Identification and Characterization of a Novel Di-(2-ethylhexyl) Phthalate Hydrolase from a Marine Bacterial Strain Mycolicibacterium phocaicum RL-HY01
Abstract
1. Introduction
2. Results
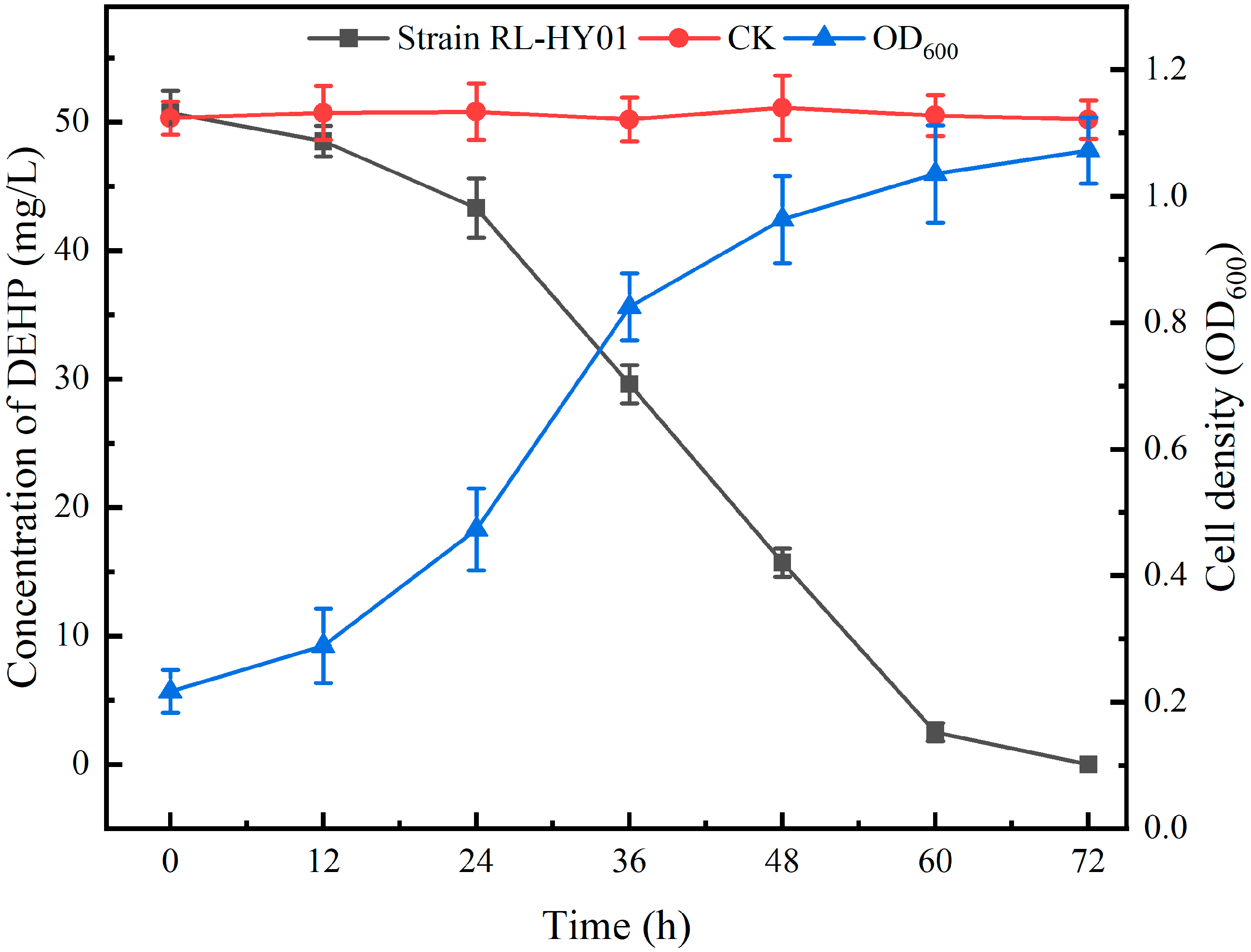
2.1. Biodegradability of PAEs by Strain RL-HY01
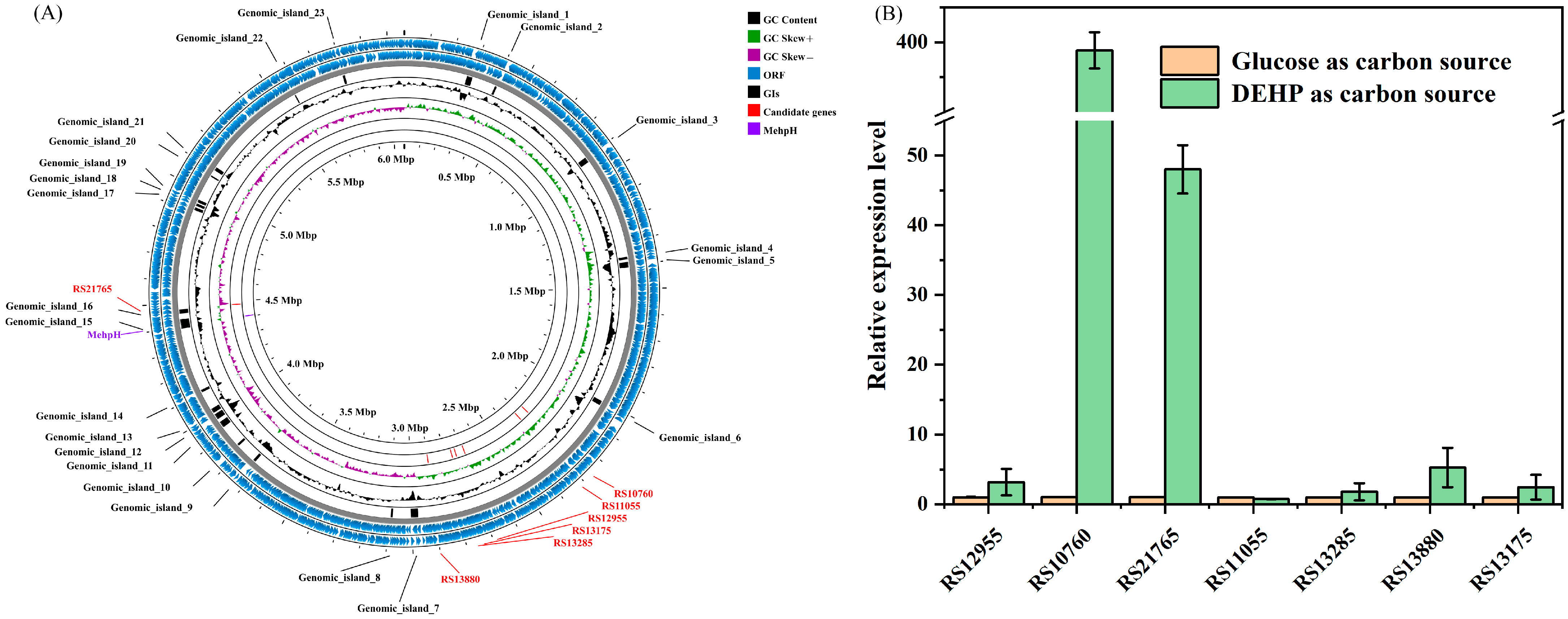
2.2. Genome Annotation and Bioinformatics Analysis
2.3. RT-qPCR Analyses of Candidate Genes
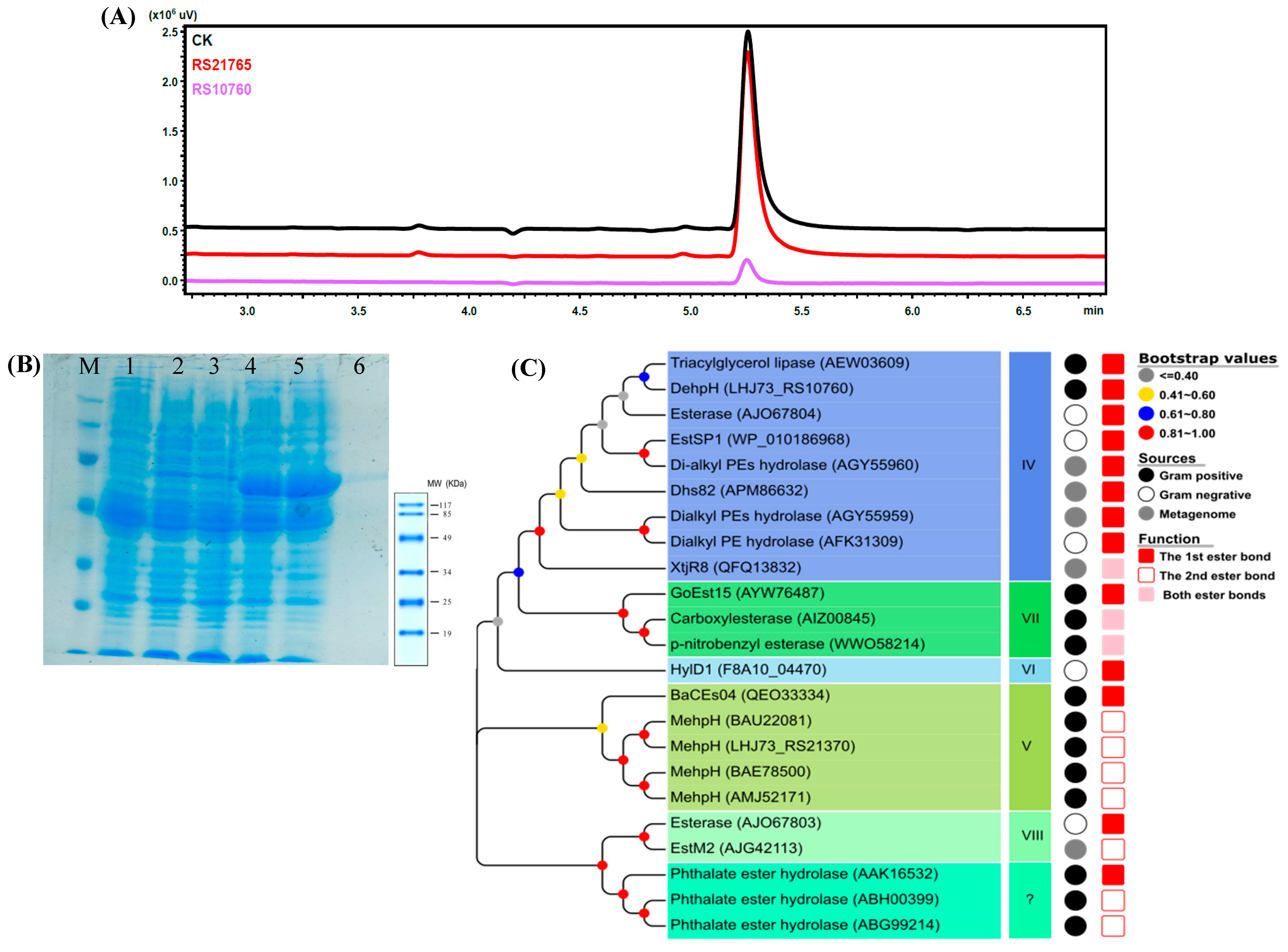
2.4. Cloning and Heterologous Expression of Candidate Genes
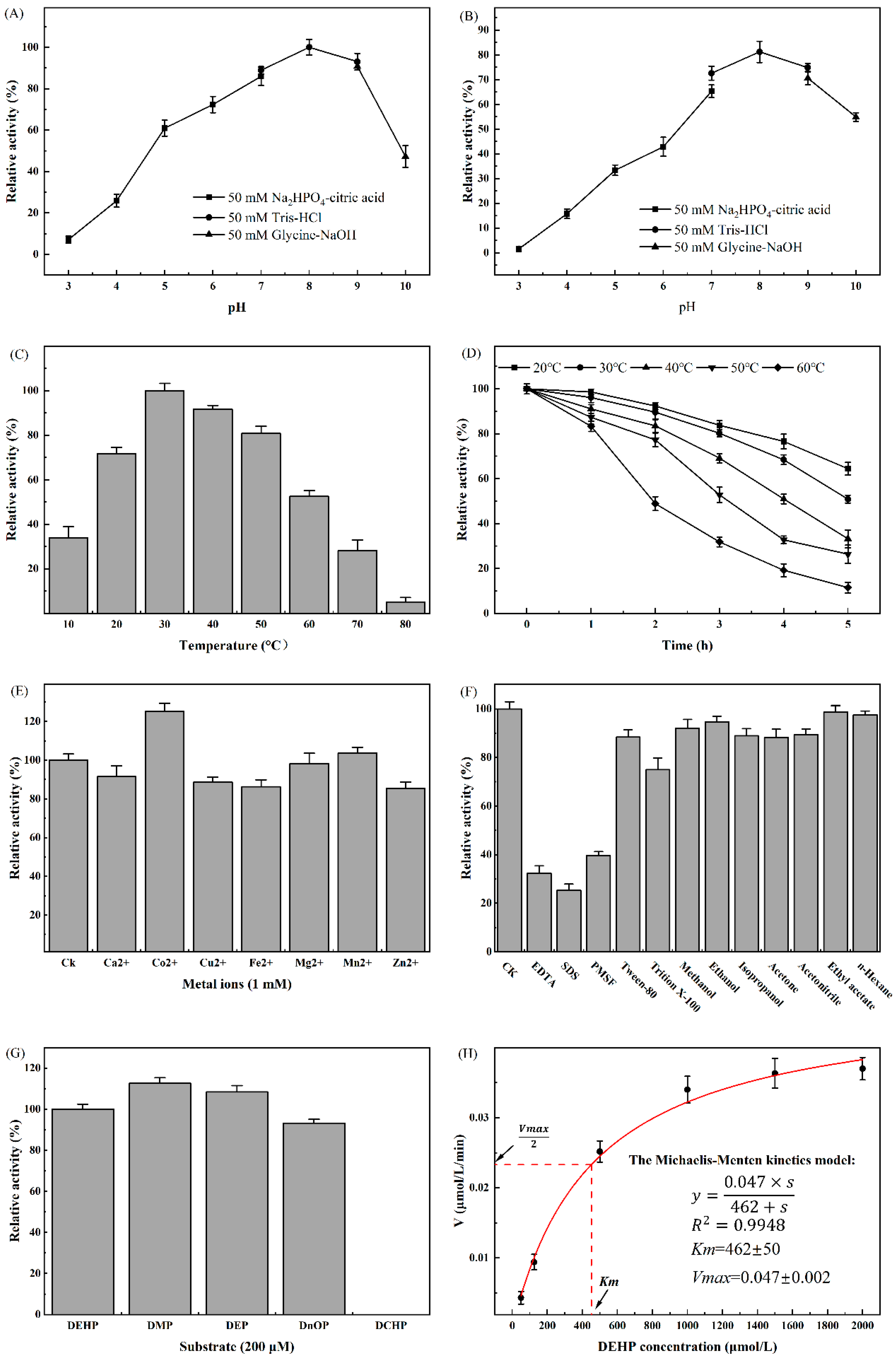
2.5. Biochemical Characterization of DehpH
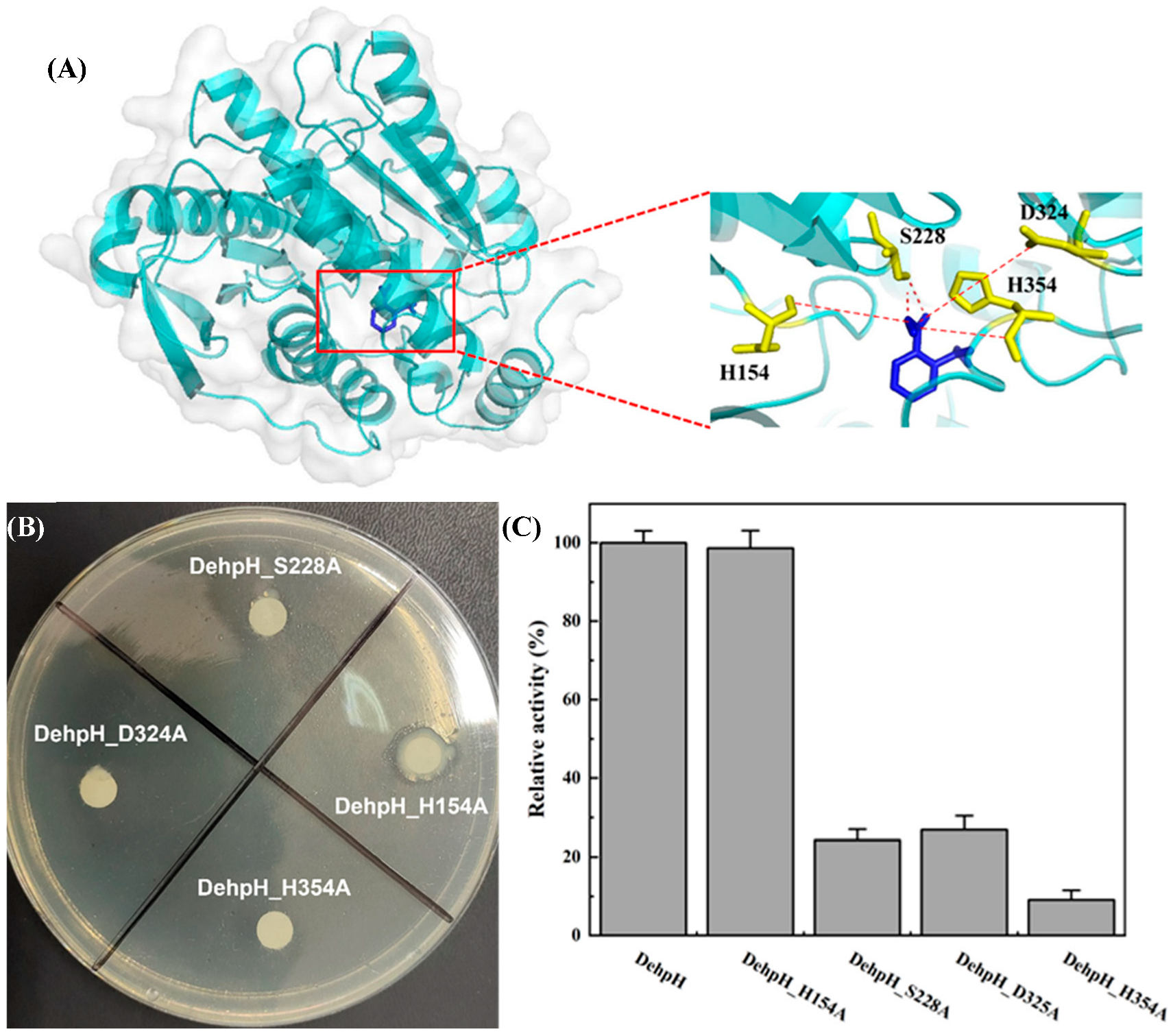
2.6. Structural Modeling and Molecular Docking of DehpH
2.7. Site-Directed Mutagenesis of the Active-Site Residues
3. Discussion
4. Materials and Methods
4.1. Chemicals
4.2. Stains, Plasmids, and Media
4.3. Degradation Test
4.4. Genomic DNA Extraction, Sequencing, and Annotation
4.5. RNA Isolation and RT-qPCR Analyses
4.6. Cloning and Heterologous Expression of Candidate Genes
4.7. Sequence Analysis and Purification of Recombinant DehpH
4.8. Biochemical Characterization of DehpH
4.9. Structural Modeling and Molecular Docking of DehpH
4.10. Site-Directed Mutagenesis of the Active-Site Residues
4.11. Accession Number
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Katsikantami, I.; Sifakis, S.; Tzatzarakis, M.N.; Vakonaki, E.; Kalantzi, O.; Tsatsakis, A.M.; Rizos, A.K. A global assessment of phthalates burden and related links to health effects. Environ. Int. 2016, 97, 212–236. [Google Scholar] [CrossRef]
- Ren, L.; Lin, Z.; Liu, H.; Hu, H. Bacteria-mediated phthalic acid esters degradation and related molecular mechanisms. Appl. Microbiol. Biotechnol. 2018, 102, 1085–1096. [Google Scholar] [CrossRef]
- Fan, Y.; Li, T.; Zhang, Z.; Song, X.; Cun, D.; Cui, B.; Wang, Y. Uptake, accumulation, and degradation of dibutyl phthalate by three wetland plants. Water Sci. Technol. 2023, 88, 1508–1517. [Google Scholar] [CrossRef]
- Feng, N.; Pan, B.; Huang, H.; Huang, Y.; Lyu, H.; Xiang, L.; Zhao, H.; Liu, B.; Li, Y.; Cai, Q.; et al. Uptake, translocation, and biotransformation of phthalate acid esters in crop plants: A comprehensive review. J. Hazard. Mater. 2025, 489, 137580. [Google Scholar] [CrossRef]
- Zhang, J.; Li, Y.; Wang, Y.; Li, Z.; Li, X.; Bao, H.; Li, J.; Zhou, D. Transcriptome sequencing and metabolite analysis revealed the single and combined effects of microplastics and di-(2-ethylhexyl) phthalate on mouse liver. Int. J. Mol. Sci. 2025, 26, 4943. [Google Scholar] [CrossRef]
- Mondal, S.; Bandyopadhyay, A. Antioxidants in mitigating phthalate-induced male reproductive toxicity: A comprehensive review. Chemosphere 2024, 364, 143297. [Google Scholar] [CrossRef] [PubMed]
- Stojanoska, M.M.; Milosevic, N.; Milic, N.; Abenavoli, L. The influence of phthalates and bisphenol A on the obesity development and glucose metabolism disorders. Endocrine 2017, 55, 666–681. [Google Scholar] [CrossRef] [PubMed]
- EPA, U. Code of Federal Regulations, 40 CFR, Part 1992; Office of the Federal Register: Washington, DC, USA, 1992. Available online: https://www.ecfr.gov/current/title-40/chapter-I/subchapter-N/part-423/appendix-Appendix%20A%20to%20Part%20423 (accessed on 10 March 2025).
- European, U. Council Regulation (EEC), No 793/93 of 23 March 1993 on the Evaluation and Control of the Risks of Existing Substances (OJ L84, 5 April 1993). European Union, Brussels 1993. Available online: https://eur-lex.europa.eu/legal-content/EN/TXT/PDF/?uri=CELEX:31993R0793 (accessed on 10 March 2025).
- Zhang, H.; Lin, Z.; Liu, B.; Wang, G.; Weng, L.; Zhou, J.; Hu, H.; He, H.; Huang, Y.; Chen, J.; et al. Bioremediation of di-(2-ethylhexyl) phthalate contaminated red soil by Gordonia terrae RL-JC02: Characterization, metabolic pathway and kinetics. Sci. Total Environ. 2020, 733, 139138. [Google Scholar] [CrossRef] [PubMed]
- Fenner, K.; Canonica, S.; Wackett, L.P.; Elsner, M. Evaluating pesticide degradation in the environment: Blind spots and emerging opportunities. Science 2013, 341, 752–758. [Google Scholar] [CrossRef]
- Feng, N.; Feng, Y.; Liang, Q.; Chen, X.; Xiang, L.; Zhao, H.; Liu, B.; Cao, G.; Li, Y.; Li, H.; et al. Complete biodegradation of di-n-butyl phthalate (DBP) by a novel Pseudomonas sp. YJB6. Sci. Total Environ. 2021, 761, 143208. [Google Scholar] [CrossRef]
- Kamaraj, Y.; Jayathandar, R.S.; Dhayalan, S.; Subramaniyan, S.; Punamalai, G. Biodegradation of di-(2-ethylhexyl) phthalate by novel Rhodococcus sp. PFS1 strain isolated from paddy field soil. Arch. Microbiol. 2022, 204, 21. [Google Scholar] [CrossRef]
- Ren, L.; Jia, Y.; Ruth, N.; Qiao, C.; Wang, J.; Zhao, B.; Yan, Y. Biodegradation of phthalic acid esters by a newly isolated Mycobacterium sp. YC-RL4 and the bioprocess with environmental samples. Environ. Sci. Pollut. Res. 2016, 23, 16609–16619. [Google Scholar] [CrossRef]
- Zeng, P.; Moy, B.Y.; Song, Y.; Tay, J. Biodegradation of dimethyl phthalate by Sphingomonas sp. isolated from phthalic-acid-degrading aerobic granules. Appl. Microbiol. Biotechnol. 2008, 80, 899–905. [Google Scholar] [CrossRef]
- Durante Rodríguez, G.; de Francisco Polanco, S.; Fernández Arévalo, U.; Díaz, E. Engineering bacterial biocatalysts for the degradation of phthalic acid esters. Microb. Biotechnol. 2024, 17, e70024. [Google Scholar] [CrossRef]
- Fuchs, G.; Boll, M.; Heider, J. Microbial degradation of aromatic compounds—From one strategy to four. Nat. Rev. Microbiol. 2011, 9, 803–816. [Google Scholar] [CrossRef]
- Ren, L.; Wang, G.; Huang, Y.; Guo, J.; Li, C.; Jia, Y.; Chen, S.; Zhou, J.L.; Hu, H. Phthalic acid esters degradation by a novel marine bacterial strain Mycolicibacterium phocaicum RL-HY01: Characterization, metabolic pathway and bioaugmentation. Sci. Total Environ. 2021, 791, 148303. [Google Scholar] [CrossRef]
- Gupta, R.S.; Lo, B.; Son, J. Phylogenomics and comparative genomic studies robustly support division of the genus mycobacterium into an emended genus Mycobacterium and four novel genera. Front. Microbiol. 2018, 9, 67. [Google Scholar] [CrossRef]
- Bhattacharyya, M.; Dhar, R.; Basu, S.; Das, A.; Reynolds, D.M.; Dutta, T.K. Molecular evaluation of the metabolism of estrogenic di(2-ethylhexyl) phthalate in Mycolicibacterium sp. Microb. Cell. Fact. 2023, 22, 82. [Google Scholar] [CrossRef]
- Sakdapetsiri, C.; Jeerasantikul, C.; Ningthoujam, R.; Rungsihiranrut, A.; Pinyakong, O. Biodegradation pathways of phthalate esters by Mycolicibacterium parafortuitum J101 and its ability to enhance bioremediation. Int. Biodeterior. Biodegrad. 2025, 202, 106085. [Google Scholar] [CrossRef]
- Yan, Z.; Ding, L.; Zou, D.; Wang, L.; Tan, Y.; Guo, S.; Zhang, Y.; Xin, Z. Identification and characterization of a novel carboxylesterase EstQ7 from a soil metagenomic library. Arch. Microbiol. 2021, 203, 4113–4125. [Google Scholar] [CrossRef]
- Jia, Y.; Wang, J.; Ren, C.; Nahurira, R.; Khokhar, I.; Wang, J.; Fan, S.; Yan, Y. Identification and characterization of a meta-cleavage product hydrolase involved in biphenyl degradation from Arthrobacter sp. YC-RL1. Appl. Microbiol. Biotechnol. 2019, 103, 6825–6836. [Google Scholar] [CrossRef]
- Wu, S.; Nan, F.; Jiang, J.; Qiu, J.; Zhang, Y.; Qiao, B.; Li, S.; Xin, Z. Molecular cloning, expression and characterization of a novel feruloyl esterase from a soil metagenomic library with phthalate-degrading activity. Biotechnol. Lett. 2019, 41, 995–1006. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, Y.; Wen, H.; Liu, X.; Fan, X. Cloning and rational modification of a cold-adapted esterase for phthalate esters and parabens degradation. Chemosphere 2023, 325, 138393. [Google Scholar] [CrossRef]
- Arpigny, J.L.; Jaeger, K.E. Bacterial lipolytic enzymes: Classification and properties. Biochem. J. 1999, 343 Pt 1, 177–183. [Google Scholar] [CrossRef]
- Qiu, J.; Yang, H.; Yan, Z.; Shi, Y.; Zou, D.; Ding, L.; Shao, Y.; Li, L.; Khan, U.; Sun, S.; et al. Characterization of XtjR8: A novel esterase with phthalate-hydrolyzing activity from a metagenomic library of lotus pond sludge. Int. J. Biol. Macromol. 2020, 164, 1510–1518. [Google Scholar] [CrossRef]
- Zhang, X.; Fan, X.; Qiu, Y.; Li, C.; Xing, S.; Zheng, Y.; Xu, J. Newly identified thermostable esterase from Sulfobacillus acidophilus: Properties and performance in phthalate ester degradation. Appl. Environ. Microbiol. 2014, 80, 6870–6878. [Google Scholar] [CrossRef]
- Hong, D.K.; Jang, S.; Lee, C. Gene cloning and characterization of a psychrophilic phthalate esterase with organic solvent tolerance from an Arctic bacterium Sphingomonas glacialis PAMC 26605. J. Mol. Catal. B Enzym. 2016, 133, S337–S345. [Google Scholar] [CrossRef]
- Jiao, Y.; Chen, X.; Wang, X.; Liao, X.; Xiao, L.; Miao, A.; Wu, J.; Yang, L. Identification and characterization of a cold-active phthalate esters hydrolase by screening a metagenomic library derived from biofilms of a wastewater treatment plant. PLoS ONE 2013, 8, e75977. [Google Scholar] [CrossRef]
- Wu, J.; Liao, X.; Yu, F.; Wei, Z.; Yang, L. Cloning of a dibutyl phthalate hydrolase gene from Acinetobacter sp. strain M673 and functional analysis of its expression product in Escherichia coli. Appl. Microbiol. Biotechnol. 2013, 97, 2483–2491. [Google Scholar] [CrossRef]
- Wang, Y.; Deng, C.; Wang, X. Characterization of a novel salt- and solvent-tolerant esterase Dhs82 from soil metagenome capable of hydrolyzing estrogenic phthalate esters. Biophys. Chem. 2025, 316, 107348. [Google Scholar] [CrossRef]
- Jiang, L.; Wang, C.; Xu, L.; Pei, R. Metal Ion-mediated optical biosensors with signal amplification: Recent advances. Anal. Sens. 2024, 4, e202400014. [Google Scholar] [CrossRef]
- Gopinath, K.P.; Kathiravan, M.N.; Srinivasan, R.; Sankaranarayanan, S. Evaluation and elimination of inhibitory effects of salts and heavy metal ions on biodegradation of Congo red by Pseudomonas sp. mutant. Bioresour. Technol. 2011, 102, 3687–3693. [Google Scholar] [CrossRef]
- Feng, J.; Ni, H. Effects of heavy metals and metalloids on the biodegradation of organic contaminants. Environ. Res. 2024, 246, 118069. [Google Scholar] [CrossRef]
- Wang, W.; Li, Q.; Zhang, L.; Cui, J.; Yu, H.; Wang, X.; Ouyang, X.; Tao, F.; Xu, P.; Tang, H. Genetic mapping of highly versatile and solvent-tolerant Pseudomonas putida B6-2 (ATCC BAA-2545) as a ‘superstar’ for mineralization of PAHs and dioxin-like compounds. Environ. Microbiol. 2021, 23, 4309–4325. [Google Scholar] [CrossRef]
- Koren, S.; Walenz, B.P.; Berlin, K.; Miller, J.R.; Bergman, N.H.; Phillippy, A.M. Canu: Scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 2017, 27, 722–736. [Google Scholar] [CrossRef]
- Brettin, T.; Davis, J.J.; Disz, T.; Edwards, R.A.; Gerdes, S.; Olsen, G.J.; Olson, R.; Overbeek, R.; Parrello, B.; Pusch, G.D.; et al. RASTtk: A modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci. Rep. 2015, 5, 8365. [Google Scholar] [CrossRef]
- Overbeek, R.; Olson, R.; Pusch, G.D.; Olsen, G.J.; Davis, J.J.; Disz, T.; Edwards, R.A.; Gerdes, S.; Parrello, B.; Shukla, M.; et al. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic. Acids. Res. 2013, 42, D206–D214. [Google Scholar] [CrossRef]
- Bertelli, C.; Laird, M.R.; Williams, K.P.; Lau, B.Y.; Hoad, G.; Winsor, G.L.; Brinkman, F. IslandViewer 4: Expanded prediction of genomic islands for larger-scale datasets. Nucleic. Acids. Res. 2017, 45, W30–W35. [Google Scholar] [CrossRef]
- Grant, J.R.; Enns, E.; Marinier, E.; Mandal, A.; Herman, E.K.; Chen, C.; Graham, M.; Van Domselaar, G.; Stothard, P. Proksee: In-depth characterization and visualization of bacterial genomes. Nucleic. Acids. Res. 2023, 51, W484–W492. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Robert, X.; Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucleic. Acids. Res. 2014, 42, W320–W324. [Google Scholar] [CrossRef]
- Hu, J.; Zhang, Y.; Wu, Y.; Zheng, J.; Yu, Z.; Qian, H.; Yu, J.; Cheng, Z.; Chen, J. Heterologous expression of bacterial cytochrome P450 from Microbacterium keratanolyticum ZY and its application in dichloromethane dechlorination. Environ. Pollut. 2021, 287, 117597. [Google Scholar] [CrossRef]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef]
- Eaton, R.W. Plasmid-encoded phthalate catabolic pathway in Arthrobacter keyseri 12B. J. Bacteriol. 2001, 183, 3689–3703. [Google Scholar] [CrossRef]
- Whangsuk, W.; Sungkeeree, P.; Nakasiri, M.; Thiengmag, S.; Mongkolsuk, S.; Loprasert, S. Two endocrine disrupting dibutyl phthalate degrading esterases and their compensatory gene expression in Sphingobium sp. SM42. Int. Biodeterior. Biodegrad. 2015, 99, 45–54. [Google Scholar] [CrossRef]
- Nishioka, T.; Iwata, M.; Imaoka, T.; Mutoh, M.; Egashira, Y.; Nishiyama, T.; Shin, T.; Fujii, T. A mono-2-ethylhexyl phthalate hydrolase from a Gordonia sp. that is able to dissimilate di-2-ethylhexyl phthalate. Appl. Environ. Microbiol. 2006, 72, 2394–2399. [Google Scholar] [CrossRef]
- Hara, H.; Stewart, G.R.; Mohn, W.W. Involvement of a novel ABC transporter and monoalkyl phthalate ester hydrolase in phthalate ester catabolism by Rhodococcus jostii RHA1. Appl. Environ. Microbiol. 2010, 76, 1516–1523. [Google Scholar] [CrossRef]
- Nahurira, R.; Ren, L.; Song, J.; Jia, Y.; Wang, J.; Fan, S.; Wang, H.; Yan, Y. Degradation of di(2-ethylhexyl) phthalate by a novel Gordonia alkanivorans strain YC-RL2. Curr. Microbiol. 2017, 74, 309–319. [Google Scholar] [CrossRef]
- Iwata, M.; Imaoka, T.; Nishiyama, T.; Fujii, T. Re-characterization of mono-2-ethylhexyl phthalate hydrolase belonging to the serine hydrolase family. J. Biosci. Bioeng. 2016, 122, 140–145. [Google Scholar] [CrossRef]
- Ding, J.; Wang, C.; Xie, Z.; Li, J.; Yang, Y.; Mu, Y.; Tang, X.; Xu, B.; Zhou, J.; Huang, Z. Properties of a newly identified esterase from Bacillus sp. K91 and its novel function in diisobutyl phthalate degradation. PLoS ONE. 2015, 10, e119216. [Google Scholar] [CrossRef]
- Huang, H.; Zhang, X.; Chen, T.; Zhao, Y.; Xu, D.; Bai, Y. Biodegradation of structurally diverse phthalate esters by a newly identified esterase with catalytic activity toward di(2-ethylhexyl) phthalate. J. Agric. Food. Chem. 2019, 67, 8548–8558. [Google Scholar] [CrossRef]
- Mu, B.; Sadowski, P.; Te’O, J.; Patel, B.; Pathiraja, N.; Dudley, K. Identification and characterization of moderately thermostable diisobutyl phthalate degrading esterase from a Great Artesian Basin Bacillus velezensis NP05. Biotechnol. Rep. 2024, 42, e840. [Google Scholar] [CrossRef]
- Wang, N.; Zhang, N.; Sun, M.; Sun, Y.; Dong, Q.; Wang, Y.; Gu, Z.; Ding, H.; Qin, Q.; Jiang, Y.; et al. Molecular insights into the catalytic mechanism of a phthalate ester hydrolase. J. Hazard. Mater. 2024, 476, 135191. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Meng, D.; Tian, Q.; Yang, S.; Deng, H.; Guan, Z.; Cai, Y.; Liao, X. Characterization of a novel carboxylesterase from Bacillus velezensis SYBC H47 and its application in degradation of phthalate esters. J. Biosci. Bioeng. 2020, 129, 588–594. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, J.; Dutta, A.; Pal Chowdhury, P.; Chakraborty, J.; Dutta, T.K. Characterization of a novel family VIII esterase EstM2 from soil metagenome capable of hydrolyzing estrogenic phthalates. Microb. Cell Fact. 2020, 19, 77. [Google Scholar] [CrossRef] [PubMed]
| Target Fragments | Forward Primer (5′–3′) | Reverse Primer (5′–3′) | Objectives |
|---|---|---|---|
| RS10760 | CCGATGCTGGTCTTCTTCCA | CCAGAGATAGGTGGCGTAGC | RT-qPCR |
| RS11055 | GACCAGTACGTCCCCAACAC | GGATGGCTCCGTAGTACAGC | |
| RS12955 | GACGGACTCACGCTGAAAGA | ATGGTGTCGGTGACGAGTTC | |
| RS13175 | CCATCACCGTCGAGGATCTG | GTCGACTGGCTGTAGGTCAG | |
| RS13285 | GGAAGCGGTTCCCTTCCAAT | TGGAAGAACAGCACGACCG | |
| RS13880 | TATCGACAGCGGAACCATCG | ACGCTCAGCATCGTGTACTT | |
| RS21765 | CAGAAGCTGGTCGGGATCG | ATGTGGGCGGAACGTAGTG | |
| 16S rRNA gene | AGAGTTTGATCCTGGCTCAG | GGTTACCTTGTTACGACTT | |
| RS10760 | GCGC GAATTC ACCATGAGCATGCCAGGCGA | GCGC AAGCTT GCGGGTGAGGTGCGCCCGG | Gene expression |
| RS21765 | GCGC GGATCC ACTGCGAGTCTGCCAGCTGACG | GCGC GAATTC GACGCCGCGCAGACGGGTGCGCAGC | |
| H154up | GCTGATATCG GATCC ACCATGAGCATGCCAGG | GACGAATCCGCCGCC GGC GAAGAAGACCAGCAT | Site-directed mutagenesis (H154A) |
| H154down | ATGCTGGTCTTCTTC GCC GGCGGCGGATTCGTC | GAGTGCGGCCGCAAGCTT GCGGGTGAGGTGCGCCC | |
| S228up | GCTGATATCG GATCC ACCATGAGCATGCCAGG | GCCGCCGGC GGC GTCGCCGCCGACTG | Site-directed mutagenesis (S228A) |
| S228down | GCGACGCC GCCGGCGGCAACCTGGCG | GAGTGCGGCCGC AAGCTT GCGGGTGAGGTGCGCCC | |
| D324up | GCTGATATCGGATCC ACCATGAGCATGCCAGG | GTCGCGCAGCGG GGC GAACCCACCGGT | Site-directed mutagenesis (D324A) |
| D324down | ACCGGTGGGTTC GCC CCGCTGCGCGAC | GAGTGCGGCCGCAAGCTT GCGGGTGAGGTGCGCCC | |
| H354up | GCTGATATCGGATCC ACCATGAGCATGCCAGG | ATTCGCGAAGGC GGC GATCAGGGACCC | Site-directed mutagenesis (H354A) |
| H354down | GGGTCCCTGATC GCC GCCTTCGCGAAT | GAGTGCGGCCGCAAGCTT GCGGGTGAGGTGCGCCC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ren, L.; Kuang, C.; Wang, H.; Zhou, J.L.; Shi, M.; Xu, D.; Hu, H.; Wang, Y. Identification and Characterization of a Novel Di-(2-ethylhexyl) Phthalate Hydrolase from a Marine Bacterial Strain Mycolicibacterium phocaicum RL-HY01. Int. J. Mol. Sci. 2025, 26, 8141. https://doi.org/10.3390/ijms26178141
Ren L, Kuang C, Wang H, Zhou JL, Shi M, Xu D, Hu H, Wang Y. Identification and Characterization of a Novel Di-(2-ethylhexyl) Phthalate Hydrolase from a Marine Bacterial Strain Mycolicibacterium phocaicum RL-HY01. International Journal of Molecular Sciences. 2025; 26(17):8141. https://doi.org/10.3390/ijms26178141
Chicago/Turabian StyleRen, Lei, Caiyu Kuang, Hongle Wang, John L. Zhou, Min Shi, Danting Xu, Hanqiao Hu, and Yanyan Wang. 2025. "Identification and Characterization of a Novel Di-(2-ethylhexyl) Phthalate Hydrolase from a Marine Bacterial Strain Mycolicibacterium phocaicum RL-HY01" International Journal of Molecular Sciences 26, no. 17: 8141. https://doi.org/10.3390/ijms26178141
APA StyleRen, L., Kuang, C., Wang, H., Zhou, J. L., Shi, M., Xu, D., Hu, H., & Wang, Y. (2025). Identification and Characterization of a Novel Di-(2-ethylhexyl) Phthalate Hydrolase from a Marine Bacterial Strain Mycolicibacterium phocaicum RL-HY01. International Journal of Molecular Sciences, 26(17), 8141. https://doi.org/10.3390/ijms26178141






