierckxsens, N.; Mardulyn, P.; Smits, G. NOVOPlasty: De novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 2016, 45, e18. [Google Scholar]
- Shi, L.; Chen, H.; Jiang, M.; Wang, L.; Wu, X.; Huang, L.; Liu, C. CPGAVAS2, an integrated plastome sequence annotator and analyzer. Nucleic Acids Res. 2019, 47, W65–W73. [Google Scholar] [CrossRef]
- Greiner, S.; Lehwark, P.; Bock, R. OrganellarGenomeDRAW (OGDRAW) version 1.3.1: Expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 2019, 47, W59–W64. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Rozas, J.; Ferrer-Mata, A.; Sánchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sánchez-Gracia, A. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef]
- Peden, J.F. Analysis of Codon Usage. Ph.D. Thesis, University of Nottingham, Nottingham, UK, 1999. Volume 90. pp. 73–74. [Google Scholar]
- Benson, G. Tandem repeats finder: A program to analyze DNA sequences. Nucleic Acids Res. 1999, 27, 573–580. [Google Scholar] [CrossRef]
- Thiel, T.; Michalek, W.; Varshney, R.; Graner, A. Exploiting EST databases for the development and characterization of gene-derived SSR-markers in barley (Hordeum vulgare L.). Theor. Appl. Genet. 2003, 106, 411–422. [Google Scholar] [CrossRef]
- Kurtz, S.; Choudhuri, J.V.; Ohlebusch, E.; Schleiermacher, C.; Stoye, J.; Giegerich, R. REPuter: The manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 2001, 29, 4633–4642. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z. PAML 4: Phylogenetic Analysis by Maximum Likelihood. Mol. Biol. Evol. 2007, 24, 1586–1591. [Google Scholar] [CrossRef]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; von Haeseler, A.; Lanfear, R. IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic Era. Mol. Biol. Evol. 2020, 37, 1530–1534, Erratum in Mol. Biol. Evol. 2020, 37, 2461. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Teslenko, M.; Van Der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef] [PubMed]
- Swofford, D. PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods), version 4, Sinauer Associates: Sunderland, MA, USA, 2003.
- Nylander, J. MrModeltest V2. Program Distributed by the Author. Bioinformatics 2004, 24, 581–583. [Google Scholar] [CrossRef]
- Rambaut, A.; Drummond, A.J.; Xie, D.; Baele, G.; Suchard, M.A. Posterior summarization in bayesian phylogenetics using Tracer 1.7. Syst. Biol. 2018, 67, 901–904. [Google Scholar] [CrossRef]
- Bradbury, P.J.; Zhang, Z.; Kroon, D.E.; Casstevens, T.M.; Ramdoss, Y.; Buckler, E.S. TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics 2007, 23, 2633–2635. [Google Scholar] [CrossRef]
- Leigh, J.W.; Bryant, D.; Nakagawa, S. POPART: Full-feature software for haplotype network construction. Methods Ecol. Evol. 2015, 6, 1110–1116. [Google Scholar] [CrossRef]
- Clement, M.; Posada, D.; Crandall, K.A. TCS: A computer program to estimate gene genealogies. Mol. Ecol. 2000, 9, 1657–1659. [Google Scholar] [CrossRef]
- Pons, O.; Petit, R.J. Measwring and testing genetic differentiation with ordered versus unordered alleles. Genetics 1996, 144, 1237–1245. [Google Scholar] [CrossRef] [PubMed]
- Excoffier, L.; Lischer, H.E. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 2010, 10, 564–567. [Google Scholar] [CrossRef]
- Hijmans, R.J.; Williams, E.; Vennes, C.; Hijmans, M.R.J. Package ‘geosphere’. Spherical Trigonometry 2017, 1, 1–45. [Google Scholar]
- Fick, S.E.; Hijmans, R.J. WorldClim 2: New 1-km spatial resolution climate surfaces for global land areas. Int. J. Climatol. 2017, 37, 4302–4315. [Google Scholar] [CrossRef]
- Oksanen, J.; Kindt, R.; Legendre, P.; O’Hara, B.; Stevens, M.H.H.; Oksanen, M.J.; Suggests, M. The vegan package. Community Ecol. Package 2007, 10, 719. [Google Scholar]
- Phillips, S.J.; Dudík, M. Modeling of species distributions with Maxent: New extensions and a comprehensive evaluation. Echography 2008, 31, 161–175. [Google Scholar] [CrossRef]
- Hijmans, R.J.; Cameron, S.E.; Parra, J.L.; Jones, P.G.; Jarvis, A. Very highresolution interpolated climate surfaces for global land areas. Int. J. Climatol. 2005, 25, 1965–1978. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lin, N.; He, Y.; Wang, X.; Wang, Y.; Wang, J.; Li, Y. Pan-Plastome Analysis Reveals the Genetic Diversity and Genetic Divergence of Adenocaulon himalaicum (Asteraceae). Int. J. Mol. Sci. 2025, 26, 8594. https://doi.org/10.3390/ijms26178594
Lin N, He Y, Wang X, Wang Y, Wang J, Li Y. Pan-Plastome Analysis Reveals the Genetic Diversity and Genetic Divergence of Adenocaulon himalaicum (Asteraceae). International Journal of Molecular Sciences. 2025; 26(17):8594. https://doi.org/10.3390/ijms26178594
Chicago/Turabian StyleLin, Nan, Yuxuan He, Xiankun Wang, Yakun Wang, Jinhao Wang, and Yang Li. 2025. "Pan-Plastome Analysis Reveals the Genetic Diversity and Genetic Divergence of Adenocaulon himalaicum (Asteraceae)" International Journal of Molecular Sciences 26, no. 17: 8594. https://doi.org/10.3390/ijms26178594
APA StyleLin, N., He, Y., Wang, X., Wang, Y., Wang, J., & Li, Y. (2025). Pan-Plastome Analysis Reveals the Genetic Diversity and Genetic Divergence of Adenocaulon himalaicum (Asteraceae). International Journal of Molecular Sciences, 26(17), 8594. https://doi.org/10.3390/ijms26178594





