Genomic Features and Predicted 3D Structures of the CcWOX Transcription Factors in Cinnamomum camphora
Abstract
1. Introduction
2. Results
2.1. Identification of WOX Genes in C. camphora
2.2. Phylogenetic and Conserved Domain Analysis
2.3. Synteny Analysis
2.4. Expression Patterns of CcWOX Genes in Different Tissues
2.5. CcWOX Three-Dimensional Structure and Interaction Prediction
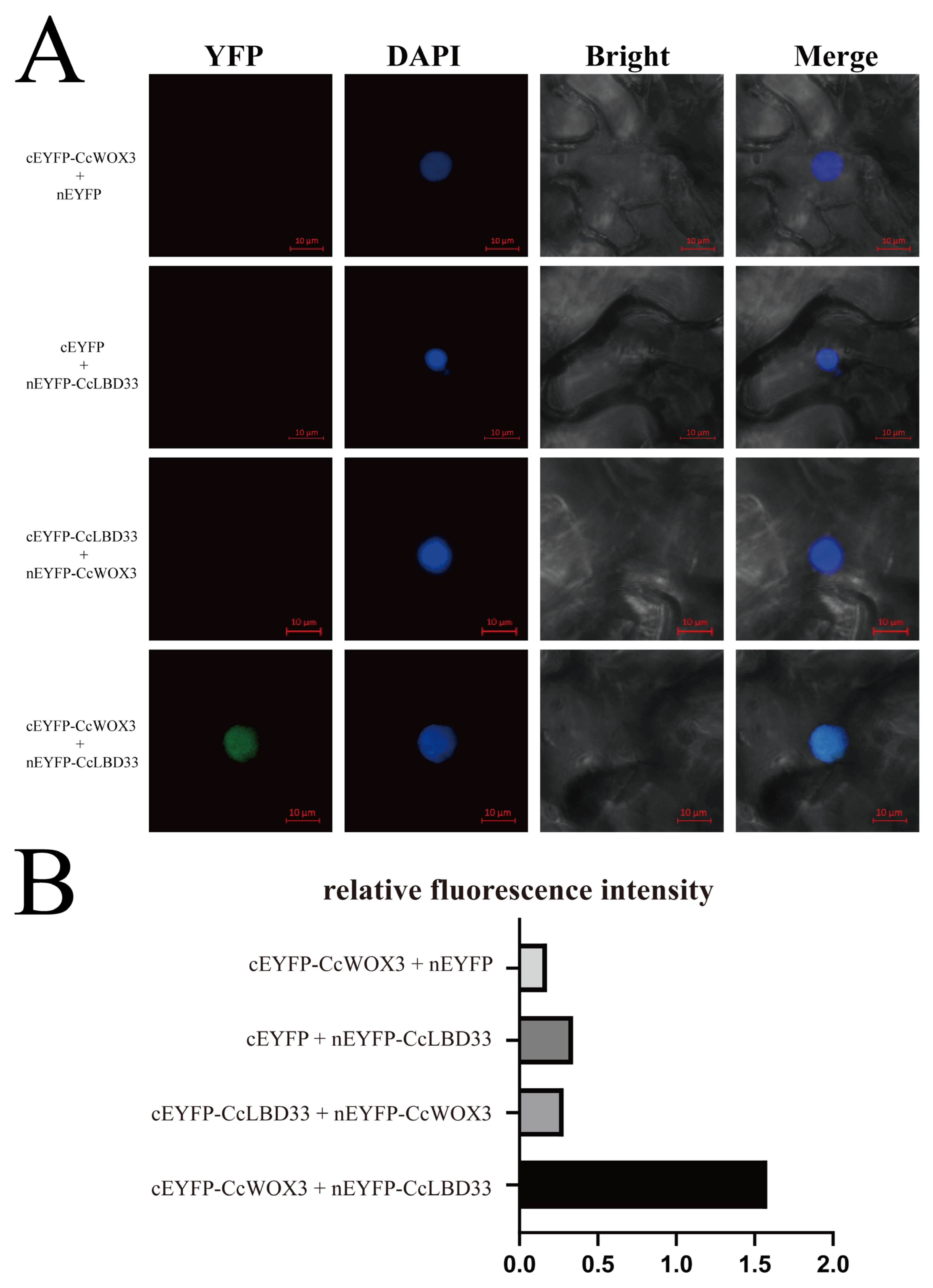
2.6. Validation of CcWOX3 and CcLBD33 Interaction Using BiFC Assay
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Bioinformatics Analysis
4.3. Quantitative Expression Analysis
4.4. Three-Dimensional Structures and Interaction Prediction
4.5. Bimolecular Fluorescence Complementation (BiFC)
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| WOX | WUSCHEL-related homeobox |
| C. camphora | Cinnamomum camphora |
| qRT-PCR | Quantitative Reverse Transcription Polymerase Chain Reaction |
| BiFC | Bimolecular Fluorescence Complementation |
| EYFP | Enhanced Yellow Fluorescent Protein |
| AI | Artificial Intelligence |
| LBD | lateral organ boundary (LOB) domain |
| MYB | v-myb avian myeloblastosis viral oncogene homolog |
| HD | homeodomain |
| WUS | WUSCHEL |
| OC | organizing center |
| SAM | shoot apical meristem |
| CLV | CLAVATA |
| HMM | Hidden Markov Model |
| GRAVY | grand average of hydropathicity |
| PI | isoelectric points |
| TPM | transcripts per million |
| PH | trunk phloem |
| FL | flowers |
| FR | fruits |
| LE | leaves |
| XY | developing xylem |
| R | roots |
| ST | young stems |
| DAPI | 4′,6-diamidino-2-phenylindole |
| pLDDT | Predicted local-distance difference test |
| CDS | coding sequences |
References
- Bellini, C.; Pacurar, D.I.; Perrone, I. Adventitious Roots and Lateral Roots: Similarities and Differences. Annu. Rev. Plant Biol. 2014, 65, 639–666. [Google Scholar] [CrossRef]
- Lakehal, A.; Bellini, C. Control of adventitious root formation: Insights into synergistic and antagonistic hormonal interactions. Physiol. Plant. 2018, 165, 90–100. [Google Scholar] [CrossRef] [PubMed]
- Druege, U.; Franken, P.; Hajirezaei, M.R. Plant Hormone Homeostasis, Signaling, and Function during Adventitious Root Formation in Cuttings. Front. Plant Sci. 2016, 7, 381. [Google Scholar] [CrossRef]
- Geng, L.; Li, Q.; Jiao, L.; Xiang, Y.; Deng, Q.; Zhou, D.; Zhao, Y. WOX11 and CRL1 act synergistically to promote crown root development by maintaining cytokinin homeostasis in rice. New Phytol. 2022, 237, 204–216. [Google Scholar] [CrossRef]
- Liu, J.; Chen, T.; Zhang, J.; Li, C.; Xu, Y.; Zheng, H.; Zhou, J.; Zha, L.; Jiang, C.; Jin, Y.; et al. Ginsenosides regulate adventitious root formation in Panax ginseng via a CLE45–WOX11 regulatory module. J. Exp. Bot. 2020, 71, 6396–6407. [Google Scholar] [CrossRef]
- Liu, W.; Xu, L. Recruitment of IC-WOX Genes in Root Evolution. Trends Plant Sci. 2018, 23, 490–496. [Google Scholar] [CrossRef]
- Zhang, T.; Ge, Y.; Cai, G.; Pan, X.; Xu, L. WOX-ARF modules initiate different types of roots. Cell Rep. 2023, 42, 112966. [Google Scholar] [CrossRef]
- Liu, W.; Yu, J.; Ge, Y.; Qin, P.; Xu, L. Pivotal role of LBD16 in root and root-like organ initiation. Cell. Mol. Life Sci. 2018, 75, 3329–3338. [Google Scholar] [CrossRef] [PubMed]
- Alahakoon, D.; Fennell, A. Genetic analysis of grapevine root system architecture and loci associated gene networks. Front. Plant Sci. 2023, 13, 1083374. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.; Park, O.-S.; Seo, P.J. ATXR2 as a core regulator of de novo root organogenesis. Plant Signal. Behav. 2018, 13, e1449543-5. [Google Scholar] [CrossRef]
- Zhu, L.; Zheng, C.; Liu, R.; Song, A.; Zhang, Z.; Xin, J.; Jiang, J.; Chen, S.; Zhang, F.; Fang, W.; et al. Chrysanthemum transcription factor CmLBD1 direct lateral root formation in Arabidopsis thaliana. Sci. Rep. 2016, 6, 20009. [Google Scholar] [CrossRef] [PubMed]
- Ai, Y.; Qian, X.; Wang, X.; Chen, Y.; Zhang, T.; Chao, Y.; Zhao, Y. Uncovering early transcriptional regulation during adventitious root formation in Medicago sativa. BMC Plant Biol. 2023, 23, 176. [Google Scholar] [CrossRef]
- Wang, H.; Pak, S.; Yang, J.; Wu, Y.; Li, W.; Feng, H.; Yang, J.; Wei, H.; Li, C. Two high hierarchical regulators, PuMYB40 and PuWRKY75, control the low phosphorus driven adventitious root formation in Populus ussuriensis. Plant Biotechnol. J. 2022, 20, 1561–1577. [Google Scholar] [CrossRef]
- Sheng, L.; Hu, X.; Du, Y.; Zhang, G.; Huang, H.; Scheres, B.; Xu, L. Non-canonical WOX11-mediated root branching contributes to plasticity in Arabidopsis root system architecture. Development 2017, 144, 3126–3133. [Google Scholar] [CrossRef]
- Zhao, Y.; Cheng, S.; Song, Y.; Huang, Y.; Zhou, S.; Liu, X.; Zhou, D.-X. The Interaction between Rice ERF3 and WOX11 Promotes Crown Root Development by Regulating Gene Expression Involved in Cytokinin Signaling. Plant Cell 2015, 27, 2469–2483. [Google Scholar] [CrossRef]
- Geng, L.; Tan, M.; Deng, Q.; Wang, Y.; Zhang, T.; Hu, X.; Ye, M.; Lian, X.; Zhou, D.-X.; Zhao, Y. Transcription factors WOX11 and LBD16 function with histone demethylase JMJ706 to control crown root development in rice. Plant Cell 2024, 36, 1777–1790. [Google Scholar] [CrossRef]
- Liu, B.; Wang, L.; Zhang, J.; Li, J.; Zheng, H.; Chen, J.; Lu, M. WUSCHEL-related Homeobox genes in Populus tomentosa: Diversified expression patterns and a functional similarity in adventitious root formation. BMC Genom. 2014, 15, 296. [Google Scholar] [CrossRef]
- Liu, J.; Sheng, L.; Xu, Y.; Li, J.; Yang, Z.; Huang, H.; Xu, L. WOX11 and 12 Are Involved in the First-Step Cell Fate Transition during de Novo Root Organogenesis in Arabidopsis. Plant Cell 2014, 26, 1081–1093. [Google Scholar] [CrossRef]
- Kamiya, N.; Nagasaki, H.; Morikami, A.; Sato, Y.; Matsuoka, M. Isolation and characterization of a rice WUSCHEL-type homeobox gene that is specifically expressed in the central cells of a quiescent center in the root apical meristem. Plant J. 2003, 35, 429–441. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, K.; Brocchieri, L.; Bürglin, T.R. A Comprehensive Classification and Evolutionary Analysis of Plant Homeobox Genes. Mol. Biol. Evol. 2009, 26, 2775–2794. [Google Scholar] [CrossRef] [PubMed]
- van der Graaff, E.; Laux, T.; Rensing, S.A. The WUS homeobox-containing (WOX) protein family. Genome Biol. 2009, 10, 248. [Google Scholar] [CrossRef]
- Ikeda, M.; Mitsuda, N.; Ohme-Takagi, M. Arabidopsis WUSCHEL Is a Bifunctional Transcription Factor That Acts as a Repressor in Stem Cell Regulation and as an Activator in Floral Patterning. Plant Cell 2009, 21, 3493–3505. [Google Scholar] [CrossRef]
- Dolzblasz, A.; Nardmann, J.; Clerici, E.; Causier, B.; van der Graaff, E.; Chen, J.; Davies, B.; Werr, W.; Laux, T. Stem Cell Regulation by Arabidopsis WOX Genes. Mol. Plant 2016, 9, 1028–1039. [Google Scholar] [CrossRef]
- Bueno, N.; Cuesta, C.; Centeno, M.L.; Ordás, R.J.; Alvarez, J.M. In Vitro Plant Regeneration in Conifers: The Role of WOX and KNOX Gene Families. Genes 2021, 12, 438. [Google Scholar] [CrossRef]
- Kong, D.; Hao, Y.; Cui, H. The WUSCHEL Related Homeobox Protein WOX7 Regulates the Sugar Response of Lateral Root Development in Arabidopsis thaliana. Mol. Plant 2016, 9, 261–270. [Google Scholar] [CrossRef]
- Cheng, S.; Tan, F.; Lu, Y.; Liu, X.; Li, T.; Yuan, W.; Zhao, Y.; Zhou, D.-X. WOX11 recruits a histone H3K27me3 demethylase to promote gene expression during shoot development in rice. Nucleic Acids Res. 2018, 46, 2356–2369. [Google Scholar] [CrossRef] [PubMed]
- Park, S.O.; Zheng, Z.; Oppenheimer, D.G.; Hauser, B.A. The PRETTY FEW SEEDS2 gene encodes an Arabidopsis homeodomain protein that regulates ovule development. Development 2005, 132, 841–849. [Google Scholar] [CrossRef] [PubMed]
- Etchells, J.P.; Provost, C.M.; Mishra, L.; Turner, S.R. WOX4 and WOX14 act downstream of the PXY receptor kinase to regulate plant vascular proliferation independently of any role in vascular organisation. Development 2013, 140, 2224–2234. [Google Scholar] [CrossRef] [PubMed]
- Denis, E.; Kbiri, N.; Mary, V.; Claisse, G.; e Silva, N.C.; Kreis, M.; Deveaux, Y. WOX14 promotes bioactive gibberellin synthesis and vascular cell differentiation in Arabidopsis. Plant J. 2017, 90, 560–572. [Google Scholar] [CrossRef]
- Wu, C.-C.; Li, F.-W.; Kramer, E.M.; Shiu, S.-H. Large-scale phylogenomic analysis suggests three ancient superclades of the WUSCHEL-RELATED HOMEOBOX transcription factor family in plants. PLoS ONE 2019, 14, e0223521. [Google Scholar] [CrossRef]
- Laux, T.; Mayer, K.F.; Berger, J.; Jürgens, G. The WUSCHEL gene is required for shoot and floral meristem integrity in Arabidopsis. Development 1996, 122, 87–96. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zong, J.; Liu, J.; Yin, J.; Zhang, D. Genome-Wide Analysis of WOX Gene Family in Rice, Sorghum, Maize, Arabidopsis and Poplar. J. Integr. Plant Biol. 2010, 52, 1016–1026. [Google Scholar] [CrossRef]
- Haecker, A.; Groß-Hardt, R.; Geiges, B.; Sarkar, A.; Breuninger, H.; Herrmann, M.; Laux, T. Expression dynamics of WOX genes mark cell fate decisions during early embryonic patterning in Arabidopsis thaliana. Development 2004, 131, 657–668. [Google Scholar] [CrossRef]
- Sarkar, A.K.; Luijten, M.; Miyashima, S.; Lenhard, M.; Hashimoto, T.; Nakajima, K.; Scheres, B.; Heidstra, R.; Laux, T. Conserved factors regulate signalling in Arabidopsis thaliana shoot and root stem cell organizers. Nature 2007, 446, 811–814. [Google Scholar] [CrossRef]
- Li, J.; Zhang, J.; Jia, H.; Liu, B.; Sun, P.; Hu, J.; Wang, L.; Lu, M. The WUSCHEL-related homeobox 5a (PtoWOX5a) is involved in adventitious root development in poplar. Tree Physiol. 2017, 38, 139–153. [Google Scholar] [CrossRef]
- Xu, M.; Xie, W.; Huang, M. Two WUSCHEL-related HOMEOBOX genes, PeWOX11a and PeWOX11b, are involved in adventitious root formation of poplar. Physiol. Plant. 2015, 155, 446–456. [Google Scholar] [CrossRef]
- Zhu, T.; Moschou, P.N.; Alvarez, J.M.; Sohlberg, J.J.; von Arnold, S. WUSCHEL-RELATED HOMEOBOX 2 is important for protoderm and suspensor development in the gymnosperm Norway spruce. BMC Plant Biol. 2016, 16, 19. [Google Scholar] [CrossRef]
- Jha, P.; Ochatt, S.J.; Kumar, V. WUSCHEL: A master regulator in plant growth signaling. Plant Cell Rep. 2020, 39, 431–444. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.; Jawaid, T. Cinnamomum camphora (Kapur): Review. Pharmacogn. J. 2012, 4, 1–5. [Google Scholar] [CrossRef]
- Zhong, Y.; Yang, A.; Li, Z.; Zhang, H.; Liu, L.; Wu, Z.; Li, Y.; Liu, T.; Xu, M.; Yu, F. Genetic Diversity and Population Genetic Structure of Cinnamomum camphora in South China Revealed by EST-SSR Markers. Forests 2019, 10, 1019. [Google Scholar] [CrossRef]
- Hou, J.; Sun, Y.; Wang, L.; Jiang, Y.; Chen, N.; Tong, S. Genome-Wide Analysis of the Homeobox Gene Family and Identification of Drought-Responsive Members in Populus trichocarpa. Plants 2021, 10, 2284. [Google Scholar] [CrossRef]
- Yu, Y.; Yang, M.; Liu, X.; Xia, Y.; Hu, R.; Xia, Q.; Jing, D.; Guo, Q. Genome-wide analysis of the WOX gene family and the role of EjWUSa in regulating flowering in loquat (Eriobotrya japonica). Front. Plant Sci. 2022, 13, 1024515. [Google Scholar] [CrossRef] [PubMed]
- Khan, F.S.; Zeng, R.-F.; Gan, Z.-M.; Zhang, J.-Z.; Hu, C.-G. Genome-Wide Identification and Expression Profiling of the WOX Gene Family in Citrus sinensis and Functional Analysis of a CsWUS Member. Int. J. Mol. Sci. 2021, 22, 4919. [Google Scholar] [CrossRef]
- Shen, T.; Qi, H.; Luan, X.; Xu, W.; Yu, F.; Zhong, Y.; Xu, M. The chromosome-level genome sequence of the camphor tree provides insights into Lauraceae evolution and terpene biosynthesis. Plant Biotechnol. J. 2021, 20, 244–246. [Google Scholar] [CrossRef]
- Bürglin, T.R.; Affolter, M. Homeodomain proteins: An update. Chromosoma 2015, 125, 497–521. [Google Scholar] [CrossRef] [PubMed]
- Takai, Y.; Sasaki, T.; Matozaki, T. Small GTP-Binding Proteins. Physiol. Rev. 2001, 81, 153–208. [Google Scholar] [CrossRef]
- Zhu, Y.; Wu, N.; Song, W.; Yin, G.; Qin, Y.; Yan, Y.; Hu, Y. Soybean (Glycine max) expansin gene superfamily origins: Segmental and tandem duplication events followed by divergent selection among subfamilies. BMC Plant Biol. 2014, 14, 93. [Google Scholar] [CrossRef]
- Cannon, S.B.; Mitra, A.; Baumgarten, A.; Young, N.D.; May, G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004, 4, 10. [Google Scholar] [CrossRef]
- Papageorgiou, S. Physical Laws Shape Up HOX Gene Collinearity. J. Dev. Biol. 2021, 9, 17. [Google Scholar] [CrossRef]
- Твoрoгoва, В.Е.; Краснoперoва, Е.Ю.; Кудряшoв, А.А.; Пoценкoвская, Э.А.; Дoдуева, И.Е.; Лутoва, Л.А. Чтo мoгут рассказать белки WOX? Обзoр мишеней, регулятoрoв, партнерoв. Mol. Biol. 2021, 55, 362–391. [Google Scholar] [CrossRef]
- Costanzo, E.; Trehin, C.; Vandenbussche, M. The role of WOX genes in flower development. Ann. Bot. 2014, 114, 1545–1553. [Google Scholar] [CrossRef]
- Van Zundert, G.C.P.; Rodrigues, J.P.G.L.M.; Trellet, M.; Schmitz, C.; Kastritis, P.L.; Karaca, E.; Melquiond, A.S.J.; van Dijk, M.; De Vries, S.J.; Bonvin, A.M.J.J. The HADDOCK2.2 Web Server: User-Friendly Integrative Modeling of Biomolecular Complexes. J. Mol. Biol. 2016, 428, 720–725. [Google Scholar] [CrossRef] [PubMed]
- Honorato, R.V.; Koukos, P.I.; Jiménez-García, B.; Tsaregorodtsev, A.; Verlato, M.; Giachetti, A.; Rosato, A.; Bonvin, A.M.J.J. Structural Biology in the Clouds: The WeNMR-EOSC Ecosystem. Front. Mol. Biosci. 2021, 8, 729513. [Google Scholar] [CrossRef] [PubMed]
- Valencia-Lozano, E.; Herrera-Isidrón, L.; Flores-López, J.A.; Recoder-Meléndez, O.S.; Barraza, A.; Cabrera-Ponce, J.L. Solanum tuberosum Microtuber Development under Darkness Unveiled through RNAseq Transcriptomic Analysis. Int. J. Mol. Sci. 2022, 23, 13835. [Google Scholar] [CrossRef] [PubMed]
- Schorderet, M.; Muni, R.R.D.; Fiebig, A.; Reinhardt, D. Deregulation of MADS-box transcription factor genes in a mutant defective in the WUSCHEL-LIKE HOMEOBOX gene EVERGREEN of Petunia hybrida. Plant Signal. Behav. 2018, 13, e1471299-14. [Google Scholar] [CrossRef] [PubMed]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef]
- Chen, C.J.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.H.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Wilkins, M.R.; Gasteiger, E.; Bairoch, A.; Sanchez, J.C.; Williams, K.L.; Appel, R.D.; Hochstrasser, D.F. Protein identification and analysis tools in the ExPASy server. Methods Mol. Biol. 1999, 112, 531–552. [Google Scholar] [CrossRef]
- Pečenková, T.; Pleskot, R.; Žárský, V. Subcellular Localization of Arabidopsis Pathogenesis-Related 1 (PR1) Protein. Int. J. Mol. Sci. 2017, 18, 825. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME Suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef]
- Kim, K.-C.; Lai, Z.; Fan, B.; Chen, Z. Arabidopsis WRKY38 and WRKY62 Transcription Factors Interact with Histone Deacetylase 19 in Basal Defense. Plant Cell 2008, 20, 2357–2371. [Google Scholar] [CrossRef]
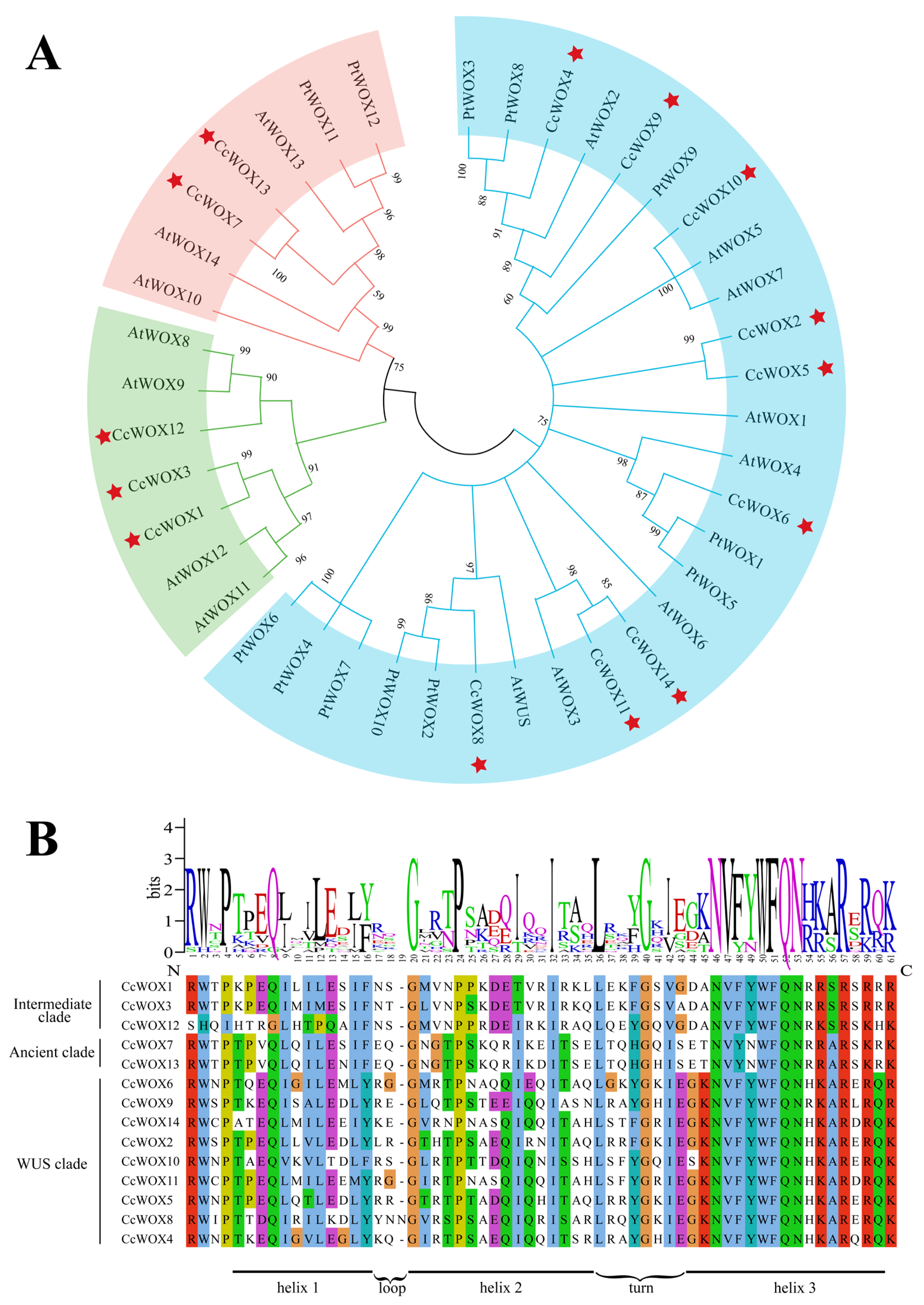
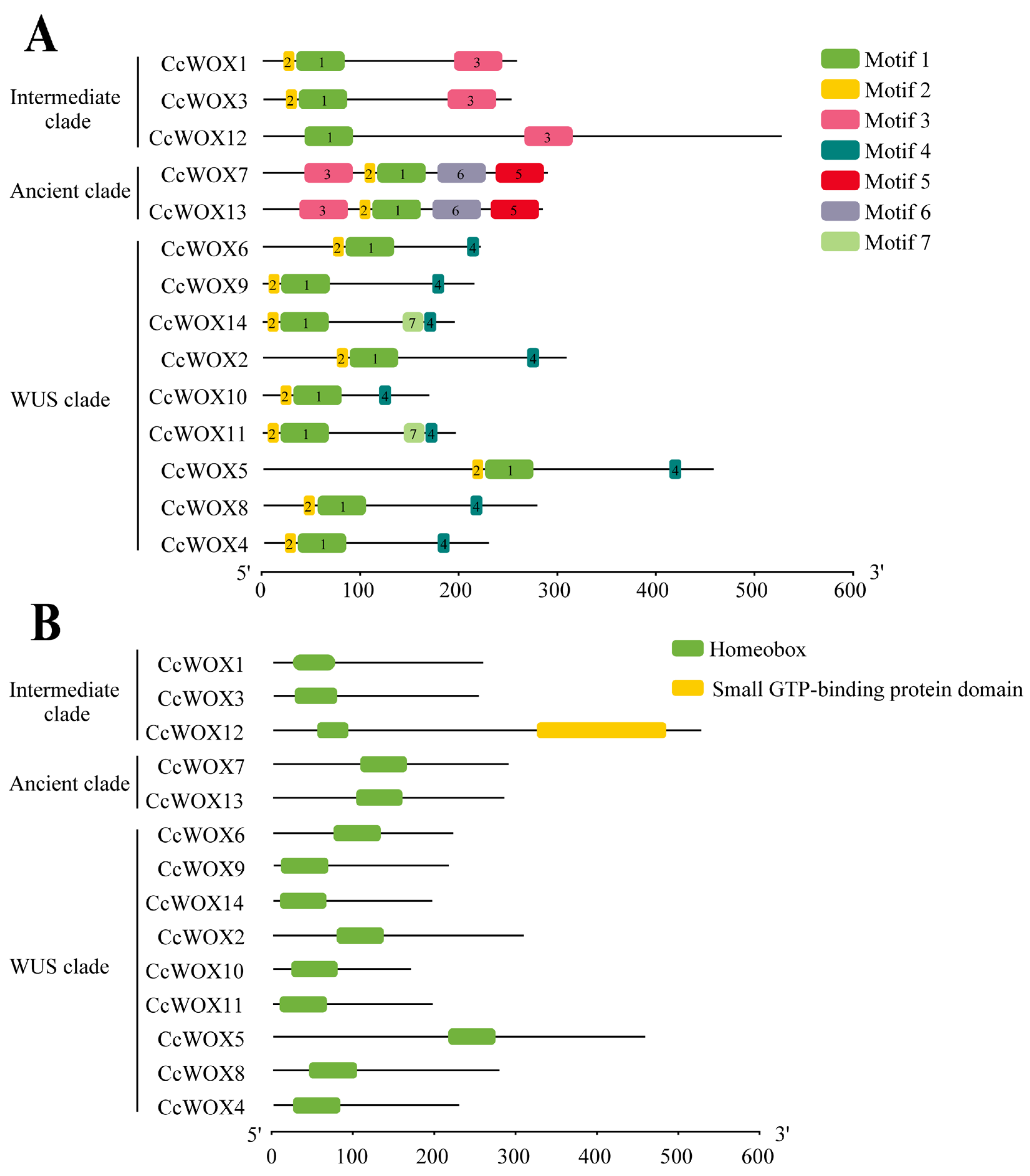

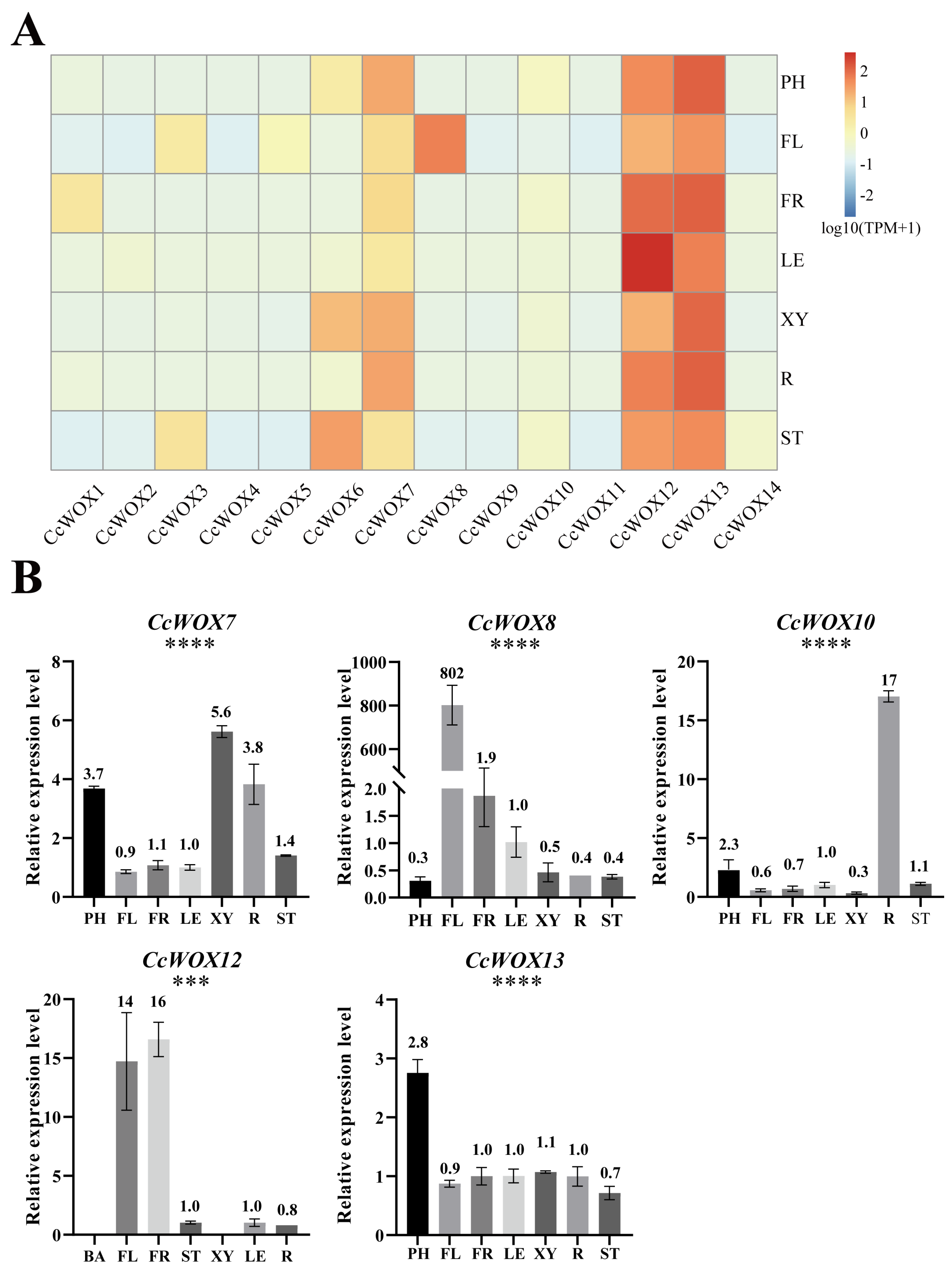

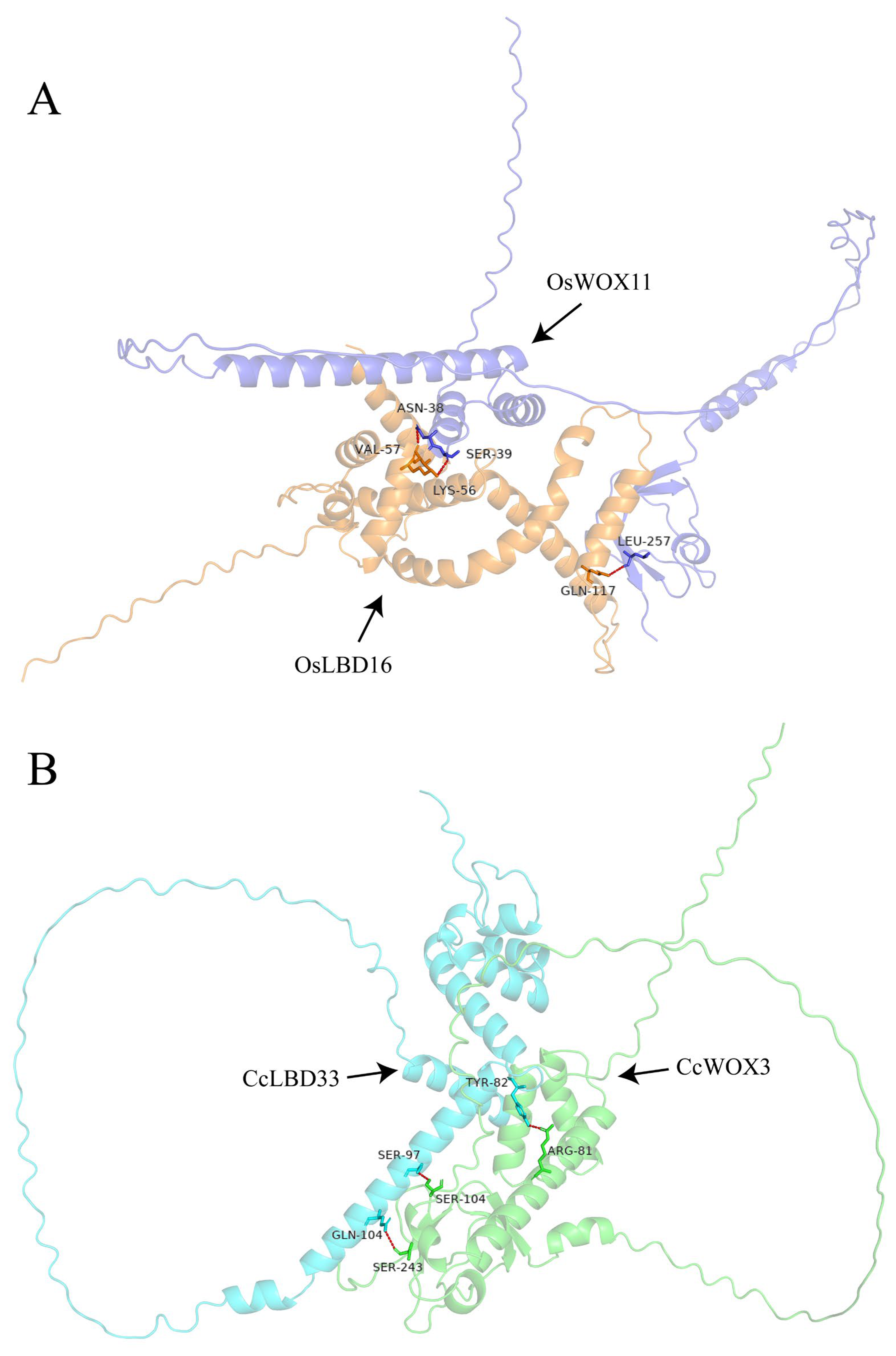
| Gene ID | Name | Protein Lengths (aa) | Molecular Weight (kDa) | Isoelectric Points (pI) | Instability Indices | Hydrophilicity | Predicted Subcellular Locations |
|---|---|---|---|---|---|---|---|
| Ccam01T003400 | CcWOX1 | 256 | 27.89 | 5.11 | 81.23 | −0.251 | Nucleus |
| Ccam01T003479 | CcWOX2 | 306 | 34.97 | 6.27 | 73.33 | −0.709 | Nucleus |
| Ccam02T000641 | CcWOX3 | 250 | 27.75 | 5.65 | 73.29 | −0.429 | Nucleus |
| Ccam02T001857 | CcWOX4 | 226 | 25.47 | 6.71 | 70.43 | −0.781 | Nucleus |
| Ccam02T002266 | CcWOX5 | 455 | 51.59 | 9.54 | 63.26 | −0.592 | Nucleus |
| Ccam02T002701 | CcWOX6 | 219 | 24.70 | 8.57 | 57.28 | −0.911 | Nucleus |
| Ccam03T000173 | CcWOX7 | 287 | 32.58 | 6.08 | 56.33 | −0.959 | Nucleus |
| Ccam03T000852 | CcWOX8 | 276 | 31.42 | 8.26 | 64.5 | −0.97 | Nucleus |
| Ccam04T003014 | CcWOX9 | 213 | 23.98 | 5.54 | 70.83 | −0.463 | Nucleus |
| Ccam04T003125 | CcWOX10 | 167 | 19.29 | 7.94 | 61.46 | −0.787 | Nucleus |
| Ccam05T000584 | CcWOX11 | 194 | 22.62 | 8.96 | 52.55 | −0.831 | Nucleus |
| Ccam07T000210 | CcWOX12 | 524 | 56.83 | 6.51 | 41.86 | −0.323 | Nucleus and Cytoplasm |
| Ccam09T000192 | CcWOX13 | 282 | 31.82 | 6.1 | 64.51 | −0.909 | Nucleus |
| Ccam11T000566 | CcWOX14 | 193 | 22.82 | 8.9 | 61.01 | −0.956 | Nucleus |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cui, F.; Wang, K.; Qi, H.; Shen, T.; Chen, C.; Zhong, Y.; Xu, M. Genomic Features and Predicted 3D Structures of the CcWOX Transcription Factors in Cinnamomum camphora. Int. J. Mol. Sci. 2025, 26, 8204. https://doi.org/10.3390/ijms26178204
Cui F, Wang K, Qi H, Shen T, Chen C, Zhong Y, Xu M. Genomic Features and Predicted 3D Structures of the CcWOX Transcription Factors in Cinnamomum camphora. International Journal of Molecular Sciences. 2025; 26(17):8204. https://doi.org/10.3390/ijms26178204
Chicago/Turabian StyleCui, Fengshuo, Kang Wang, Haoran Qi, Tengfei Shen, Caihui Chen, Yongda Zhong, and Meng Xu. 2025. "Genomic Features and Predicted 3D Structures of the CcWOX Transcription Factors in Cinnamomum camphora" International Journal of Molecular Sciences 26, no. 17: 8204. https://doi.org/10.3390/ijms26178204
APA StyleCui, F., Wang, K., Qi, H., Shen, T., Chen, C., Zhong, Y., & Xu, M. (2025). Genomic Features and Predicted 3D Structures of the CcWOX Transcription Factors in Cinnamomum camphora. International Journal of Molecular Sciences, 26(17), 8204. https://doi.org/10.3390/ijms26178204






