Optimizing Tissue Sampling Timing for Accurate Gene Expression Analysis
Abstract
1. Introduction
2. Results
2.1. Exploratory Dataset Analysis: Uncovering Insights into Gene Expression Changes
2.2. Validation Dataset Analysis: Confirming Gene Expression Patterns
3. Discussion
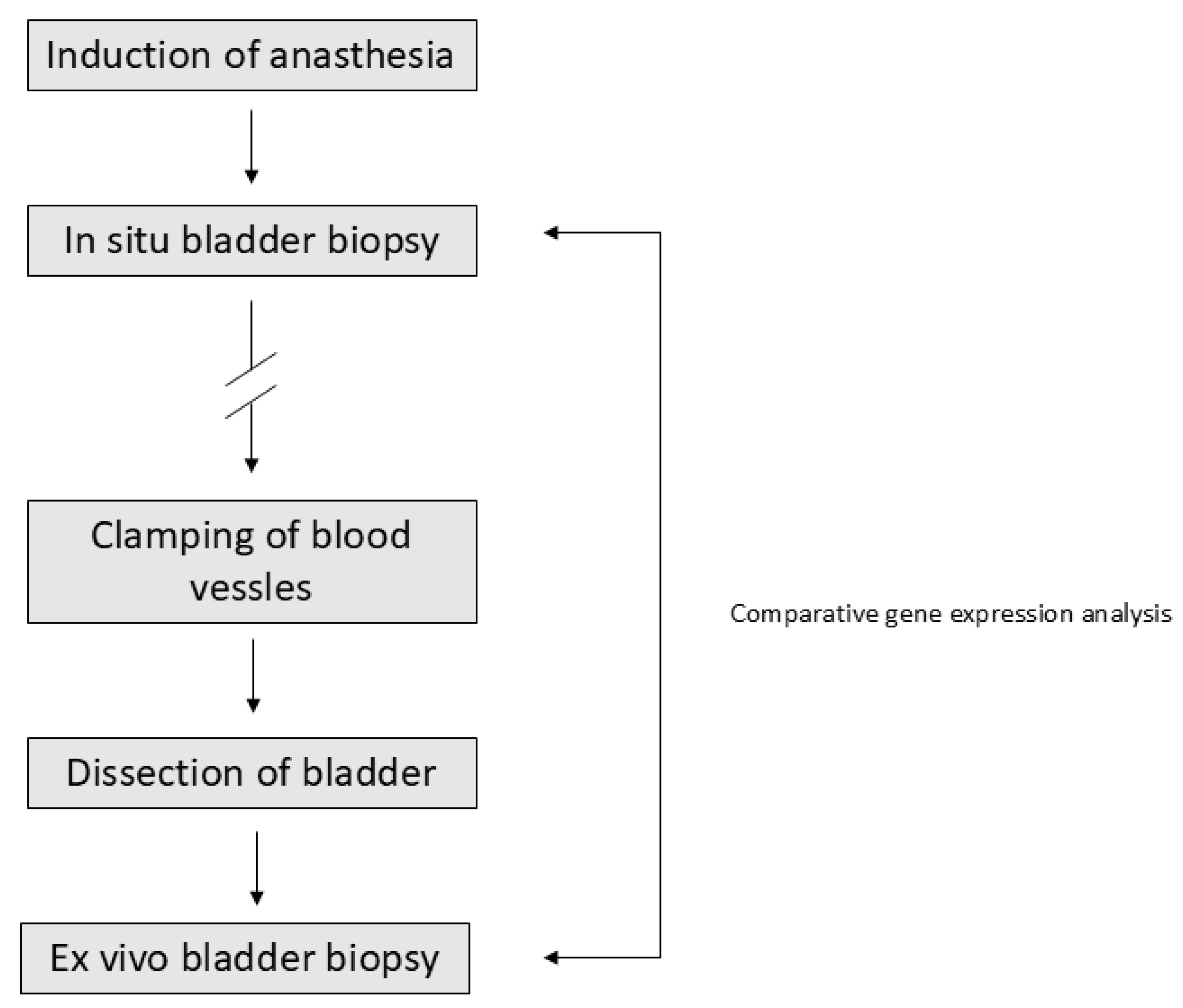
4. Materials and Methods
4.1. Patient Samples
4.2. Gene Expression Analysis
4.2.1. Explorative Dataset
4.2.2. Validation Dataset
4.3. Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| FC | Fold Change |
| RIN | RNA Integrity Number |
| sd | Standard Deviation |
| UBC | Urinary Bladder Cancer |
References
- Zhou, J.H.; Sahin, A.A.; Myers, J.N. Biobanking in genomic medicine. Arch. Pathol. Lab. Med. 2015, 139, 812–818. [Google Scholar] [CrossRef]
- Vaught, J.; Lockhart, N.C. The evolution of biobanking best practices. Clin. Chim. Acta 2012, 413, 1569–1575. [Google Scholar] [CrossRef]
- Ransohoff, D.F.; Gourlay, M.L. Sources of bias in specimens for research about molecular markers for cancer. J. Clin. Oncol. 2010, 28, 698–704. [Google Scholar] [CrossRef] [PubMed]
- Lippi, G.; Chance, J.J.; Church, S.; Dazzi, P.; Fontana, R.; Giavarina, D.; Grankvist, K.; Huisman, W.; Kouri, T.; Palicka, V.; et al. Preanalytical quality improvement: From dream to reality. Clin. Chem. Lab. Med. 2011, 49, 1113–1126. [Google Scholar] [CrossRef] [PubMed]
- Spruessel, A.; Steimann, G.; Jung, M.; Lee, S.A.; Carr, T.; Fentz, A.K.; Spangenberg, J.; Zornig, C.; Juhl, H.H.; David, K.A. Tissue ischemia time affects gene and protein expression patterns within minutes following surgical tumor excision. Biotechniques 2004, 36, 1030–1037. [Google Scholar] [CrossRef] [PubMed]
- Neumeister, V.M.; Juhl, H. Tumor Pre-Analytics in Molecular Pathology: Impact on Protein Expression and Analysis. Curr. Pathobiol. Rep. 2018, 6, 265–274. [Google Scholar] [CrossRef]
- True, L.D. Methodological requirements for valid tissue-based biomarker studies that can be used in clinical practice. Virchows Arch. 2014, 464, 257–263. [Google Scholar] [CrossRef][Green Version]
- Lucchinetti, E.; Hofer, C.; Bestmann, L.; Hersberger, M.; Feng, J.; Zhu, M.; Furrer, L.; Schaub, M.C.; Tavakoli, R.; Genoni, M.; et al. Gene regulatory control of myocardial energy metabolism predicts postoperative cardiac function in patients undergoing off-pump coronary artery bypass graft surgery: Inhalational versus intravenous anesthetics. Anesthesiology 2007, 106, 444–457. [Google Scholar] [CrossRef]
- Lin, D.W.; Coleman, I.M.; Hawley, S.; Huang, C.Y.; Dumpit, R.; Gifford, D.; Kezele, P.; Hung, H.; Knudsen, B.S.; Kristal, A.R.; et al. Influence of surgical manipulation on prostate gene expression: Implications for molecular correlates of treatment effects and disease prognosis. J. Clin. Oncol. 2007, 24, 3763–3770, Erratum in J. Clin. Oncol. 2007, 25, 2334. [Google Scholar] [CrossRef]
- Pedersen, I.S.; Thomassen, M.; Tan, Q.; Kruse, T.; Thorlacius-Ussing, O.; Garne, J.P.; Krarup, H.B. Differential effect of surgical manipulation on gene expression in normal breast tissue and breast tumor tissue. Mol. Med. 2018, 24, 57. [Google Scholar] [CrossRef]
- Huang, T.T.; Deoghare, H.V.; Smith, B.K.; Beaver, T.M.; Baker, H.V.; Mehinto, A.C.; Martin, A.D. Gene expression changes in the human diaphragm after cardiothoracic surgery. J. Thorac. Cardiovasc. Surg. 2011, 142, 1214–1222, 22 e1-20. [Google Scholar] [CrossRef]
- Freidin, M.B.; Bhudia, N.; Lim, E.; Nicholson, A.G.; Cookson, W.O.; Moffatt, M.F. Impact of collection and storage of lung tumor tissue on whole genome expression profiling. J. Mol. Diagn. 2012, 14, 140–148. [Google Scholar] [CrossRef]
- Jewell, S.D.; Srinivasan, M.; McCart, L.M.; Williams, N.; Grizzle, W.H.; LiVolsi, V.; MacLennan, G.; Sedmak, D.D. Analysis of the molecular quality of human tissues: An experience from the Cooperative Human Tissue Network. Am. J. Clin. Pathol. 2002, 118, 733–741. [Google Scholar] [CrossRef]
- Huang, J.; Qi, R.; Quackenbush, J.; Dauway, E.; Lazaridis, E.; Yeatman, T. Effects of ischemia on gene expression. J. Surg. Res. 2001, 99, 222–227. [Google Scholar] [CrossRef] [PubMed]
- Martin, N.M.; Cooke, K.M.; Radford, C.C.; Perley, L.E.; Silasi, M.; Flannery, C.A. Time course analysis of RNA quality in placenta preserved by RNAlater or flash freezing. Am. J. Reprod. Immunol. 2017, 77, e12637. [Google Scholar] [CrossRef] [PubMed]
- Lindner, M.; Morresi-Hauf, A.; Stowasser, A.; Hapfelmeier, A.; Hatz, R.A.; Koch, I. Quality assessment of tissue samples stored in a specialized human lung biobank. PLoS ONE 2019, 14, e0203977. [Google Scholar] [CrossRef] [PubMed]
- Mutter, G.L.; Zahrieh, D.; Liu, C.; Neuberg, D.; Finkelstein, D.; Baker, H.E.; Warrington, J.A. Comparison of frozen and RNALater solid tissue storage methods for use in RNA expression microarrays. BMC Genom. 2004, 5, 88. [Google Scholar] [CrossRef]
- Li, G.; Wang, S.; Fan, Z. Oxidative stress in intestinal ischemia-reperfusion. Front. Med. 2022, 14, 750731. [Google Scholar] [CrossRef]
- Lin, Y.M.; Li, F.; Shi, X.Z. Mechanical stress is a pro-inflammatory stimulus in the gut: In vitro, in vivo and ex vivo evidence. PLoS ONE 2014, 9, e106242. [Google Scholar] [CrossRef]
- Chen, K.; Henn, D.; Sivaraj, D.; Bonham, C.A.; Griffin, M.; Kussie, H.C.; Padmanabhan, J.; Trotsyuk, A.A.; Wan, D.C.; Januszyk, M.; et al. Mechanical strain drives myeloid cell differentiation toward proinflammatory subpopulation. Adv. Wound Care 2022, 11, 466–478. [Google Scholar] [CrossRef]
- Fagundes, T.R.; Coradi, C.; Sotomayor, M.R.; Campos, A.G.H.; da Silva, L.C.F.; Ferneda, H.A.; Junior, W.d.S.P.; Bellandi, G.B.; Simonato, M.E.P.; Steffanello, V.V.; et al. Mechanisms of anesthetic-induced immune dysregulation. Anesthesiol. Perioper. Sci. 2025, 3, 37. [Google Scholar] [CrossRef]
- Kurosawa, S.; Kato, M. Anesthetics, immune cells, and immune reponses. J. Anesth. 2008, 22, 263–277. [Google Scholar] [CrossRef] [PubMed]
- Andersen, C.L.; Jensen, J.L.; Orntoft, T.F. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef]
- Pfaffl, M.W.; Tichopad, A.; Prgomet, C.; Neuvians, T.P. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper—Excel-based tool using pair-wise correlations. Biotechnol. Lett. 2004, 26, 509–515. [Google Scholar] [CrossRef]
- Gentleman, R.C.; Carey, V.J.; Bates, D.M.; Bolstad, B.; Dettling, M.; Dudoit, S.; Ellis, B.; Gautier, L.; Ge, Y.; Gentry, J.; et al. Bioconductor: Open software development for computational biology and bioinformatics. Genome Biol. 2004, 5, R80. [Google Scholar] [CrossRef]
- Caraux, G.; Pinloche, S. PermutMatrix: A graphical environment to arrange gene expression profiles in optimal linear order. Bioinformatics 2005, 21, 1280–1281. [Google Scholar] [CrossRef]


| Gene | Raw p-Value | Corrected p-Value a | FC b |
|---|---|---|---|
| FOS | 7.73 × 10−7 | 0.0002 | 32.71 |
| NFX1 | 1.89 × 10−5 | 0.0033 | 0.49 |
| GPI | 4.57 × 10−5 | 0.0049 | 0.43 |
| IL6 | 5.56 × 10−5 | 0.0049 | 73.57 |
| ADORA1 | 9.08 × 10−5 | 0.0065 | 0.21 |
| OSM | 0.0002 | 0.012 | 13.25 |
| CXCL2 | 0.0005 | 0.026 | 26.31 |
| VEGFA | 0.0006 | 0.028 | 4.30 |
| IFNA2 | 0.0007 | 0.028 | 5.11 |
| IL10 | 0.0007 | 0.028 | 2.53 |
| IL2RG | 0.001 | 0.042 | 0.41 |
| IL17RB | 0.001 | 0.042 | 0.32 |
| TNFSF15 | 0.002 | 0.046 | 0.26 |
| IFNA14 | 0.002 | 0.049 | 6.07 |
| FASLG | 0.002 | 0.049 | 0.18 |
| NFATC3 | 0.002 | 0.049 | 0.40 |
| CEBPB | 0.003 | 0.049 | 4.31 |
| INHBB | 0.003 | 0.049 | 2.86 |
| IL23R | 0.003 | 0.049 | 0.41 |
| NAMPT | 0.003 | 0.049 | 3.65 |
| PTGS2 | 0.003 | 0.049 | 21.12 |
| F3 | 0.003 | 0.049 | 4.70 |
| CD70 | 0.004 | 0.049 | 0.45 |
| IL13RA1 | 0.004 | 0.049 | 0.56 |
| IFNAR1 | 0.004 | 0.049 | 0.55 |
| IRF4 | 0.004 | 0.049 | 2.02 |
| PARP4 | 0.004 | 0.049 | 0.41 |
| Gene | FC a | Up/Down | No. of Patients | p-Value | Corrected p-Value b |
|---|---|---|---|---|---|
| IL6 | 5.6 × 10−5 | 0.005 | |||
| 125.7 | ↑ | 10 | |||
| ADORA1 | 9.1 × 10−5 | 0.006 | |||
| 1.8 | ↑ | 1 | |||
| 0.4 | ↓ | 9 | |||
| GPI | 4.6× 10−5 | 0.005 | |||
| 1.4 | ↑ | 6 | |||
| 0.6 | ↓ | 4 | |||
| NFX1 | 1.9 × 10−5 | 0.003 | |||
| 1.3 | ↑ | 6 | |||
| 0.6 | ↓ | 4 | |||
| FOS | 7.7 × 10−7 | 0.0003 | |||
| 34.6 | ↑ | 10 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Davidsson, S.; Jerlström, T.; Carlsson, J. Optimizing Tissue Sampling Timing for Accurate Gene Expression Analysis. Int. J. Mol. Sci. 2025, 26, 8581. https://doi.org/10.3390/ijms26178581
Davidsson S, Jerlström T, Carlsson J. Optimizing Tissue Sampling Timing for Accurate Gene Expression Analysis. International Journal of Molecular Sciences. 2025; 26(17):8581. https://doi.org/10.3390/ijms26178581
Chicago/Turabian StyleDavidsson, Sabina, Tomas Jerlström, and Jessica Carlsson. 2025. "Optimizing Tissue Sampling Timing for Accurate Gene Expression Analysis" International Journal of Molecular Sciences 26, no. 17: 8581. https://doi.org/10.3390/ijms26178581
APA StyleDavidsson, S., Jerlström, T., & Carlsson, J. (2025). Optimizing Tissue Sampling Timing for Accurate Gene Expression Analysis. International Journal of Molecular Sciences, 26(17), 8581. https://doi.org/10.3390/ijms26178581





