Unraveling the Epigenetic Landscape of Mature B Cell Neoplasia: Mechanisms, Biomarkers, and Therapeutic Opportunities
Abstract
1. Introduction
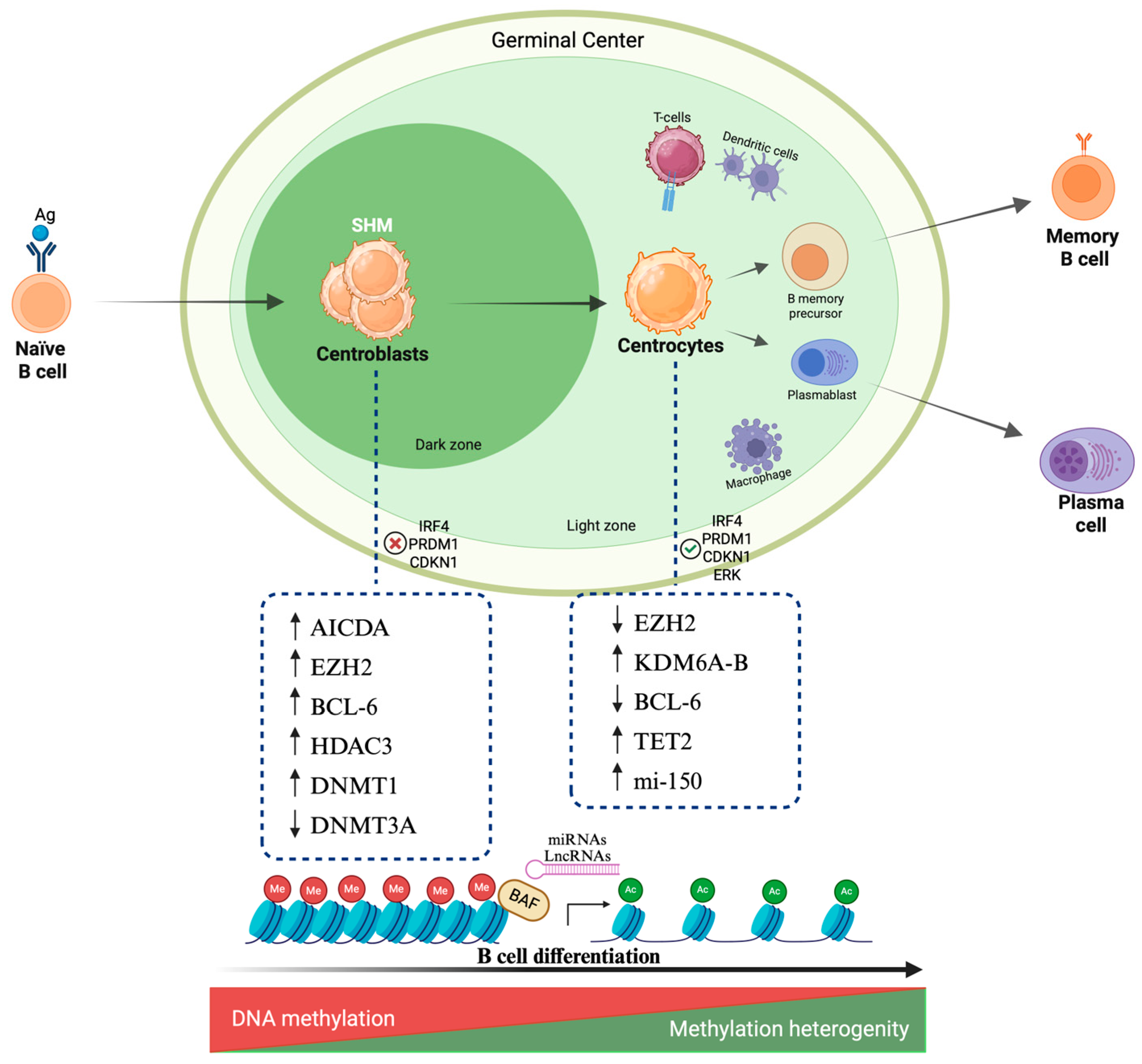
2. Epigenetic Mechanisms in B Cell Maturation
2.1. DNA Methylation
2.2. Histone Modifications
2.3. Chromatin Remodeling
2.4. Noncoding RNA
3. Epigenetic Landscape in B Cell Neoplasia
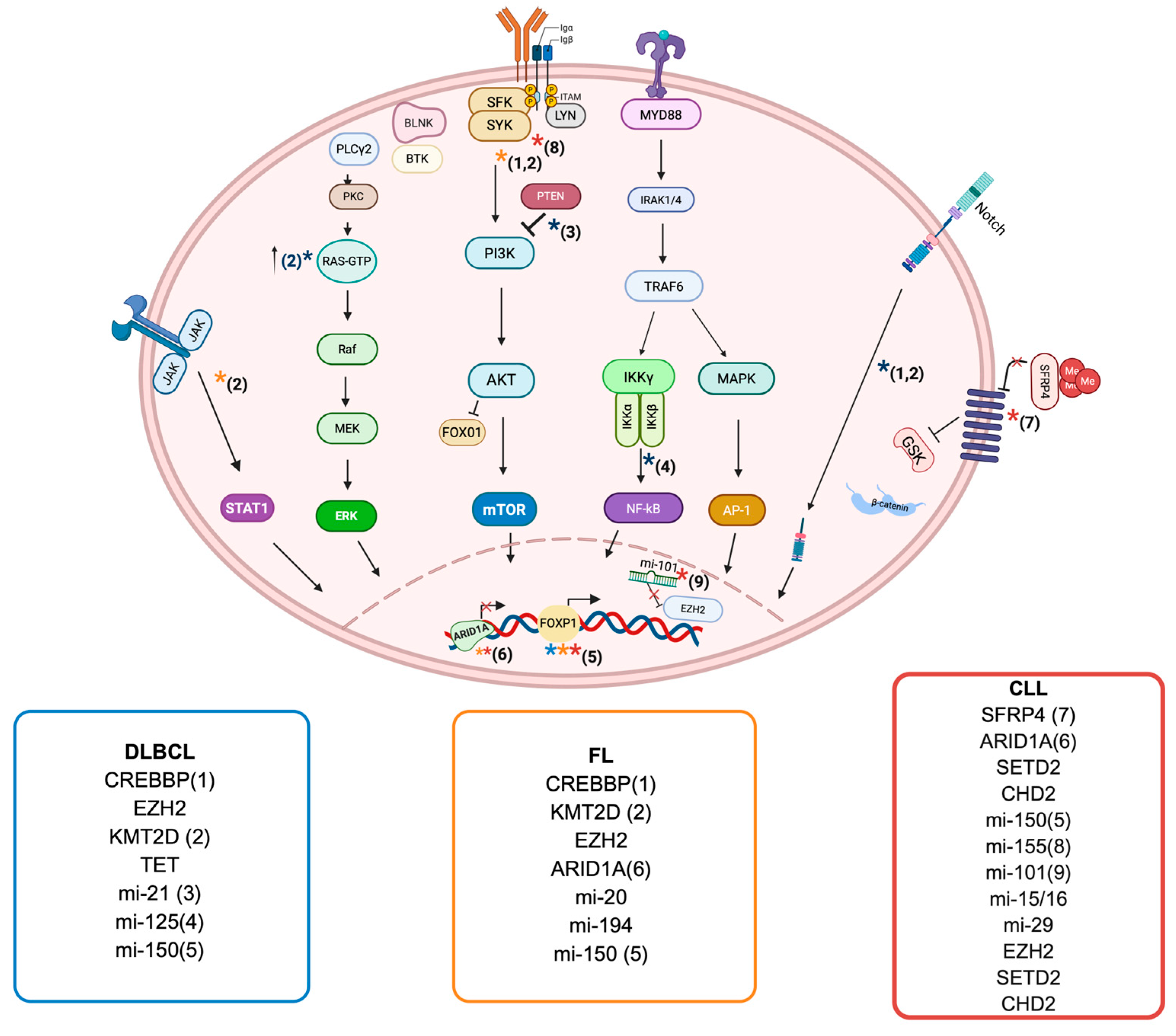
3.1. Diffuse Large B Cell Lymphoma
3.1.1. DNA Methylation in DLBCL
3.1.2. Histone Modifications and Chromain Remodeling in DLBCL
3.1.3. Noncoding RNA in DLBCL
3.2. Follicular Lymphoma
3.2.1. Histone Modifications and Chromatin Remodeling in FL
3.2.2. DNA Methylation in FL
3.2.3. Noncoding RNA in FL
3.3. Chronic Lymphocytic Leukemia
3.3.1. DNA Methylation in CLL
3.3.2. Histone Modifications and Chromatin Remodeling in CLL
3.3.3. Noncoding RNA in CLL
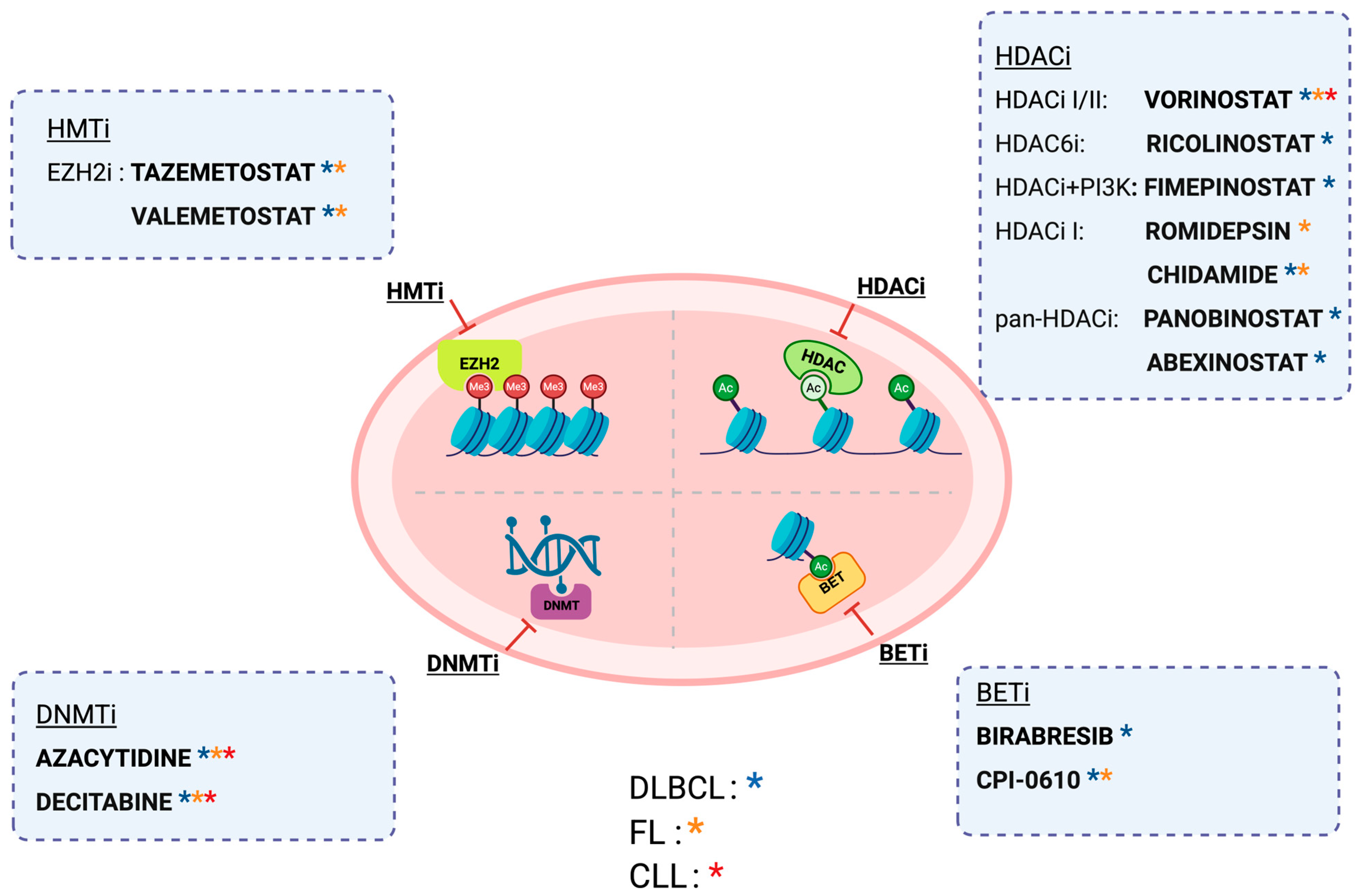
4. Therapeutic Strategies Targeting Epigenetic Alterations
4.1. Demethylating Agents
4.2. HMT Inhibitors
4.3. HDACs Inhibitors
4.4. BET Inhibitor
5. Discussion and Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Alaggio, R.; Amador, C.; Anagnostopoulos, I.; Attygalle, A.D.; Araujo, I.B.d.O.; Berti, E.; Bhagat, G.; Borges, A.M.; Boyer, D.; Calaminici, M.; et al. The 5th edition of the World Health Organization Classification of Haematolymphoid Tumours: Lymphoid Neoplasms. Leukemia 2022, 36, 1720–1748. [Google Scholar] [CrossRef] [PubMed]
- Campo, E.; Jaffe, E.S.; Cook, J.R.; Quintanilla-Martinez, L.; Swerdlow, S.H.; Anderson, K.C.; Brousset, P.; Cerroni, L.; de Leval, L.; Dirnhofer, S.; et al. The International Consensus Classification of Mature Lymphoid Neoplasms: A report from the Clinical Advisory Committee. Blood 2022, 140, 1229–1253. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Zhao, H.; Wang, R.; Chen, Y.; Ouyang, X.; Li, W.; Sun, Y.; Peng, A. Cancer epigenetics: From laboratory studies and clinical trials to precision medicine. Cell Death Discov. 2024, 10, 28. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Deng, Y.; Feng, Y.; Long, D.; Ma, K.; Wang, X.; Zhao, M.; Lu, L.; Lu, Q. Epigenetic regulation in B-cell maturation and its dysregulation in autoimmunity. Cell. Mol. Immunol. 2018, 15, 676–684. [Google Scholar] [CrossRef]
- Schmitz, R.; Wright, G.W.; Huang, D.W.; Johnson, C.A.; Phelan, J.D.; Wang, J.Q.; Roulland, S.; Kasbekar, M.; Young, R.M.; Shaffer, A.L.; et al. Genetics and Pathogenesis of Diffuse Large B-Cell Lymphoma. N. Engl. J. Med. 2018, 378, 1396–1407. [Google Scholar] [CrossRef]
- Jiang, Y.; Melnick, A. The epigenetic basis of diffuse large B-cell lymphoma. Semin. Hematol. 2015, 52, 86–96. [Google Scholar] [CrossRef]
- Allis, C.D.; Jenuwein, T. The molecular hallmarks of epigenetic control. Nat. Rev. Genet. 2016, 17, 487–500. [Google Scholar] [CrossRef]
- Yang, H.; Green, M.R. Harnessing lymphoma epigenetics to improve therapies. Blood 2020, 136, 2386–2391. [Google Scholar] [CrossRef]
- Stomper, J.; Rotondo, J.C.; Greve, G.; Lübbert, M. Hypomethylating agents (HMA) for the treatment of acute myeloid leukemia and myelodysplastic syndromes: Mechanisms of resistance and novel HMA-based therapies. Leukemia 2021, 35, 1873–1889. [Google Scholar] [CrossRef]
- Cooper, M.D. The early history of B cells. Nat. Rev. Immunol. 2015, 15, 191–197. [Google Scholar] [CrossRef]
- Matthias, P.; Rolink, A.G. Transcriptional networks in developing and mature B cells. Nat. Rev. Immunol. 2005, 5, 497–508. [Google Scholar] [CrossRef]
- Bernstein, B.E.; Meissner, A.; Lander, E.S. The mammalian epigenome. Cell 2007, 128, 669–681. [Google Scholar] [CrossRef]
- Chi, P.; Allis, C.D.; Wang, G.G. Covalent histone modifications—Miswritten, misinterpreted and mis-erased in human cancers. Nat. Rev. Cancer 2010, 10, 457–469. [Google Scholar] [CrossRef] [PubMed]
- Saxonov, S.; Berg, P.; Brutlag, D.L. A genome-wide analysis of CpG dinucleotides in the human genome distinguishes two distinct classes of promoters. Proc. Natl. Acad. Sci. USA 2006, 103, 1412–1417. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Zhang, Y. TET-mediated active DNA demethylation: Mechanism, function and beyond. Nat. Rev. Genet. 2017, 18, 517–534. [Google Scholar] [CrossRef]
- Solary, E.; Bernard, O.A.; Tefferi, A.; Fuks, F.; Vainchenker, W. The Ten-Eleven Translocation-2 (TET2) gene in hematopoiesis and hematopoietic diseases. Leukemia 2014, 28, 485–496. [Google Scholar] [CrossRef]
- Kulis, M.; Merkel, A.; Heath, S.; Queirós, A.C.; Schuyler, R.P.; Castellano, G.; Beekman, R.; Raineri, E.; Esteve, A.; Clot, G.; et al. Whole-genome fingerprint of the DNA methylome during human B cell differentiation. Nat. Genet. 2015, 47, 746–756. [Google Scholar] [CrossRef] [PubMed]
- Barwick, B.G.; Scharer, C.D.; Martinez, R.J.; Price, M.J.; Wein, A.N.; Haines, R.R.; Bally, A.P.R.; Kohlmeier, J.E.; Boss, J.M. B cell activation and plasma cell differentiation are inhibited by de novo DNA methylation. Nat. Commun. 2018, 9, 1900. [Google Scholar] [CrossRef]
- Shaknovich, R.; Cerchietti, L.; Tsikitas, L.; Kormaksson, M.; De, S.; Figueroa, M.E.; Ballon, G.; Yang, S.N.; Weinhold, N.; Reimers, M.; et al. DNA methyltransferase 1 and DNA methylation patterning contribute to germinal center B-cell differentiation. Blood 2011, 118, 3559–3569. [Google Scholar] [CrossRef]
- Lio, C.W.; Zhang, J.; González-Avalos, E.; Hogan, P.G.; Chang, X.; Rao, A. Tet2 and Tet3 cooperate with B-lineage transcription factors to regulate DNA modification and chromatin accessibility. Elife 2016, 5, e18290. [Google Scholar] [CrossRef]
- Kornberg, R.D.; Lorch, Y. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell 1999, 98, 285–294. [Google Scholar] [CrossRef]
- Zhao, Z.; Shilatifard, A. Epigenetic modifications of histones in cancer. Genome Biol. 2019, 20, 245. [Google Scholar] [CrossRef]
- Audia, J.E.; Campbell, R.M. Histone Modifications and Cancer. Cold Spring Harb. Perspect. Biol. 2016, 8, a019521. [Google Scholar] [CrossRef]
- Cheung, K.L.; Kim, C.; Zhou, M.M. The Functions of BET Proteins in Gene Transcription of Biology and Diseases. Front. Mol. Biosci. 2021, 8, 728777. [Google Scholar] [CrossRef]
- Greer, E.L.; Shi, Y. Histone methylation: A dynamic mark in health, disease and inheritance. Nat. Rev. Genet. 2012, 13, 343–357. [Google Scholar] [CrossRef]
- Liu, Y.; Yang, Q. The roles of EZH2 in cancer and its inhibitors. Med. Oncol. 2023, 40, 167. [Google Scholar] [CrossRef]
- Béguelin, W.; Popovic, R.; Teater, M.; Jiang, Y.; Bunting, K.L.; Rosen, M.; Shen, H.; Yang, S.N.; Wang, L.; Ezponda, T.; et al. EZH2 is required for germinal center formation and somatic EZH2 mutations promote lymphoid transformation. Cancer Cell 2013, 23, 677–692. [Google Scholar] [CrossRef]
- Béguelin, W.; Rivas, M.A.; Calvo Fernández, M.T.; Teater, M.; Purwada, A.; Redmond, D.; Shen, H.; Challman, M.F.; Elemento, O.; Singh, A.; et al. EZH2 enables germinal centre formation through epigenetic silencing of CDKN1A and an Rb-E2F1 feedback loop. Nat. Commun. 2017, 8, 877. [Google Scholar] [CrossRef] [PubMed]
- Aslam, M.A.; Alemdehy, M.F.; Kwesi-Maliepaard, E.M.; Muhaimin, F.I.; Caganova, M.; Pardieck, I.N.; van den Brand, T.; van Welsem, T.; de Rink, I.; Song, J.Y.; et al. Histone methyltransferase DOT1L controls state-specific identity during B cell differentiation. EMBO Rep. 2021, 22, e51184. [Google Scholar] [CrossRef] [PubMed]
- Cobaleda, C.; Schebesta, A.; Delogu, A.; Busslinger, M. Pax5: The guardian of B cell identity and function. Nat. Immunol. 2007, 8, 463–470. [Google Scholar] [CrossRef] [PubMed]
- Hatzi, K.; Melnick, A. Breaking bad in the germinal center: How deregulation of BCL6 contributes to lymphomagenesis. Trends Mol. Med. 2014, 20, 343–352. [Google Scholar] [CrossRef] [PubMed]
- Mochizuki, K.; Nishiyama, A.; Jang, M.K.; Dey, A.; Ghosh, A.; Tamura, T.; Natsume, H.; Yao, H.; Ozato, K. The bromodomain protein Brd4 stimulates G1 gene transcription and promotes progression to S phase. J. Biol. Chem. 2008, 283, 9040–9048. [Google Scholar] [CrossRef]
- McGinty, R.K.; Tan, S. Histone, Nucleosome, and Chromatin Structure. In Fundamentals of Chromatin; Workman, J.L., Abmayr, S.M., Eds.; Springer: New York, NY, USA, 2014; pp. 1–28. [Google Scholar]
- Bell, O.; Tiwari, V.K.; Thomä, N.H.; Schübeler, D. Determinants and dynamics of genome accessibility. Nat. Rev. Genet. 2011, 12, 554–564. [Google Scholar] [CrossRef]
- Klemm, S.L.; Shipony, Z.; Greenleaf, W.J. Chromatin accessibility and the regulatory epigenome. Nat. Rev. Genet. 2019, 20, 207–220. [Google Scholar] [CrossRef]
- Choi, J.; Jeon, S.; Choi, S.; Park, K.; Seong, R.H. The SWI/SNF chromatin remodeling complex regulates germinal center formation by repressing Blimp-1 expression. Proc. Natl. Acad. Sci. USA 2015, 112, E718–E727. [Google Scholar] [CrossRef]
- Vilarrasa-Blasi, R.; Soler-Vila, P.; Verdaguer-Dot, N.; Russiñol, N.; Di Stefano, M.; Chapaprieta, V.; Clot, G.; Farabella, I.; Cuscó, P.; Kulis, M.; et al. Dynamics of genome architecture and chromatin function during human B cell differentiation and neoplastic transformation. Nat. Commun. 2021, 12, 651. [Google Scholar] [CrossRef]
- Schmiedel, D.; Hezroni, H.; Hamburg, A.; Shulman, Z. Brg1 Supports B Cell Proliferation and Germinal Center Formation Through Enhancer Activation. Front. Immunol. 2021, 12, 705848. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Yin, Y.; Wang, X. The epigenetic regulation of the germinal center response. Biochim. Biophys. Acta Gene Regul. Mech. 2022, 1865, 194828. [Google Scholar] [CrossRef]
- Sun, W.; Yang, Y.; Xu, C.; Guo, J. Regulatory mechanisms of long noncoding RNAs on gene expression in cancers. Cancer Genet. 2017, 216, 105–110. [Google Scholar] [CrossRef]
- Koues, O.I.; Oltz, E.M.; Payton, J.E. Short-Circuiting Gene Regulatory Networks: Origins of B Cell Lymphoma. Trends Genet. 2015, 31, 720–731. [Google Scholar] [CrossRef] [PubMed]
- Mohammadian, M.; Salimi, M.; Farhadihosseinabadi, B.; Noorazar, L.; Dehghani Ghorbi, M.; Mohammadali, F.; Mirfakhraie, R.; Roshandel, E.; Hajifathali, A. B-Cell Chronic Lymphocytic Leukemia and B-Cell Lymphomas: The Key Role of Micro and Long Noncoding RNAs. J. Clin. Pharm. Ther. 2024, 2024, 3091760. [Google Scholar] [CrossRef]
- Attaway, M.; Chwat-Edelstein, T.; Vuong, B.Q. Regulatory Non-Coding RNAs Modulate Transcriptional Activation During B Cell Development. Front. Genet. 2021, 12, 678084. [Google Scholar] [CrossRef]
- Shi, Y.; Xu, Y.; Shen, H.; Jin, J.; Tong, H.; Xie, W. Advances in biology, diagnosis and treatment of DLBCL. Ann. Hematol. 2024, 103, 3315–3334. [Google Scholar] [CrossRef] [PubMed]
- Pasqualucci, L.; Dalla-Favera, R. Genetics of diffuse large B-cell lymphoma. Blood 2018, 131, 2307–2319. [Google Scholar] [CrossRef]
- Melchardt, T.; Egle, A.; Greil, R. How I treat diffuse large B-cell lymphoma. ESMO Open 2023, 8, 100750. [Google Scholar] [CrossRef]
- Caloian, A.D.; Cristian, M.; Calin, E.; Pricop, A.R.; Mociu, S.I.; Seicaru, L.; Deacu, S.; Ciufu, N.; Suceveanu, A.I.; Suceveanu, A.P.; et al. Epigenetic Symphony in Diffuse Large B-Cell Lymphoma: Orchestrating the Tumor Microenvironment. Biomedicines 2025, 13, 853. [Google Scholar] [CrossRef]
- Chapuy, B.; Stewart, C.; Dunford, A.J.; Kim, J.; Kamburov, A.; Redd, R.A.; Lawrence, M.S.; Roemer, M.G.M.; Li, A.J.; Ziepert, M.; et al. Molecular subtypes of diffuse large B cell lymphoma are associated with distinct pathogenic mechanisms and outcomes. Nat. Med. 2018, 24, 679–690. [Google Scholar] [CrossRef]
- Wright, G.W.; Huang, D.W.; Phelan, J.D.; Coulibaly, Z.A.; Roulland, S.; Young, R.M.; Wang, J.Q.; Schmitz, R.; Morin, R.D.; Tang, J.; et al. A Probabilistic Classification Tool for Genetic Subtypes of Diffuse Large B Cell Lymphoma with Therapeutic Implications. Cancer Cell 2020, 37, 551–568.e14. [Google Scholar] [CrossRef]
- Moia, R.; Talotta, D.; Terzi Di Bergamo, L.; Almasri, M.; Dondolin, R.; Salehi, M.; Cosentino, C.; Soscia, R.; Della Starza, I.; Bruscaggin, A.; et al. Molecular clustering on ctDNA improves the prognostic stratification of patients with DLBCL compared with ctDNA levels. Blood Adv. 2025, 9, 1692–1701. [Google Scholar] [CrossRef] [PubMed]
- Pan, H.; Jiang, Y.; Boi, M.; Tabbò, F.; Redmond, D.; Nie, K.; Ladetto, M.; Chiappella, A.; Cerchietti, L.; Shaknovich, R.; et al. Epigenomic evolution in diffuse large B-cell lymphomas. Nat. Commun. 2015, 6, 6921. [Google Scholar] [CrossRef] [PubMed]
- Clozel, T.; Yang, S.; Elstrom, R.L.; Tam, W.; Martin, P.; Kormaksson, M.; Banerjee, S.; Vasanthakumar, A.; Culjkovic, B.; Scott, D.W.; et al. Mechanism-based epigenetic chemosensitization therapy of diffuse large B-cell lymphoma. Cancer Discov. 2013, 3, 1002–1019. [Google Scholar] [CrossRef]
- Bakhshi, T.J.; Georgel, P.T. Genetic and epigenetic determinants of diffuse large B-cell lymphoma. Blood Cancer J. 2020, 10, 123. [Google Scholar] [CrossRef] [PubMed]
- Hashwah, H.; Schmid, C.A.; Kasser, S.; Bertram, K.; Stelling, A.; Manz, M.G.; Müller, A. Inactivation of CREBBP expands the germinal center B cell compartment, down-regulates MHCII expression and promotes DLBCL growth. Proc. Natl. Acad. Sci. USA 2017, 114, 9701–9706. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.H.; Cai, K.; Xu, P.P.; Wang, L.; Huang, C.X.; Fang, Y.; Cheng, S.; Sun, X.J.; Liu, F.; Huang, J.Y.; et al. CREBBP/EP300 mutations promoted tumor progression in diffuse large B-cell lymphoma through altering tumor-associated macrophage polarization via FBXW7-NOTCH-CCL2/CSF1 axis. Signal Transduct. Target. Ther. 2021, 6, 10. [Google Scholar] [CrossRef]
- Pasqualucci, L.; Dominguez-Sola, D.; Chiarenza, A.; Fabbri, G.; Grunn, A.; Trifonov, V.; Kasper, L.H.; Lerach, S.; Tang, H.; Ma, J.; et al. Inactivating mutations of acetyltransferase genes in B-cell lymphoma. Nature 2011, 471, 189–195. [Google Scholar] [CrossRef]
- Morin, R.D.; Johnson, N.A.; Severson, T.M.; Mungall, A.J.; An, J.; Goya, R.; Paul, J.E.; Boyle, M.; Woolcock, B.W.; Kuchenbauer, F.; et al. Somatic mutations altering EZH2 (Tyr641) in follicular and diffuse large B-cell lymphomas of germinal-center origin. Nat. Genet. 2010, 42, 181–185. [Google Scholar] [CrossRef]
- Caganova, M.; Carrisi, C.; Varano, G.; Mainoldi, F.; Zanardi, F.; Germain, P.L.; George, L.; Alberghini, F.; Ferrarini, L.; Talukder, A.K.; et al. Germinal center dysregulation by histone methyltransferase EZH2 promotes lymphomagenesis. J. Clin. Investig. 2013, 123, 5009–5022. [Google Scholar] [CrossRef]
- Velichutina, I.; Shaknovich, R.; Geng, H.; Johnson, N.A.; Gascoyne, R.D.; Melnick, A.M.; Elemento, O. EZH2-mediated epigenetic silencing in germinal center B cells contributes to proliferation and lymphomagenesis. Blood 2010, 116, 5247–5255. [Google Scholar] [CrossRef] [PubMed]
- Froimchuk, E.; Jang, Y.; Ge, K. Histone H3 lysine 4 methyltransferase KMT2D. Gene 2017, 627, 337–342. [Google Scholar] [CrossRef]
- Zhang, J.; Dominguez-Sola, D.; Hussein, S.; Lee, J.E.; Holmes, A.B.; Bansal, M.; Vlasevska, S.; Mo, T.; Tang, H.; Basso, K.; et al. Disruption of KMT2D perturbs germinal center B cell development and promotes lymphomagenesis. Nat. Med. 2015, 21, 1190–1198. [Google Scholar] [CrossRef]
- Liu, Q.X.; Zhu, Y.; Yi, H.M.; Shen, Y.G.; Wang, L.; Cheng, S.; Xu, P.P.; Xu, H.M.; Zhou, L.T.; Huang, Y.H.; et al. KMT2D mutations promoted tumor progression in diffuse large B-cell lymphoma through altering tumor-induced regulatory T cell trafficking via FBXW7-NOTCH-MYC/TGF-β1 axis. Int. J. Biol. Sci. 2024, 20, 3972–3985. [Google Scholar] [CrossRef]
- Bowman, R.L.; Levine, R.L. TET2 in Normal and Malignant Hematopoiesis. Cold Spring Harb. Perspect. Med. 2017, 7, a026518. [Google Scholar] [CrossRef] [PubMed]
- Rosikiewicz, W.; Chen, X.; Dominguez, P.M.; Ghamlouch, H.; Aoufouchi, S.; Bernard, O.A.; Melnick, A.; Li, S. TET2 deficiency reprograms the germinal center B cell epigenome and silences genes linked to lymphomagenesis. Sci. Adv. 2020, 6, eaay5872. [Google Scholar] [CrossRef] [PubMed]
- Shukla, V.; Samaniego-Castruita, D.; Dong, Z.; González-Avalos, E.; Yan, Q.; Sarma, K.; Rao, A. TET deficiency perturbs mature B cell homeostasis and promotes oncogenesis associated with accumulation of G-quadruplex and R-loop structures. Nat. Immunol. 2022, 23, 99–108. [Google Scholar] [CrossRef] [PubMed]
- Go, H.; Jang, J.Y.; Kim, P.J.; Kim, Y.G.; Nam, S.J.; Paik, J.H.; Kim, T.M.; Heo, D.S.; Kim, C.W.; Jeon, Y.K. MicroRNA-21 plays an oncogenic role by targeting FOXO1 and activating the PI3K/AKT pathway in diffuse large B-cell lymphoma. Oncotarget 2015, 6, 15035–15049. [Google Scholar] [CrossRef]
- Due, H.; Schönherz, A.A.; Ryø, L.; Primo, M.N.; Jespersen, D.S.; Thomsen, E.A.; Roug, A.S.; Xiao, M.; Tan, X.; Pang, Y.; et al. MicroRNA-155 controls vincristine sensitivity and predicts superior clinical outcome in diffuse large B-cell lymphoma. Blood Adv. 2019, 3, 1185–1196. [Google Scholar] [CrossRef]
- Kim, S.W.; Ramasamy, K.; Bouamar, H.; Lin, A.P.; Jiang, D.; Aguiar, R.C. MicroRNAs miR-125a and miR-125b constitutively activate the NF-κB pathway by targeting the tumor necrosis factor alpha-induced protein 3 (TNFAIP3, A20). Proc. Natl. Acad. Sci. USA 2012, 109, 7865–7870. [Google Scholar] [CrossRef]
- Larrabeiti-Etxebarria, A.; Bilbao-Aldaiturriaga, N.; Arzuaga-Mendez, J.; Martin-Arruti, M.; Cozzuto, L.; Gaafar, A.; Ruiz-Diaz, I.; Guerra, I.; Martin-Guerrero, I.; Lopez-Lopez, E.; et al. microRNA sequencing for biomarker detection in the diagnosis, classification and prognosis of Diffuse Large B Cell Lymphoma. Sci. Rep. 2023, 13, 12159. [Google Scholar] [CrossRef]
- Swerdlow, S.H.; Campo, E.; Pileri, S.A.; Harris, N.L.; Stein, H.; Siebert, R.; Advani, R.; Ghielmini, M.; Salles, G.A.; Zelenetz, A.D.; et al. The 2016 revision of the World Health Organization classification of lymphoid neoplasms. Blood 2016, 127, 2375–2390. [Google Scholar] [CrossRef]
- Maher, N.; Mouhssine, S.; Matti, B.F.; Alwan, A.F.; Gaidano, G. Molecular Mechanisms in the Transformation from Indolent to Aggressive B Cell Malignancies. Cancers 2025, 17, 907. [Google Scholar] [CrossRef]
- Green, M.R. Chromatin modifying gene mutations in follicular lymphoma. Blood 2018, 131, 595–604. [Google Scholar] [CrossRef]
- Hellmuth, J.C.; Louissaint, A., Jr.; Szczepanowski, M.; Haebe, S.; Pastore, A.; Alig, S.; Staiger, A.M.; Hartmann, S.; Kridel, R.; Ducar, M.D.; et al. Duodenal-type and nodal follicular lymphomas differ by their immune microenvironment rather than their mutation profiles. Blood 2018, 132, 1695–1702. [Google Scholar] [CrossRef]
- Green, M.R.; Gentles, A.J.; Nair, R.V.; Irish, J.M.; Kihira, S.; Liu, C.L.; Kela, I.; Hopmans, E.S.; Myklebust, J.H.; Ji, H.; et al. Hierarchy in somatic mutations arising during genomic evolution and progression of follicular lymphoma. Blood 2013, 121, 1604–1611. [Google Scholar] [CrossRef]
- Okosun, J.; Bödör, C.; Wang, J.; Araf, S.; Yang, C.Y.; Pan, C.; Boller, S.; Cittaro, D.; Bozek, M.; Iqbal, S.; et al. Integrated genomic analysis identifies recurrent mutations and evolution patterns driving the initiation and progression of follicular lymphoma. Nat. Genet. 2014, 46, 176–181. [Google Scholar] [CrossRef]
- Béguelin, W.; Teater, M.; Meydan, C.; Hoehn, K.B.; Phillip, J.M.; Soshnev, A.A.; Venturutti, L.; Rivas, M.A.; Calvo-Fernández, M.T.; Gutierrez, J.; et al. Mutant EZH2 Induces a Pre-malignant Lymphoma Niche by Reprogramming the Immune Response. Cancer Cell 2020, 37, 655–673.e11. [Google Scholar] [CrossRef] [PubMed]
- Ortega-Molina, A.; Boss, I.; Pan, H.; Jiang, Y.; Hu, D.; Gao, X.; Shaknovich, R.; Shilatifard, A.; Melnick, A.M.; Wendel, H.-G. Abstract LB-064: Characterization of the tumor suppressor function of the lysine-specific methyltransferase KMT2D in follicular lymphoma. Cancer Res. 2015, 75, LB-064. [Google Scholar] [CrossRef]
- Margueron, R.; Li, G.; Sarma, K.; Blais, A.; Zavadil, J.; Woodcock, C.L.; Dynlacht, B.D.; Reinberg, D. Ezh1 and Ezh2 maintain repressive chromatin through different mechanisms. Mol. Cell 2008, 32, 503–518. [Google Scholar] [CrossRef]
- Shen, X.; Liu, Y.; Hsu, Y.J.; Fujiwara, Y.; Kim, J.; Mao, X.; Yuan, G.C.; Orkin, S.H. EZH1 mediates methylation on histone H3 lysine 27 and complements EZH2 in maintaining stem cell identity and executing pluripotency. Mol. Cell 2008, 32, 491–502. [Google Scholar] [CrossRef]
- Zhang, J.; Vlasevska, S.; Wells, V.A.; Nataraj, S.; Holmes, A.B.; Duval, R.; Meyer, S.N.; Mo, T.; Basso, K.; Brindle, P.K.; et al. The CREBBP Acetyltransferase Is a Haploinsufficient Tumor Suppressor in B-cell Lymphoma. Cancer Discov. 2017, 7, 322–337. [Google Scholar] [CrossRef] [PubMed]
- Schroers-Martin, J.G.; Soo, J.; Brisou, G.; Scherer, F.; Kurtz, D.M.; Sworder, B.J.; Khodadoust, M.S.; Jin, M.C.; Bru, A.; Liu, C.L.; et al. Tracing Founder Mutations in Circulating and Tissue-Resident Follicular Lymphoma Precursors. Cancer Discov. 2023, 13, 1310–1323. [Google Scholar] [CrossRef]
- Korfi, K.; Ali, S.; Heward, J.A.; Fitzgibbon, J. Follicular lymphoma, a B cell malignancy addicted to epigenetic mutations. Epigenetics 2017, 12, 370–377. [Google Scholar] [CrossRef] [PubMed]
- Takeshima, H.; Suetake, I.; Tajima, S. Mouse Dnmt3a preferentially methylates linker DNA and is inhibited by histone H1. J. Mol. Biol. 2008, 383, 810–821. [Google Scholar] [CrossRef]
- Alhejaily, A.; Day, A.G.; Feilotter, H.E.; Baetz, T.; Lebrun, D.P. Inactivation of the CDKN2A tumor-suppressor gene by deletion or methylation is common at diagnosis in follicular lymphoma and associated with poor clinical outcome. Clin. Cancer Res. 2014, 20, 1676–1686. [Google Scholar] [CrossRef]
- De, S.; Shaknovich, R.; Riester, M.; Elemento, O.; Geng, H.; Kormaksson, M.; Jiang, Y.; Woolcock, B.; Johnson, N.; Polo, J.M.; et al. Aberration in DNA methylation in B-cell lymphomas has a complex origin and increases with disease severity. PLoS Genet. 2013, 9, e1003137. [Google Scholar] [CrossRef]
- Kretzmer, H.; Bernhart, S.H.; Wang, W.; Haake, A.; Weniger, M.A.; Bergmann, A.K.; Betts, M.J.; Carrillo-de-Santa-Pau, E.; Doose, G.; Gutwein, J.; et al. DNA methylome analysis in Burkitt and follicular lymphomas identifies differentially methylated regions linked to somatic mutation and transcriptional control. Nat. Genet. 2015, 47, 1316–1325. [Google Scholar] [CrossRef]
- Barisic, D.; Chin, C.; Meydan, C.; Teater, M.; Tsialta, I.; Mlynarczyk, C.; Chadburn, A.; Xia, M.; Steidl, C.; Scott, D.W.; et al. SWI/SNF Chromatin Remodeling Complex Orchestrates Sequential Binding of Key Transcription Factors in B Cells and Restricts Aggressive Lymphoma. Blood 2023, 142, 718. [Google Scholar] [CrossRef]
- Pastore, A.; Jurinovic, V.; Kridel, R.; Hoster, E.; Staiger, A.M.; Szczepanowski, M.; Pott, C.; Kopp, N.; Murakami, M.; Horn, H.; et al. Integration of gene mutations in risk prognostication for patients receiving first-line immunochemotherapy for follicular lymphoma: A retrospective analysis of a prospective clinical trial and validation in a population-based registry. Lancet Oncol. 2015, 16, 1111–1122. [Google Scholar] [CrossRef]
- Roisman, A.; Castellano, G.; Navarro, A.; Gonzalez-Farre, B.; Pérez-Galan, P.; Esteve-Codina, A.; Dabad, M.; Heath, S.; Gut, M.; Bosio, M.; et al. Differential expression of long non-coding RNAs are related to proliferation and histological diversity in follicular lymphomas. Br. J. Haematol. 2019, 184, 373–383. [Google Scholar] [CrossRef]
- Wang, W.; Corrigan-Cummins, M.; Hudson, J.; Maric, I.; Simakova, O.; Neelapu, S.S.; Kwak, L.W.; Janik, J.E.; Gause, B.; Jaffe, E.S.; et al. MicroRNA profiling of follicular lymphoma identifies microRNAs related to cell proliferation and tumor response. Haematologica 2012, 97, 586–594. [Google Scholar] [CrossRef]
- Artemaki, P.I.; Letsos, P.A.; Zoupa, I.C.; Katsaraki, K.; Karousi, P.; Papageorgiou, S.G.; Pappa, V.; Scorilas, A.; Kontos, C.K. The Multifaceted Role and Utility of MicroRNAs in Indolent B-Cell Non-Hodgkin Lymphomas. Biomedicines 2021, 9, 333. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zou, J.; Wan, X.; Sun, C.; Peng, F.; Chu, Z.; Hu, Y. The Role of Noncoding RNAs in B-Cell Lymphoma. Front. Oncol. 2020, 10, 577890. [Google Scholar] [CrossRef]
- Musilova, K.; Devan, J.; Cerna, K.; Seda, V.; Pavlasova, G.; Sharma, S.; Oppelt, J.; Pytlik, R.; Prochazka, V.; Prouzova, Z.; et al. miR-150 downregulation contributes to the high-grade transformation of follicular lymphoma by upregulating FOXP1 levels. Blood 2018, 132, 2389–2400. [Google Scholar] [CrossRef] [PubMed]
- Hallek, M. Chronic Lymphocytic Leukemia: 2025 Update on the Epidemiology, Pathogenesis, Diagnosis, and Therapy. Am. J. Hematol. 2025, 100, 450–480. [Google Scholar] [CrossRef] [PubMed]
- Moia, R.; Gaidano, G. Prognostication in chronic lymphocytic leukemia. Semin. Hematol. 2024, 61, 83–90. [Google Scholar] [CrossRef]
- Mouhssine, S.; Maher, N.; Kogila, S.; Cerchione, C.; Martinelli, G.; Gaidano, G. Current Therapeutic Sequencing in Chronic Lymphocytic Leukemia. Hematol. Rep. 2024, 16, 270–282. [Google Scholar] [CrossRef] [PubMed]
- Kulis, M.; Heath, S.; Bibikova, M.; Queirós, A.C.; Navarro, A.; Clot, G.; Martínez-Trillos, A.; Castellano, G.; Brun-Heath, I.; Pinyol, M.; et al. Epigenomic analysis detects widespread gene-body DNA hypomethylation in chronic lymphocytic leukemia. Nat. Genet. 2012, 44, 1236–1242. [Google Scholar] [CrossRef]
- Cahill, N.; Bergh, A.C.; Kanduri, M.; Göransson-Kultima, H.; Mansouri, L.; Isaksson, A.; Ryan, F.; Smedby, K.E.; Juliusson, G.; Sundström, C.; et al. 450K-array analysis of chronic lymphocytic leukemia cells reveals global DNA methylation to be relatively stable over time and similar in resting and proliferative compartments. Leukemia 2013, 27, 150–158. [Google Scholar] [CrossRef]
- Oakes, C.C.; Seifert, M.; Assenov, Y.; Gu, L.; Przekopowitz, M.; Ruppert, A.S.; Wang, Q.; Imbusch, C.D.; Serva, A.; Koser, S.D.; et al. DNA methylation dynamics during B cell maturation underlie a continuum of disease phenotypes in chronic lymphocytic leukemia. Nat. Genet. 2016, 48, 253–264. [Google Scholar] [CrossRef]
- Queirós, A.C.; Villamor, N.; Clot, G.; Martinez-Trillos, A.; Kulis, M.; Navarro, A.; Penas, E.M.; Jayne, S.; Majid, A.; Richter, J.; et al. A B-cell epigenetic signature defines three biologic subgroups of chronic lymphocytic leukemia with clinical impact. Leukemia 2015, 29, 598–605. [Google Scholar] [CrossRef]
- Puente, X.S.; Beà, S.; Valdés-Mas, R.; Villamor, N.; Gutiérrez-Abril, J.; Martín-Subero, J.I.; Munar, M.; Rubio-Pérez, C.; Jares, P.; Aymerich, M.; et al. Non-coding recurrent mutations in chronic lymphocytic leukaemia. Nature 2015, 526, 519–524. [Google Scholar] [CrossRef]
- Bhoi, S.; Ljungström, V.; Baliakas, P.; Mattsson, M.; Smedby, K.E.; Juliusson, G.; Rosenquist, R.; Mansouri, L. Prognostic impact of epigenetic classification in chronic lymphocytic leukemia: The case of subset #2. Epigenetics 2016, 11, 449–455. [Google Scholar] [CrossRef]
- Giacopelli, B.; Zhao, Q.; Ruppert, A.S.; Agyeman, A.; Weigel, C.; Wu, Y.Z.; Gerber, M.M.; Rabe, K.G.; Larson, M.C.; Lu, J.; et al. Developmental subtypes assessed by DNA methylation-iPLEX forecast the natural history of chronic lymphocytic leukemia. Blood 2019, 134, 688–698. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, X. Targeting the Wnt/β-catenin signaling pathway in cancer. J. Hematol. Oncol. 2020, 13, 165. [Google Scholar] [CrossRef]
- Pawar, N.M.; Rao, P. Secreted frizzled related protein 4 (sFRP4) update: A brief review. Cell Signal 2018, 45, 63–70. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.H.; Raval, A.; Chen, S.S.; Matkovic, J.J.; Byrd, J.C.; Plass, C. CpG island methylation and expression of the secreted frizzled-related protein gene family in chronic lymphocytic leukemia. Cancer Res. 2006, 66, 653–658. [Google Scholar] [CrossRef] [PubMed]
- Barrow, T.M.; Nakjang, S.; Lafta, F.; Bilotkach, K.; Woodhouse, L.; Junge, G.; Tudhope, S.J.; Wallis, J.P.; Marr, H.; Marshall, S.; et al. Epigenome-wide analysis reveals functional modulators of drug sensitivity and post-treatment survival in chronic lymphocytic leukaemia. Br. J. Cancer 2021, 124, 474–483. [Google Scholar] [CrossRef]
- Terzi Di Bergamo, L.; Forestieri, G.; Loh, J.W.; Singh, A.; Spina, V.; Jauk, F.; Zucchetto, A.; Condoluci, A.; Bruscaggin, A.; Pini, K.; et al. Adaptation of Chronic Lymphocytic Leukemia to Ibrutinib Is Mediated By Epigenetic Plasticity of Residual Disease and By-Pass Signaling Via MAPK Pathway. Blood 2024, 144, 73. [Google Scholar] [CrossRef]
- Chapaprieta, V.; Maiques-Diaz, A.; Nadeu, F.; Clot, G.; Massoni-Badosa, R.; Mozas, P.; Mateos-Jaimez, J.; Vidal, A.; Charalampopoulou, S.; Duran-Ferrer, M.; et al. Dual biological role and clinical impact of de novo chromatin activation in chronic lymphocytic leukemia. Blood 2025, 145, 2473–2487. [Google Scholar] [CrossRef] [PubMed]
- Beekman, R.; Chapaprieta, V.; Russiñol, N.; Vilarrasa-Blasi, R.; Verdaguer-Dot, N.; Martens, J.H.A.; Duran-Ferrer, M.; Kulis, M.; Serra, F.; Javierre, B.M.; et al. The reference epigenome and regulatory chromatin landscape of chronic lymphocytic leukemia. Nat. Med. 2018, 24, 868–880. [Google Scholar] [CrossRef]
- Mansouri, L.; Wierzbinska, J.A.; Plass, C.; Rosenquist, R. Epigenetic deregulation in chronic lymphocytic leukemia: Clinical and biological impact. Semin. Cancer Biol. 2018, 51, 1–11. [Google Scholar] [CrossRef]
- Nadeu, F.; Diaz-Navarro, A.; Delgado, J.; Puente, X.S.; Campo, E. Genomic and Epigenomic Alterations in Chronic Lymphocytic Leukemia. Annu. Rev. Pathol. 2020, 15, 149–177. [Google Scholar] [CrossRef]
- Rosenthal, A.C.; Munoz, J.L.; Villasboas, J.C. Clinical advances in epigenetic therapies for lymphoma. Clin. Epigenetics 2023, 15, 39. [Google Scholar] [CrossRef]
- Mallm, J.P.; Iskar, M.; Ishaque, N.; Klett, L.C.; Kugler, S.J.; Muino, J.M.; Teif, V.B.; Poos, A.M.; Großmann, S.; Erdel, F.; et al. Linking aberrant chromatin features in chronic lymphocytic leukemia to transcription factor networks. Mol. Syst. Biol. 2019, 15, e8339. [Google Scholar] [CrossRef] [PubMed]
- Papakonstantinou, N.; Ntoufa, S.; Chartomatsidou, E.; Kotta, K.; Agathangelidis, A.; Giassafaki, L.; Karamanli, T.; Bele, P.; Moysiadis, T.; Baliakas, P.; et al. The histone methyltransferase EZH2 as a novel prosurvival factor in clinically aggressive chronic lymphocytic leukemia. Oncotarget 2016, 7, 35946. [Google Scholar] [CrossRef]
- Rodríguez, D.; Bretones, G.; Quesada, V.; Villamor, N.; Arango, J.R.; López-Guillermo, A.; Ramsay, A.J.; Baumann, T.; Quirós, P.M.; Navarro, A.; et al. Mutations in CHD2 cause defective association with active chromatin in chronic lymphocytic leukemia. Blood 2015, 126, 195–202. [Google Scholar] [CrossRef] [PubMed]
- Parker, H.; Rose-Zerilli, M.J.J.; Larrayoz, M.; Clifford, R.; Edelmann, J.; Blakemore, S.; Gibson, J.; Wang, J.; Ljungström, V.; Wojdacz, T.K.; et al. Genomic disruption of the histone methyltransferase SETD2 in chronic lymphocytic leukaemia. Leukemia 2016, 30, 2179–2186. [Google Scholar] [CrossRef] [PubMed]
- Pfister, S.X.; Ahrabi, S.; Zalmas, L.P.; Sarkar, S.; Aymard, F.; Bachrati, C.Z.; Helleday, T.; Legube, G.; La Thangue, N.B.; Porter, A.C.; et al. SETD2-dependent histone H3K36 trimethylation is required for homologous recombination repair and genome stability. Cell Rep. 2014, 7, 2006–2018. [Google Scholar] [CrossRef] [PubMed]
- Cimmino, A.; Calin, G.A.; Fabbri, M.; Iorio, M.V.; Ferracin, M.; Shimizu, M.; Wojcik, S.E.; Aqeilan, R.I.; Zupo, S.; Dono, M.; et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc. Natl. Acad. Sci. USA 2005, 102, 13944–13949. [Google Scholar] [CrossRef]
- Pekarsky, Y.; Croce, C.M. Role of miR-15/16 in CLL. Cell Death Differ. 2015, 22, 6–11. [Google Scholar] [CrossRef]
- Corney, D.C.; Flesken-Nikitin, A.; Godwin, A.K.; Wang, W.; Nikitin, A.Y. MicroRNA-34b and MicroRNA-34c are targets of p53 and cooperate in control of cell proliferation and adhesion-independent growth. Cancer Res. 2007, 67, 8433–8438. [Google Scholar] [CrossRef]
- Deneberg, S.; Kanduri, M.; Ali, D.; Bengtzen, S.; Karimi, M.; Qu, Y.; Kimby, E.; Mansouri, L.; Rosenquist, R.; Lennartsson, A.; et al. microRNA-34b/c on chromosome 11q23 is aberrantly methylated in chronic lymphocytic leukemia. Epigenetics 2014, 9, 910–917. [Google Scholar] [CrossRef]
- Eis, P.S.; Tam, W.; Sun, L.; Chadburn, A.; Li, Z.; Gomez, M.F.; Lund, E.; Dahlberg, J.E. Accumulation of miR-155 and BIC RNA in human B cell lymphomas. Proc. Natl. Acad. Sci. USA 2005, 102, 3627–3632. [Google Scholar] [CrossRef]
- Cui, B.; Chen, L.; Zhang, S.; Mraz, M.; Fecteau, J.F.; Yu, J.; Ghia, E.M.; Zhang, L.; Bao, L.; Rassenti, L.Z.; et al. MicroRNA-155 influences B-cell receptor signaling and associates with aggressive disease in chronic lymphocytic leukemia. Blood 2014, 124, 546–554. [Google Scholar] [CrossRef]
- Ferrajoli, A.; Shanafelt, T.D.; Ivan, C.; Shimizu, M.; Rabe, K.G.; Nouraee, N.; Ikuo, M.; Ghosh, A.K.; Lerner, S.; Rassenti, L.Z.; et al. Prognostic value of miR-155 in individuals with monoclonal B-cell lymphocytosis and patients with B chronic lymphocytic leukemia. Blood 2013, 122, 1891–1899. [Google Scholar] [CrossRef]
- Moia, R.; Terzi di Bergamo, L.; Talotta, D.; Bomben, R.; Forestieri, G.; Spina, V.; Bruscaggin, A.; Cosentino, C.; Almasri, M.; Dondolin, R.; et al. XPO1 mutations identify early-stage CLL characterized by shorter time to first treatment and enhanced BCR signalling. Br. J. Haematol. 2023, 203, 416–425. [Google Scholar] [CrossRef]
- Sharma, S.; Pavlasova, G.M.; Seda, V.; Cerna, K.A.; Vojackova, E.; Filip, D.; Ondrisova, L.; Sandova, V.; Kostalova, L.; Zeni, P.F.; et al. miR-29 modulates CD40 signaling in chronic lymphocytic leukemia by targeting TRAF4: An axis affected by BCR inhibitors. Blood 2021, 137, 2481–2494. [Google Scholar] [CrossRef]
- Balatti, V.; Tomasello, L.; Rassenti, L.Z.; Veneziano, D.; Nigita, G.; Wang, H.Y.; Thorson, J.A.; Kipps, T.J.; Pekarsky, Y.; Croce, C.M. miR-125a and miR-34a expression predicts Richter syndrome in chronic lymphocytic leukemia patients. Blood 2018, 132, 2179–2182. [Google Scholar] [CrossRef] [PubMed]
- Mraz, M.; Chen, L.; Rassenti, L.Z.; Ghia, E.M.; Li, H.; Jepsen, K.; Smith, E.N.; Messer, K.; Frazer, K.A.; Kipps, T.J. miR-150 influences B-cell receptor signaling in chronic lymphocytic leukemia by regulating expression of GAB1 and FOXP1. Blood 2014, 124, 84–95. [Google Scholar] [CrossRef]
- Malik, A.; Shoukier, M.; Garcia-Manero, G.; Wierda, W.; Cortes, J.; Bickel, S.; Keating, M.J.; Estrov, Z. Azacitidine in fludarabine-refractory chronic lymphocytic leukemia: A phase II study. Clin. Lymphoma Myeloma Leuk. 2013, 13, 292–295. [Google Scholar] [CrossRef] [PubMed]
- Martin, P.; Bartlett, N.L.; Chavez, J.C.; Reagan, J.L.; Smith, S.M.; LaCasce, A.S.; Jones, J.; Drew, J.; Wu, C.; Mulvey, E.; et al. Phase 1 study of oral azacitidine (CC-486) plus R-CHOP in previously untreated intermediate- to high-risk DLBCL. Blood 2022, 139, 1147–1159. [Google Scholar] [CrossRef] [PubMed]
- Nieto, Y.; Valdez, B.C.; Thall, P.F.; Jones, R.B.; Wei, W.; Myers, A.; Hosing, C.; Ahmed, S.; Popat, U.; Shpall, E.J.; et al. Double epigenetic modulation of high-dose chemotherapy with azacitidine and vorinostat for patients with refractory or poor-risk relapsed lymphoma. Cancer 2016, 122, 2680–2688. [Google Scholar] [CrossRef] [PubMed]
- Coiffier, B.; Pro, B.; Prince, H.M.; Foss, F.; Sokol, L.; Greenwood, M.; Caballero, D.; Borchmann, P.; Morschhauser, F.; Wilhelm, M.; et al. Results From a Pivotal, Open-Label, Phase II Study of Romidepsin in Relapsed or Refractory Peripheral T-Cell Lymphoma After Prior Systemic Therapy. J. Clin. Oncol. 2012, 30, 631–636. [Google Scholar] [CrossRef]
- Falchi, L.; Lue, J.K.; Amengual, J.E.; Sawas, A.; Deng, C.; Marchi, E.; Lichtenstein, E.; Khan, K.; Kim, H.; Atkins, L.E.; et al. A Phase 1/2 Study of Oral 5-Azacitidine and Romidepsin in Patients with Lymphoid Malignancies Reveals Promising Activity in Heavily Pretreated Peripheral T-Cell Lymphoma (PTCL). Blood 2017, 130, 1515. [Google Scholar] [CrossRef]
- Italiano, A.; Soria, J.C.; Toulmonde, M.; Michot, J.M.; Lucchesi, C.; Varga, A.; Coindre, J.M.; Blakemore, S.J.; Clawson, A.; Suttle, B.; et al. Tazemetostat, an EZH2 inhibitor, in relapsed or refractory B-cell non-Hodgkin lymphoma and advanced solid tumours: A first-in-human, open-label, phase 1 study. Lancet Oncol. 2018, 19, 649–659. [Google Scholar] [CrossRef] [PubMed]
- Morschhauser, F.; Tilly, H.; Chaidos, A.; McKay, P.; Phillips, T.; Assouline, S.; Batlevi, C.L.; Campbell, P.; Ribrag, V.; Damaj, G.L.; et al. Tazemetostat for patients with relapsed or refractory follicular lymphoma: An open-label, single-arm, multicentre, phase 2 trial. Lancet Oncol. 2020, 21, 1433–1442. [Google Scholar] [CrossRef]
- Sarkozy, C.; Molina, T.J.; Dubois, S.; Portugues, C.; Bohers, E.; Ysebaert, L.; Houot, R.; Pica, G.M.; Ruminy, P.; Herbaux, C.; et al. Efficacy of tazemetostat in combination with R-CHOP in elderly patients newly diagnosed with diffuse large B cell lymphoma: Results of the EpiRCHOP phase II study of the LYSA. EClinicalMedicine 2025, 82, 103157. [Google Scholar] [CrossRef]
- Palomba, M.L.; Cartron, G.; Popplewell, L.; Ribrag, V.; Westin, J.; Huw, L.Y.; Agarwal, S.; Shivhare, M.; Hong, W.J.; Raval, A.; et al. Combination of Atezolizumab and Tazemetostat in Patients with Relapsed/Refractory Diffuse Large B-Cell Lymphoma: Results From a Phase Ib Study. Clin. Lymphoma Myeloma Leuk. 2022, 22, 504–512. [Google Scholar] [CrossRef]
- Huang, S.; Wang, Z.; Zhou, J.; Huang, J.; Zhou, L.; Luo, J.; Wan, Y.Y.; Long, H.; Zhu, B. EZH2 Inhibitor GSK126 Suppresses Antitumor Immunity by Driving Production of Myeloid-Derived Suppressor Cells. Cancer Res. 2019, 79, 2009–2020. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, H.; Zhang, Y.; Wang, X. Advances in epigenetic alterations of chronic lymphocytic leukemia: From pathogenesis to treatment. Clin. Exp. Med. 2024, 24, 54. [Google Scholar] [CrossRef]
- Izutsu, K.; Ishitsuka, K.; Maruyama, D.; Tsukasaki, K.; Kusumoto, S.; Kakurai, Y.; Yamauchi, H.; Inoue, A.; Tachibana, M.; Tsutsumi, S.; et al. Valemetostat for Relapsed or Refractory B-Cell Lymphomas: Primary Results from a Phase 1 Trial. Blood 2023, 142, 1731. [Google Scholar] [CrossRef]
- Chen, I.C.; Sethy, B.; Liou, J.P. Recent Update of HDAC Inhibitors in Lymphoma. Front. Cell Dev. Biol. 2020, 8, 576391. [Google Scholar] [CrossRef]
- Cui, H.; Hong, Q.; Wei, R.; Li, H.; Wan, C.; Chen, X.; Zhao, S.; Bu, H.; Zhang, B.; Yang, D.; et al. Design and synthesis of HDAC inhibitors to enhance the therapeutic effect of diffuse large B-cell lymphoma by improving metabolic stability and pharmacokinetic characteristics. Eur. J. Med. Chem. 2022, 229, 114049. [Google Scholar] [CrossRef]
- Mann, B.S.; Johnson, J.R.; Cohen, M.H.; Justice, R.; Pazdur, R. FDA approval summary: Vorinostat for treatment of advanced primary cutaneous T-cell lymphoma. Oncologist 2007, 12, 1247–1252. [Google Scholar] [CrossRef]
- El Omari, N.; Bakrim, S.; Elhrech, H.; Aanniz, T.; Balahbib, A.; Lee, L.H.; Al Abdulmonem, W.; Bouyahya, A. Clinical efficacy and mechanistic insights of FDA-approved HDAC inhibitors in the treatment of lymphoma. Eur. J. Pharm. Sci. 2025, 208, 107057. [Google Scholar] [CrossRef]
- Batlevi, C.L.; Crump, M.; Andreadis, C.; Rizzieri, D.; Assouline, S.E.; Fox, S.; van der Jagt, R.H.C.; Copeland, A.; Potvin, D.; Chao, R.; et al. A phase 2 study of mocetinostat, a histone deacetylase inhibitor, in relapsed or refractory lymphoma. Br. J. Haematol. 2017, 178, 434–441. [Google Scholar] [CrossRef]
- Amengual, J.E.; Prabhu, S.A.; Lombardo, M.; Zullo, K.; Johannet, P.M.; Gonzalez, Y.; Scotto, L.; Serrano, X.J.; Wei, Y.; Duong, J.; et al. Mechanisms of Acquired Drug Resistance to the HDAC6 Selective Inhibitor Ricolinostat Reveals Rational Drug-Drug Combination with Ibrutinib. Clin. Cancer Res. 2017, 23, 3084–3096. [Google Scholar] [CrossRef]
- Cosenza, M.; Civallero, M.; Marcheselli, L.; Sacchi, S.; Pozzi, S. Ricolinostat, a selective HDAC6 inhibitor, shows anti-lymphoma cell activity alone and in combination with bendamustine. Apoptosis 2017, 22, 827–840. [Google Scholar] [CrossRef] [PubMed]
- Amengual, J.E.; Lue, J.K.; Ma, H.; Lichtenstein, R.; Shah, B.; Cremers, S.; Jones, S.; Sawas, A. First-in-Class Selective HDAC6 Inhibitor (ACY-1215) Has a Highly Favorable Safety Profile in Patients with Relapsed and Refractory Lymphoma. Oncologist 2021, 26, 184–e366. [Google Scholar] [CrossRef]
- Landsburg, D.J.; Barta, S.K.; Ramchandren, R.; Batlevi, C.; Iyer, S.; Kelly, K.; Micallef, I.N.; Smith, S.M.; Stevens, D.A.; Alvarez, M.; et al. Fimepinostat (CUDC-907) in patients with relapsed/refractory diffuse large B cell and high-grade B-cell lymphoma: Report of a phase 2 trial and exploratory biomarker analyses. Br. J. Haematol. 2021, 195, 201–209. [Google Scholar] [CrossRef] [PubMed]
- Ribrag, V.; Kim, W.S.; Bouabdallah, R.; Lim, S.T.; Coiffier, B.; Illes, A.; Lemieux, B.; Dyer, M.J.S.; Offner, F.; Felloussi, Z.; et al. Safety and efficacy of abexinostat, a pan-histone deacetylase inhibitor, in non-Hodgkin lymphoma and chronic lymphocytic leukemia: Results of a phase II study. Haematologica 2017, 102, 903–909. [Google Scholar] [CrossRef] [PubMed]
- Amorim, S.; Stathis, A.; Gleeson, M.; Iyengar, S.; Magarotto, V.; Leleu, X.; Morschhauser, F.; Karlin, L.; Broussais, F.; Rezai, K.; et al. Bromodomain inhibitor OTX015 in patients with lymphoma or multiple myeloma: A dose-escalation, open-label, pharmacokinetic, phase 1 study. Lancet Haematol. 2016, 3, e196–e204. [Google Scholar] [CrossRef] [PubMed]
- Blum, K.A.; Supko, J.G.; Maris, M.B.; Flinn, I.W.; Goy, A.; Younes, A.; Bobba, S.; Senderowicz, A.M.; Efuni, S.; Rippley, R.; et al. A Phase I Study of Pelabresib (CPI-0610), a Small-Molecule Inhibitor of BET Proteins, in Patients with Relapsed or Refractory Lymphoma. Cancer Res. Commun. 2022, 2, 795–805. [Google Scholar] [CrossRef] [PubMed]
- Dickinson, M.; Briones, J.; Herrera, A.F.; González-Barca, E.; Ghosh, N.; Cordoba, R.; Rutherford, S.C.; Bournazou, E.; Labriola-Tompkins, E.; Franjkovic, I.; et al. Phase 1b study of the BET protein inhibitor RO6870810 with venetoclax and rituximab in patients with diffuse large B-cell lymphoma. Blood Adv. 2021, 5, 4762–4770. [Google Scholar] [CrossRef]
- Sarkozy, C.; Tessoulin, B.; Chiron, D. Unraveling MCL biology to understand resistance and identify vulnerabilities. Blood 2025, 145, 696–707. [Google Scholar] [CrossRef]
- Abu-Alghayth, M.H. Molecular carcinogenesis in Hodgkin lymphoma: Interplay between B lymphocyte mutations and NF-κB pathway dysregulation. Pathol. Res. Pr. 2025, 273, 156145. [Google Scholar] [CrossRef]
- Lyu, L.; Li, Q.; Wang, C. EBV Latency Programs: Molecular and Epigenetic Regulation and Its Role in Disease Pathogenesis. J. Med. Virol. 2025, 97, e70501. [Google Scholar] [CrossRef] [PubMed]
| Drug | Epigeneitc Target | Phase | Setting | Disease | Primary Outcome | Identifier |
|---|---|---|---|---|---|---|
| Decitabine + anti PD-1 | DNMT | Phase II | R/R | DLBCL | ORR | NCT05816746 |
| ASTX727 + Nivolumab | DNMT | Phase I | R/R | NHL and HL | Safety | NCT05272384 |
| Tazemetostat + Mosunetuzumab | EZH2 | Phase II | First-line | FL | CR | NCT05994235 |
| Tulmimetostat DZR123 (CPI-0209) | EZH2 | Phase I/II | R/R | B and T lymphomas, and solid tumors | Safety ORR | NCT04104776 |
| Tazemetostat + CART | EZH2 | Phase I | R/R | DLBCL, FL, and MCL | Safety | NCT05934838 |
| tazemetostat + lenalidomide + rituximab | EZH2 | Phase I/II | R/R | FL | Safety PFS | NCT04224493 |
| Mevrometostat (PF-06821497) | EZH2 | Phase I | R/R | FL | Safety | NCT03460977 |
| Mevrometostat | EZH2 | Phase I | R/R | FL, SCLC, and CRPC | Safety | NCT03460977 |
| Tazemetostat + Belinostat | EZH2 + HDACs | Phase I | R/R | B-NHL and T-NHL | Safety | NCT05627245 |
| HH2853 | EZH2 | Phase I/II | R/R | DLBCL, and FL | Safety ORR | NCT04390737 |
| SHR2554 + SHR1701 | EZH2 | Phase I/II | R/R | B-cell lymphoma, and Advanced/metastatic solid tumors | PFS | NCT04407741 |
| Chidamide + Decitabine | HDACs + DNMT1 | Phase I/II | R/R | NHL after CART cells | CR AE | NCT04337606 |
| Chidamide + Linperlisib | HDACs + PI3Kδ | Phase Ⅱ | R/R | FL | CR | NCT06158386 |
| Purinostat | HDAC | Phase II | R/R | DLBCL | ORR | NCT05563844 |
| Abexinostat | HDAC | Phase II | R/R | DLBCL | ORR | NCT03936153 |
| Etinostat + ZEN003694 | HDAC + BET | Phase I/II | R/R | Lymphoma, and advanced solid tumors | Safety ORR | NCT05053971 |
| Vorinostat + pembrolizumab | HDAC | Phase I | R/R | DLBCL, FL, and HL | Safety | NCT03150329 |
| Tazemetostat + epcoritamab | EZH2 | Phase II | R/R | FL | Safety CR | NCT06575686 |
| Abexinostat | HDAC | Phase II | R/R | FL | CR or PR | NCT03600441 |
| Valemetostat + Lenalidomide | EZH1/EZH2 | Phase I/II | R/R | FL | Safety | NCT05683171 |
| Valemetostat | EZH1/EZH2 | Phase II | R/R | B-cell lymphoma | ORR | NCT04842877 |
| Pevoneidstat + Ibrutinib | NEDD8-activating enzyme | Phase I | R/R | CLL + B-NHL | Safety | NCT03479268 |
| Tulmimetostat | EZH1/EZH2 | Phase I/II | R/R | Solid tumors + Lymphoma | Safety ORR | NCT04104776 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maher, N.; Maiellaro, F.; Ghanej, J.; Rasi, S.; Moia, R.; Gaidano, G. Unraveling the Epigenetic Landscape of Mature B Cell Neoplasia: Mechanisms, Biomarkers, and Therapeutic Opportunities. Int. J. Mol. Sci. 2025, 26, 8132. https://doi.org/10.3390/ijms26178132
Maher N, Maiellaro F, Ghanej J, Rasi S, Moia R, Gaidano G. Unraveling the Epigenetic Landscape of Mature B Cell Neoplasia: Mechanisms, Biomarkers, and Therapeutic Opportunities. International Journal of Molecular Sciences. 2025; 26(17):8132. https://doi.org/10.3390/ijms26178132
Chicago/Turabian StyleMaher, Nawar, Francesca Maiellaro, Joseph Ghanej, Silvia Rasi, Riccardo Moia, and Gianluca Gaidano. 2025. "Unraveling the Epigenetic Landscape of Mature B Cell Neoplasia: Mechanisms, Biomarkers, and Therapeutic Opportunities" International Journal of Molecular Sciences 26, no. 17: 8132. https://doi.org/10.3390/ijms26178132
APA StyleMaher, N., Maiellaro, F., Ghanej, J., Rasi, S., Moia, R., & Gaidano, G. (2025). Unraveling the Epigenetic Landscape of Mature B Cell Neoplasia: Mechanisms, Biomarkers, and Therapeutic Opportunities. International Journal of Molecular Sciences, 26(17), 8132. https://doi.org/10.3390/ijms26178132







