Immune Modulation Through KIR–HLA Interactions Influences Cetuximab Efficacy in Colorectal Cancer
Abstract
1. Introduction
2. Results
2.1. Baseline Patient Characteristics and Clinical Outcome
2.2. Frequencies of KIR Genes and Their Ligands
2.3. Frequency of KIR–Ligand Combinations
2.4. Frequency of KIR Genotypes and Haplotypes
2.5. Survival Analysis According to KIR Genes or Their Ligands
2.6. Survival Analysis According to KIR–Ligand Combinations
2.7. Survival Analysis According to KIR Haplotypes
2.8. Survival Analysis According to Centromeric–Telomeric KIR Haplotypes
2.9. Univariate and Multivariate Analyses
3. Discussion
4. Materials and Methods
4.1. Patients and Inclusion Criteria
4.2. Ethical Considerations
4.3. Sample Processing and DNA Isolation for Genotyping
4.4. KIR Genotyping
4.5. HLA Genotyping
4.6. KIR–HLA Interactions
4.7. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Cervantes, A.; Adam, R.; Roselló, S.; Arnold, D.; Normanno, N.; Taïeb, J.; Seligmann, J.; De Baere, T.; Osterlund, P.; Yoshino, T.; et al. Metastatic colorectal cancer: ESMO Clinical Practice Guideline for diagnosis, treatment and follow-up. Ann. Oncol. 2023, 34, 10–32. [Google Scholar] [CrossRef] [PubMed]
- Van Cutsem, E.; Cervantes, A.; Adam, R.; Sobrero, A.; Van Krieken, J.H.; Aderka, D.; Aguilar, E.A.; Bardelli, A.; Benson, A.; Bodoky, G.; et al. ESMO consensus guidelines for the management of patients with metastatic colorectal cancer. Ann. Oncol. 2016, 27, 1386–1422. [Google Scholar] [CrossRef] [PubMed]
- García-Foncillas, J.; Sunakawa, Y.; Aderka, D.; Wainberg, Z.; Ronga, P.; Witzler, P.; Stintzing, S. Distinguishing Features of Cetuximab and Panitumumab in Colorectal Cancer and Other Solid Tumors. Front. Oncol. 2019, 9, 849. [Google Scholar] [CrossRef]
- Saoudi González, N.; Ros, J.; Baraibar, I.; Salvà, F.; Rodríguez-Castells, M.; Alcaraz, A.; García, A.; Tabernero, J.; Élez, E. Cetuximab as a Key Partner in Personalized Targeted Therapy for Metastatic Colorectal Cancer. Cancers 2024, 16, 412. [Google Scholar] [CrossRef]
- Ros, J.; Vaghi, C.; Baraibar, I.; Saoudi González, N.; Rodríguez-Castells, M.; García, A.; Alcaraz, A.; Salva, F.; Tabernero, J.; Elez, E. Targeting KRAS G12C Mutation in Colorectal Cancer, A Review: New Arrows in the Quiver. Int. J. Mol. Sci. 2024, 25, 3304. [Google Scholar] [CrossRef]
- Giordano, G.; Remo, A.; Porras, A.; Pancione, M. Immune resistance and egfr antagonists in colorectal cancer. Cancers 2019, 11, 1089. [Google Scholar] [CrossRef]
- Varchetta, S.; Gibelli, N.; Oliviero, B.; Nardini, E.; Gennari, R.; Gatti, G.; Silva, L.S.; Villani, L.; Tagliabue, E.; MénArd, S.; et al. Elements related to heterogeneity of antibody-dependent cell cytotoxicity in patients under trastuzumab therapy for primary operable breast cancer overexpressing Her2. Cancer Res. 2007, 67, 11991–11999. [Google Scholar] [CrossRef]
- Kulkarni, S.; Martin, M.P.; Carrington, M. The Yin and Yang of HLA and KIR in human disease. Semin. Immunol. 2008, 20, 343–352. [Google Scholar] [CrossRef]
- Vitale, M.; Cantoni, C.; Della Chiesa, M.; Ferlazzo, G.; Carlomagno, S.; Pende, D.; Falco, M.; Pessino, A.; Muccio, L.; De Maria, A.; et al. An historical overview: The discovery of how NK cells can kill enemies, recruit defense troops, and more. Front. Immunol. 2019, 10, 1415. [Google Scholar] [CrossRef]
- Long, E.O.; Sik Kim, H.; Liu, D.; Peterson, M.E.; Rajagopalan, S. Controlling natural killer cell responses: Integration of signals for activation and inhibition. Annu. Rev. Immunol. 2013, 31, 227–258. [Google Scholar] [CrossRef] [PubMed]
- Falco, M.; Moretta, L.; Moretta, A.; Bottino, C. KIR and KIR ligand polymorphism: A new area for clinical applications? Tissue Antigens 2013, 82, 363–373. [Google Scholar] [CrossRef] [PubMed]
- Augusto, D.G. The impact of KIR polymorphism on the risk of developing cancer: Not as strong as imagined? Front. Genet. 2016, 7, 121. [Google Scholar] [CrossRef] [PubMed]
- Bontadini, A.; Testi, M.; Cuccia, M.C.; Martinetti, M.; Carcassi, C.; Chiesa, A.; Cosentini, E.; Dametto, E.; Frison, S.; Iannone, A.M.; et al. Distribution of killer cell immunoglobin-like receptors genes in the Italian Caucasian population. J. Transl. Med. 2006, 27, 4. [Google Scholar]
- Binyamin, L.; Alpaugh, R.K.; Hughes, T.L.; Lutz, C.T.; Campbell, K.S.; Weiner, L.M. Blocking NK Cell Inhibitory Self-Recognition Promotes Antibody-Dependent Cellular Cytotoxicity in a Model of Anti-Lymphoma Therapy 1. J. Immunol. 2008, 180, 6392–6401. [Google Scholar] [CrossRef]
- Wang, W.; Erbe, A.K.; Hank, J.A.; Morris, Z.S.; Sondel, P.M. NK cell-mediated antibody-dependent cellular cytotoxicity in cancer immunotherapy. Front. Immunol. 2015, 6, 368. [Google Scholar] [CrossRef]
- Morales-Estevez, C.; De la Haba-Rodriguez, J.; Manzanares-Martin, B.; Porras-Quintela, I.; Rodriguez-Ariza, A.; Moreno-Vega, A.; Ortiz-Morales, M.J.; Gomez-España, M.A.; Cano-Osuna, M.T.; Lopez-Gonzalez, J.; et al. KIR genes and their ligands predict the response to anti-EGFR monoclonal antibodies in solid tumors. Front. Immunol. 2016, 7, 561. [Google Scholar] [CrossRef]
- Liu, J.M.; Stein, M.N.; Shin, J.; Gudzowaty, O.; Bernstein, A.M. Antibody-dependent Cell Cytotoxicity to Breast Cancer Targets Despite Inhibitory KIR Signaling. Anticancer Res. 2006, 26, 1759–1763. [Google Scholar]
- Van Cutsem, E.; Peeters, M.; Siena, S.; Humblet, Y.; Hendlisz, A.; Neyns, B.; Canon, J.-L.; Van Laethem, J.-L.; Maurel, J.; Richardson, G.; et al. Open-label phase III trial of panitumumab plus best supportive care compared with best supportive care alone in patients with chemotherapy-refractory metastatic colorectal cancer. J. Clin. Oncol. 2007, 25, 1658–1664. [Google Scholar] [CrossRef]
- Misale, S.; Di Nicolantonio, F.; Sartore-Bianchi, A.; Siena, S.; Bardelli, A. Resistance to Anti-EGFR therapy in colorectal cancer: From heterogeneity to convergent evolution. Cancer Discov. 2014, 4, 1269–1280. [Google Scholar] [CrossRef]
- Bylsma, L.C.; Gillezeau, C.; Garawin, T.A.; Kelsh, M.A.; Fryzek, J.P.; Sangaré, L.; Lowe, K.A. Prevalence of RAS and BRAF mutations in metastatic colorectal cancer patients by tumor sidedness: A systematic review and meta-analysis. Cancer Med. 2020, 9, 1044–1057. [Google Scholar] [CrossRef]
- Qiu, L.X.; Mao, C.; Zhang, J.; Zhu, X.D.; Liao, R.Y.; Xue, K.; Li, J.; Chen, Q. Predictive and prognostic value of KRAS mutations in metastatic colorectal cancer patients treated with cetuximab: A meta-analysis of 22 studies. Eur. J. Cancer 2010, 46, 2781–2787. [Google Scholar] [CrossRef]
- Gonzalez-Galarza, F.F.; McCabe, A.; Santos, E.J.; Jones, J.; Takeshita, L.Y.; Ortega-Rivera, N.D.; Del Cid-Pavon, G.M.; Ramsbottom, K.; Ghattaoraya, G.S.; Alfirevic, A.; et al. Allele frequency net database (AFND) 2020 update: Gold-standard data classification, open access genotype data and new query tools. Nucleic Acid Res. 2020, 48, D783–D788. [Google Scholar] [CrossRef]
- Manzanares-Martin, B.; Cebrián Aranda, A.; Del Puerto-Nevado, L.; González, R.; Solanes, S.; Gómez-España, M.A.; García-Foncillas, J.; Aranda, E. Improving selection of patients with metastatic colorectal cancer to benefit from cetuximab based on KIR genotypes. J. Immunother. Cancer 2021, 9, e001705. [Google Scholar] [CrossRef]
- Hadjis, A.D.; McCurdy, S.R. The role and novel use of natural killer cells in graft-versus-leukemia reactions after allogeneic transplantation. Front. Immunol. 2024, 15, 1358668. [Google Scholar] [CrossRef] [PubMed]
- Younis, N.S.; AlMasoud, E.S.; Al Khawajah, F.; Alghazal, F.J.; AlMofarfesh, H.M.; Al-Khalaf, L.H.; Al Otaibi, M.S.; Alkhamis, S.M.; Al Naser, Z.A.; Al Mousa, Z.H.; et al. Potential genetic biomarker of Saudi Arabian patients with colorectal cancer. Eur. Rev. Med. Pharmacol. Sci. 2022, 26, 3109–3126. [Google Scholar] [PubMed]
- Al Omar, S.Y.; Mansour, L.; Dar, J.A.; Alwasel, S.; Alkhuriji, A.; Arafah, M.; Al Obeed, O.; Christmas, S. The Relationship between Killer Cell Immunoglobulin-Like Receptors and HLA-C Polymorphisms in Colorectal Cancer in a Saudi Population. Genet. Test. Mol. Biomark. 2015, 19, 617–622. [Google Scholar] [CrossRef] [PubMed]
- Ghanadi, K.; Shayanrad, B.; Ahmadi, S.A.Y.; Shahsavar, F.; Eliasy, H. Colorectal cancer and the KIR genes in the human genome: A meta-analysis. Genom. Data 2016, 10, 118–126. [Google Scholar] [CrossRef]
- Diaz-Peña, R.; Mondelo-Macía, P.; Molina de la Torre, A.J.; Sanz-Pamplona, R.; Moreno, V.; Martín, V. Analysis of Killer Immunoglobulin-Like Receptor Genes in Colorectal Cancer. Cells 2020, 9, 514. [Google Scholar] [CrossRef]
- Portela, P.; Merzoni, J.; Lindenau, J.D.; Damin, D.C.; Wilson, T.J.; Roesler, R.; Schwartsmann, G.; Jobim, L.F.; Jobim, M. KIR genes and HLA class I ligands in a Caucasian Brazilian population with colorectal cancer. Hum. Immunol. 2017, 78, 263–268. [Google Scholar] [CrossRef]
- Pittari, G.; Liu, X.R.; Selvakumar, A.; Zhao, Z.; Merino, E.; Huse, M.; Chewning, J.H.; Hsu, K.C.; Dupont, B. NK cell tolerance of self-specific activating receptor KIR2DS1 in individuals with cognate HLA-C2 ligand. J. Immunol. 2013, 190, 4650–4660. [Google Scholar] [CrossRef] [PubMed]
- Rivero-Juarez, A.; Gonzalez, R.; Camacho, A.; Manzanares-Martin, B.; Caruz, A.; Martinez-Peinado, A.; Torre-Cisneros, J.; Pineda, J.A.; Peña, J.; Rivero, A.; et al. Natural Killer KIR3DS1 Is Closely Associated with HCV Viral Clearance and Sustained Virological Response in HIV/HCV Patients. PLoS ONE 2013, 8, e61992. [Google Scholar] [CrossRef]
- De Re, V.; Caggiari, L.; De Zorzi, M.; Talamini, R.; Racanelli, V.; D’Andrea, M.; Buonadonna, A.; Zagonel, V.; Cecchin, E.; Innocenti, F.; et al. Genetic diversity of the KIR/HLA system and outcome of patients with metastatic colorectal cancer treated with chemotherapy. PLoS ONE 2014, 9, e84940. [Google Scholar] [CrossRef] [PubMed]
- Tanimine, N.; Ohira, M.; Kurita, E.; Nakano, R.; Sakai, H.; Tahara, H.; Ide, K.; Kobayashi, T.; Tanaka, Y.; Ohdan, H. Impact of KIR-HLA Genotype on Natural-Killer-Cell-Based Immunotherapy for Preventing Hepatocellular Carcinoma after Living-Donor Liver Transplantation. Cancers 2024, 16, 533. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.C.; Srivastava, R.M.; López-Albaitero, A.; Ferrone, S.; Ferris, R.L. Natural killer (NK): Dendritic cell (DC) cross talk induced by therapeutic monoclonal antibody triggers tumor antigen-specific T cell immunity. Immunol. Res. 2011, 50, 248–254. [Google Scholar] [CrossRef]
- Srivastava, R.M.; Lee, S.C.; Andrade Filho, P.A.; Lord, C.A.; Jie, H.B.; Davidson, H.C.; López-Albaitero, A.; Gibson, S.P.; Gooding, W.E.; Ferrone, S.; et al. Cetuximab-activated natural killer and dendritic cells collaborate to trigger tumor antigen-specific T-cell immunity in head and neck cancer patients. Clin. Cancer Res. 2013, 19, 1858–1872. [Google Scholar] [CrossRef]
- Srivastava, R.M.; Trivedi, S.; Concha-Benavente, F.; Gibson, S.P.; Reeder, C.; Ferrone, S.; Ferris, R.L. CD137 stimulation enhances cetuximab-induced natural killer: Dendritic cell priming of antitumor T-cell immunity in patients with head and neck cancer. Clin. Cancer Res. 2017, 23, 707–716. [Google Scholar] [CrossRef]
- Srivastava, R.M.; Trivedi, S.; Concha-Benavente, F.; Hyun-Bae, J.; Wang, L.; Seethala, R.R.; Branstetter, B.F., IV; Ferrone, S.; Ferris, R.L. Stat1-induced HLA class i upregulation enhances immunogenicity and clinical response to anti-EGFR mab cetuximab therapy in HNC patients. Cancer Immunol. Res. 2015, 3, 936–945. [Google Scholar] [CrossRef]
- Terszowski, G.; Klein, C.; Stern, M. KIR/HLA Interactions Negatively Affect Rituximab- but Not GA101 (Obinutuzumab)-Induced Antibody-Dependent Cellular Cytotoxicity. J. Immunol. 2014, 192, 5618–5624. [Google Scholar] [CrossRef]
- Kohrt, H.E.; Thielens, A.; Marabelle, A.; Sagiv-Barfi, I.; Sola, C.; Chanuc, F.; Fuseri, N.; Bonnafous, C.; Czerwinski, D.; Rajapaksa, A.; et al. Anti-KIR antibody enhancement of anti-lymphoma activity of natural killer cells as monotherapy and in combination with anti-CD20 antibodies. Blood 2014, 123, 678–686. [Google Scholar] [CrossRef]
- Lisovsky, I.; Kant, S.; Tremblay-McLean, A.; Isitman, G.; Kiani, Z.; Dupuy, F.P.; Gilbert, L.; Bruneau, J.; Shoukry, N.H.; Lebouché, B.; et al. Differential contribution of education through KIR2DL1, KIR2DL3, and KIR3DL1 to antibody-dependent (AD) NK cell activation and ADCC. J. Leukoc. Biol. 2019, 105, 551–563. [Google Scholar] [CrossRef]
- Nguyen, R.; Houston, J.; Chan, W.K.; Finkelstein, D.; Dyer, M.A. The role of interleukin-2, all-trans retinoic acid, and natural killer cells: Surveillance mechanisms in anti-GD2 antibody therapy in neuroblastoma. Cancer Immunol. Immunother. 2018, 67, 615–626. [Google Scholar] [CrossRef]
- Tarek, N.; Le Luduec, J.B.; Gallagher, M.M.; Zheng, J.; Venstrom, J.M.; Chamberlain, E.; Modak, S.; Heller, G.; Dupont, B.; Cheung, N.-K.V.; et al. Unlicensed NK cells target neuroblastoma following anti-GD2 antibody treatment. J. Clin. Investig. 2012, 122, 3260–3270. [Google Scholar] [CrossRef] [PubMed]
- Beelen, N.A.; Ehlers, F.A.I.; Bos, G.M.J.; Wieten, L. Inhibitory receptors for HLA class I as immune checkpoints for natural killer cell-mediated antibody-dependent cellular cytotoxicity in cancer immunotherapy. Cancer Immunol. Immunother. 2022, 72, 797–804. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Liu, S. Focusing on NK cells and ADCC: A promising immunotherapy approach in targeted therapy for HER2-positive breast cancer. Front. Immunol. 2022, 13, 1083462. [Google Scholar] [CrossRef] [PubMed]
- Muraro, E.; De Zorzi, M.; Miolo, G.; Lombardi, D.; Scalone, S.; Spazzapan, S.; Massarut, S.; Perin, T.; Dolcetti, R.; Steffan, A.; et al. KIR-HLA Functional Repertoire Influences Trastuzumab Efficiency in Patients with HER2-Positive Breast Cancer. Front. Immunol. 2022, 12, 791958. [Google Scholar] [CrossRef]
- Foley, B.; Felices, M.; Cichocki, F.; Cooley, S.; Verneris, M.R.; Miller, J.S. The biology of NK cells and their receptors affects clinical outcomes after hematopoietic cell transplantation (HCT). Immunol. Rev. 2014, 258, 45–63. [Google Scholar] [CrossRef]
- Borrero-Palacios, A.; Cebrián, A.; Gómez del Pulgar, M.T.; García-Carbonero, R.; García, P.; Aranda, E.; Elez, E.; López-López, R.; Cervantes, A.; Valladares, M.; et al. Combination of KIR2DS4 and FcγRIIa polymorphisms predicts the response to cetuximab in KRAS mutant metastatic colorectal cancer. Sci. Rep. 2019, 9, 2589. [Google Scholar]
- Cooley, S.; Weisdorf, D.J.; Guethlein, L.A.; Klein, J.P.; Wang, T.; Le, C.T.; Marsh, S.G.E.; Geraghty, D.; Spellman, S.; Haagenson, M.D.; et al. Donor selection for natural killer cell receptor genes leads to superior survival after unrelated transplantation for acute myelogenous leukemia. Blood 2010, 116, 2411–2419. [Google Scholar] [CrossRef]
- Roe, D.; Vierra-Green, C.; Pyo, C.W.; Geraghty, D.E.; Spellman, S.R.; Maiers, M.; Kuang, R. A Detailed View of KIR Haplotype Structures and Gene Families as Provided by a New Motif-Based Multiple Sequence Alignment. Front. Immunol. 2020, 11, 585731. [Google Scholar] [CrossRef]
- Agrawal, S.; Prakash, S. Significance of KIR like natural killer cell receptors in autoimmune disorders. Clin. Immunol. 2020, 216, 108449. [Google Scholar] [CrossRef]
- Pollock, N.R.; Harrison, G.F.; Norman, P.J. Immunogenomics of Killer Cell Immunoglobulin-Like Receptor (KIR) and HLA Class I: Coevolution and Consequences for Human Health. J. Allergy Clin. Immunol. Pract. 2022, 10, 1763–1775. [Google Scholar] [CrossRef]

| Variable | KRAS-WT (n = 55) (%) | KRAS-Mutant (n = 69) (%) |
|---|---|---|
| Gender (n, %) | Male: 39 (70.9) Female: 16 (29.1) | Male: 35 (50.7) Female: 34 (49.3) |
| Age (median, range) | 64 (41–84) | 64 (42–82) |
| ECOG (n, %) | 0: 24 (43.6) 1: 22 (40) 2: 3 (5.5) Unknown: 6 (10.9) | 0: 12 (17.4) 1: 51 (73.9) 2: 6 (8.7) |
| Primary site (n, %) | Right colon: 16 (29.1) Left colon: 21 (38.2) Rectum: 18 (32.7) | Right colon: 16 (23.2) Left colon: 35 (50.7) Rectum: 18 (26.1) |
| Laterality (n, %) | Right-sided: 16 (29.1) Left-sided: 39 (70.9) | Right-sided: 16 (23.2) Left-sided: 53 (76.8) |
| Number of metastatic sites (n, %) | 1: 28 (50.9) 2: 19 (34.5) 3 or more: 8 (14.5) | 1: 26 (37.7) 2: 27 (39.1) 3 or more: 16 (23.2) |
| Previous treatments (n, %) | Yes: 30 (54.5) No: 24 (43.6) Unknown: 1 (1.8) | Yes: 69 (100) No: 0 (0) |
| Treatment time, months (median, range) | 8.1 (1.9–12) | 2 (0.5–12) |
| Number of treatment cycles (median, range) | 17 (3–24) | 4 (1–24) |
| Reason for end of treatment (n, %) | Progression: 28 (73.7) Death: 1 (2.6) Toxicity: 1 (2.6) Other: 8 (21.1) | Progression: 59 (86.8) Death: 2 (2.9) Toxicity: 4 (5.9) Other: 3 (4.4) |
| Progression at 12 months (n, %) | Yes: 33 (60) No: 22 (40) | Yes: 68 (92.8) No: 1 (1.4) |
| Exit at 12 months (n, %) | Yes: 14 (25.5) No: 41 (74.5) | Yes: 51 (73.9) No: 18 (26.1) |
| Exit reason (n, %) | Progression: 8 (57.2) Adverse event: 1 (7.1) Unknown: 5 (35.7) | Progression: 42 (82.4) Adverse event: 2 (3.9) Unknown: 7 (13.7) |
| KIR Genes and HLA Ligands | KRAS-WT (n = 55) (%) | KRAS-Mutant (n = 69) (%) | p-Value (X2/Fisher) | |
|---|---|---|---|---|
| KIR gene | ||||
| 2DL1 (n, %) | Positive: 54 (98.2) Negative: 1 (1.8) | Positive: 63 (91.3) Negative: 6 (8.7) | 0.131 | (Fisher) |
| 2DL2 (n, %) | Positive: 31 (56.4) Negative: 24 (43.6) | Positive: 37 (53.6) Negative: 32 (46.4) | 0.761 | (X2) |
| 2DL3 (n, %) | Positive: 53 (96.4) Negative: 2 (3.6) | Positive: 62 (89.9) Negative: 7 (10.1) | 0.296 | (Fisher) |
| 2DL5 (n, %) | Positive: 32 (58.2) Negative: 23 (41.8) | Positive: 39 (56.5) Negative: 30 (43.5) | 0.853 | (X2) |
| 3DL1 (n, %) | Positive: 50 (90.9) Negative: 5 (9.1) | Positive: 62 (89.9) Negative: 7 (10.1) | 0.844 | (X2) |
| 2DS1 (n, %) | Positive: 25 (45.5) Negative: 30 (54.5) | Positive: 37 (53.6) Negative: 32 (46.4) | 0.366 | (X2) |
| 2DS2 (n, %) | Positive: 31 (56.4) Negative: 24 (43.6) | Positive: 36 (52.2) Negative: 33 (47.8) | 0.642 | (X2) |
| 2DS3 (n, %) | Positive: 19 (34.5) Negative: 36 (65.5) | Positive: 29 (42) Negative: 39 (56.5) Unknown: 1 (1.4) | 0.440 | (X2) |
| 2DS4 (n, %) | Positive: 50 (90.9) Negative: 5 (9.1) | Positive: 63 (91.3) Negative: 6 (8.7) | 1.000 | (Fisher) |
| 2DS5 (n, %) | Positive: 19 (34.5) Negative: 36 (65.5) | Positive: 23 (33.3) Negative: 46 (66.7) | 0.887 | (X2) |
| 3DS1 (n, %) | Positive: 25 (45.5) Negative: 30 (54.5) | Positive: 27 (39.1) Negative: 42 (60.9) | 0.478 | (X2) |
| 2DP1 (n, %) | Positive: 54 (98.2) Negative: 1 (1.8) | Positive: 62 (89.9) Negative: 7 (10.1) | 0.075 | (Fisher) |
| HLA ligand | ||||
| Bw4 (n, %) | Positive: 46 (83.6) Negative: 9 (16.4) | Positive: 46 (66.7) Negative: 21 (31.4) | 0.056 | (X2) |
| C1 (n, %) | Positive: 45 (81.8) Negative: 10 (18.2) | Positive: 54 (80.6) Negative: 13 (19.4) | 0.864 | (X2) |
| C2 (n, %) | Positive: 37 (67.3) Negative: 18 (32.7) | Positive: 48 (71.6) Negative: 19 (28.4) | 0.601 | (X2) |
| KIR–Ligand Combinations | KRAS-WT n (%) | KRAS-Mutant n (%) | p-Value (X2) |
|---|---|---|---|
| 3DL1–Bw4 | Positive: 41 (82) Negative: 9 (18) | Positive: 41 (68.3) Negative: 19 (31.7) | 0.101 |
| 2DL1–C2 | Positive: 36 (66.7) Negative: 18 (33.3) | Positive: 43 (69.4) Negative: 19 (30.6) | 0.757 |
| 2DL2–C1 | Positive: 26 (83.9) Negative: 5 (16.1) | Positive: 27 (75) Negative: 9 (25) | 0.373 |
| 2DL3–C1 | Positive: 44 (83) Negative: 9 (17) | Positive: 50 (82) Negative: 11 (18) | 0.883 |
| 2DS1–C2 | Positive: 17 (68) Negative: 8 (32) | Positive: 26 (72.2) Negative: 10 (27.8) | 0.722 |
| 3DS1–Bw4 | Positive: 20 (80) Negative: 5 (20) | Positive: 16 (59.3) Negative: 11 (40.7) | 0.105 |
| Genotypes and Haplotypes | KRAS-WT n (%) | KRAS-Mutant n (%) | p-Value (X2) |
|---|---|---|---|
| AA | Positive: 12 (21.8) Negative: 43 (78.2) | Positive: 16 (26.2) Negative: 45 (73.8) | 0.579 |
| AB | Positive: 30 (54.5) Negative: 25 (45.5) | Positive: 27 (44.3) Negative: 34 (55.7) | 0.269 |
| BB | Positive: 13 (23.6) Negative: 42 (76.4) | Positive: 18 (29.5) Negative: 43 (70.5) | 0.475 |
| CENA/TELA | Positive: 12 (21.8) Negative: 43 (78.2) | Positive: 16 (26.2) Negative: 45 (73.8) | 0.579 |
| CENA/TELB | Positive: 12 (21.8) Negative: 43 (78.2) | Positive: 12 (19.7) Negative: 49 (80.3) | 0.776 |
| CENB/TELA | Positive: 18 (32.7) Negative: 37 (67.3) | Positive: 14 (23) Negative: 47 (77) | 0.239 |
| CENB/TELB | Positive: 13 (23.6) Negative: 42 (76.4) | Positive: 19 (31.1) Negative: 42 (68.9) | 0.366 |
| KRAS-WT | KRAS-Mutant | |||
|---|---|---|---|---|
| PFS12 (IC 95%) | p-Value (Log Rank) | PFS12 (IC 95%) | p-Value (Log Rank) | |
| 2DS1 | Positive (25): 10.15 (8.11–12.19) Negative (30): 8.84 (5.56–12.12) | 0.955 | Positive (37): 1.84 (1.81–1) Negative (32): 1.84 (1.7–1.98) | 0.890 |
| 2DS2 | Positive (31): 8.71 (7.33–10.1) Negative (24): 8.43 (7.02–0.83) | 0.444 | Positive (36): 1.84 (1.62–2.07) Negative (33): 1.84 (1.79–1.89) | 0.163 |
| 2DS3 | Positive (19): 9.44 (7.96–10.92) Negative (36): 8.14 (6.88–9.42) | 0.314 | Positive (29): 1.83 (1.78–1.89) Negative (39): 1.84 (1.81–1.87) | 0.273 |
| 2DS4 | Positive (50): 8.34 (7.28–9.40) Negative (5): 11.06 (10.03–12.08) | 0.276 | Positive (63): 1.84 (1.81–1.87) Negative (6): 1.84 (0.85–2.83) | 0.800 |
| 2DS5 | Positive (19): 10.15 (7.72–12.58) Negative (36): 8.84 (6.21–11.47) | 0.751 | Positive (23): 1.84 (1.81–1.87) Negative (46): 1.84 (1.75–1.93) | 0.498 |
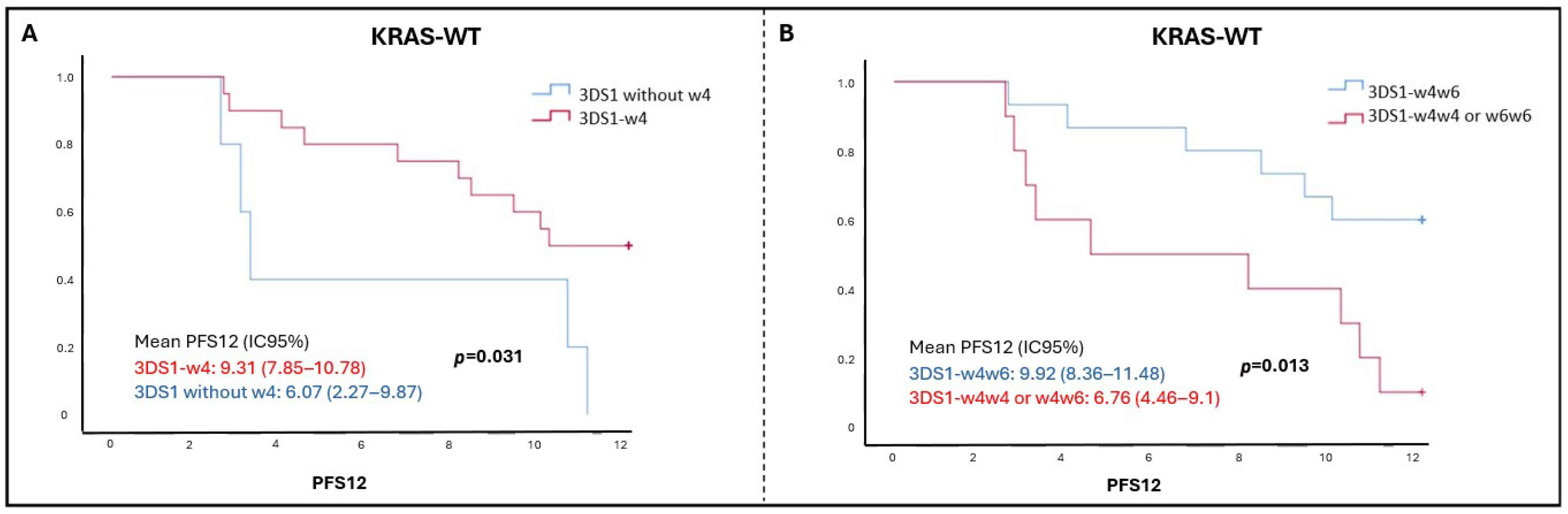
| 3DS1 | Positive (25): 10.15 (8.11–12.19) Negative (30): 8.84 (5.56–12.12) | 0.955 | Positive (27): 1.84 (1.81–1.87) Negative (42): 1.84 (1.72–1.96) | 0.968 |
| 2DL1 | Positive (54): 10.15 (7.58–12.72) Negative (1): 3.02 (0–3.02) | 0.031 | Positive (63): 1.84 (1.81–1.87) Negative (6): 2.04 (0.78–3.29) | 0.727 |
| 2DL2 | Positive (31): 8.71 (7.33–10.1) Negative (24): 8.43 (7.03–9.83) | 0.444 | Positive (37): 1.84 (1.61–2.07) Negative (32): 1.84 (1.79–1.89) | 0.232 |
| 2DL3 | Positive (53): 10.15 (8.09–12.22) Negative (2): 3.02 (1.29–13.73) | 0.944 | Positive (62): 1.84 (1.81–1.87) Negative (7): 2.04 (0.86–3.22) | 0.585 |
| 2DL5 | Positive (32): 10.15 (8.42–11.88) Negative (23): 8.84 (5.01–12.67) | 0.816 | Positive (39): 1.84 (1.61–2.07) Negative (30): 1.84 (1.77–1.91) | 0.032 |
| 3DL1 | Positive (50): 8.34 (7.28–9.40) Negative (5): 11.06 (10.03–12.84) | 0.276 | Positive (62): 1.84 (1.81–1.87) Negative (7): 1.84 (1.75–1.92) | 0.946 |
| 2DP1 | Positive (54): 10.15 (7.58–12.72) Negative (1): 3.02 (0–3.02) | 0.031 | Positive (62): 1.84 (1.81–1.87) Negative (7): 2.04 (0.86–3.22) | 0.976 |
| Bw4 | Positive (46): 8.85 (7.81–9.91) Negative (9): 7.23 (4.6–9.85) | 0.064 | Positive (46): 1.84 (1.78–1.89) Negative (21): 1.84 (1.6–2.04) | 0.620 |
| C1 | Positive (45): 9.96 (7.62–12.29) Negative (10): 10.42 (7.34–13.49) | 0.826 | Positive (54): 1.84 (1.79–1.89) Negative (13): 1.84 (1.69–1.99) | 0.198 |
| C2 | Positive (37): 10.58 (9.07–12.09) Negative (18): 4.79 (0–12.17) | 0.219 | Positive (48): 1.84 (1.75–1.94) Negative (19): 1.81 (1.75–1.86) | 0.935 |
| KRAS-WT | KRAS-Mutant | |||
|---|---|---|---|---|
| PFS12 (IC 95%) | p-Value (Log Rank) | PFS12 (IC 95%) | p-Value (Log Rank) | |
| CENA/TELA Other | (n = 12) 8.81 (8.64–8.97) (n = 43) 10.42 (8.22–12.61) | 0.571 | (n = 16) 1.74 (1.61–1.87) (n = 45) 1.84 (1.74–1.94) | 0.011 |
| CENA/TELA + CENB/TELB CENA/TELB + CENB/TELA | (n = 25) 10.15 (6.68–13.63) (n = 30) 9.96 (6.56–13.35) | 0.919 | (n = 35) 1.74 (1.51–1.97) (n = 26) 2.04 (1.74–2.33) | 0.002 |
| Univariate Analysis | Multivariate Analysis | |||||
|---|---|---|---|---|---|---|
| Variable | Frequency | HR (95% CI) | p-Value | Frequency | HR (95% CI) | p-Value |
| Gender | ||||||
| Female | 16 (29.1%) | 1 | 0.877 | 16 (29.1%) | 1 | 0.682 |
| Male | 39 (70.9%) | 0.94 (0.45–1.97) | 39 (70.9%) | 1.18 (0.54–2.58) | ||
| Median Age | ||||||
| <64 years | 25 (45.5%) | 1 | 0.788 | 25 (45.5%) | 1 | 0.477 |
| >64 years | 30 (54.5%) | 0.91 (0.46–1.81) | 30 (54.5%) | 0.12 (0.04–0.37) | ||
| ECOG | ||||||
| 0 | 24 (43.6%) | 1 | - | - | - | |
| 1 | 22 (40%) | 0.56 (0.17–1.19) | 0.136 | - | - | - |
| 2 | 3 (5.5%) | 0.46 (0.06–3.47) | 0.453 | - | - | - |
| Number of cycles | ||||||
| <6 | 5 (9.1%) | 1 | 0 | 5 (9.1%) | 1 | 0 |
| >6 | 50 (90.9%) | 0.16 (0.06–0.43) | 50 (90.9%) | 0.13 (0.04–0.39) | ||
| Laterality | ||||||
| Right-sided | 16 (29.1%) | 1 | 0.849 | - | - | - |
| Left-sided | 39 (70.9%) | 1.07 (0.51–2.26) | - | - | - | |
| Number of Metastatic sites | ||||||
| 1 | 28 (50.9%) | 1 | 0.844 0.837 | - | - | - |
| 2 | 19 (34.5%) | 1.08 (0.51–2.28) | - | - | - | |
| >3 | 8 (14.5%) | 1.11 (0.41–3.04) | - | - | - | |
| KIR2DL1 | ||||||
| No | 1 (1.8%) | 1 | 0.065 | - | - | - |
| Yes | 54 (98.2%) | 0.14 (0.02–1.97) | - | - | - | |
| KIR2DL2 | ||||||
| No | 24 (43.6%) | 1 | 0.446 | - | - | - |
| Yes | 31 (56.4%) | 0.77 (0.38–1.52) | - | - | - | |
| KIR2DL3 | ||||||
| No | 2 (3.6%) | 1 | 0.944 | - | - | - |
| Yes | 53 (96.4%) | 1.07 (0.15–7.87) | - | - | - | |
| KIR2DL5 | ||||||
| No | 23 (41.8%) | 1 | 0.816 | - | - | - |
| Yes | 32 (58.2%) | 0.92 (0.46–1.84) | - | - | - | |
| KIR 2DS1 | ||||||
| No | 30 (54.5%) | 1 | 0.955 | - | - | - |
| Yes | 25 (45.5%) | 0.98 (0.49–1.95) | - | - | - | |
| KIR 2DS2 | ||||||
| No | 24 (43.6%) | 1 | 0.446 | - | - | - |
| Yes | 31 (56.4%) | 0.77 (0.39–1.52) | - | - | - | |
| KIR 2DS3 | ||||||
| No | 36 (65.5%) | 1 | 0.317 | - | - | - |
| Yes | 19 (34.5%) | 0.68 (0.33–1.44) | - | - | - | |
| KIR 2DS4 | ||||||
| No | 5 (9.1%) | 1 | 0.289 | - | - | - |
| Yes | 50 (90.9%) | 2.17 (0.52–9.08) | - | - | - | |
| KIR 2DS5 | ||||||
| No | 36 (65.5%) | 1 | 0.751 | - | - | - |
| Yes | 19 (34.5%) | 0.89 (0.43–1.84) | - | - | - | |
| KIR 3DL1 | ||||||
| No | 5 (9.1%) | 1 | 0.289 | - | - | - |
| Yes | 50 (90.1%) | 2.17 (0.52–9.08) | - | - | - | |
| KIR 3DS1 | ||||||
| No | 30 (54.5%) | 1 | 0.955 | - | - | - |
| Yes | 25 (45.5%) | 0.98 (0.49–1.95) | - | - | - | |
| KIR 2DP1 | ||||||
| No | 1 (1.8%) | 1 | 0.065 | - | - | - |
| Yes | 54 (98.2%) | 0.14 (0.017–1.13) | - | - | - | |
| - | - | - | ||||
| KIR3DS1–Bw4 | ||||||
| No | 5 (20%) | 1 | 0.026 | 5 (20%) | 1 | 0.033 |
| Yes | 20 (80%) | 0.29 (0.1–0.86) | 20 (80%) | 0.49 (0.25–0.94) | ||
| 3DS1–HLAB | ||||||
| Heterozygous (w4w6) | 15 (27.3%) | 1 | 0.019 | - | 1 | 0.013 |
| Homozygous (w4w4 or w6w6) | 10 (18.2%) | 3.48 (1.23–9.86) | - | 2.22 (1.19–4.16) | ||
| Genotype | ||||||
| AB | 30 (54.5%) | 1 | - | - | - | |
| AA | 12 (21.8%) | 1.9 (0.81–4.48) | 0.143 | - | - | - |
| BB | 13 (23.6%) | 0.92 (0.38–2.23) | 0.849 | - | - | - |
| Genotype | ||||||
| AB | 30 (54.5%) | 1 | 0.517 | - | - | - |
| AA or BB | 25 (45.4%) | 1.26 (0.62–2.56) | - | - | - | |
| Semi-haplotype | ||||||
| CENA/TELB or CENB/TELA | 30 (54.5%) | 1 | 0.108 | - | - | - |
| CENA/TELA | 12 (21.8%) | 1.95 (0.86–4.41) | - | - | ||
| Univariate Analysis | Multivariate Analysis | |||||
|---|---|---|---|---|---|---|
| Variable | Frequency | HR (95% CI) | p-Value | Frequency | HR (95% CI) | p-Value |
| Gender | ||||||
| Female | 34 (49.3%) | 1 | 0.629 | 34 (49.3%) | 1 | 0.71 |
| Male | 35 (50.7%) | 0.89 (0.55–1.44) | 35 (50.7%) | 1.11 (0.65–1.88) | ||
| Median Age | ||||||
| <64 years | 33 (47.8%) | 1 | 0.178 | 33 (47.8%) | 1 | 0.47 |
| >64 years | 36 (52.2%) | 1.41 (0.85–2.34) | 36 (52.2%) | 1.01 (0.98–1.04) | ||
| ECOG | ||||||
| 0 | 12 (17.4%) | 1 | - | - | - | |
| 1 | 51 (73.9%) | 1.09 (0.58–2.05) | 0.792 | - | - | - |
| 2 | 6 (8.7%) | 1.26 (0.46–3.47) | 0.660 | - | - | - |
| Number of cycles | ||||||
| <6 | 55 (79.7%) | 1 | 0 | - | - | - |
| >6 | 14 (20.3%) | 0.24 (0.12–0.45) | - | - | ||
| Laterality | ||||||
| Right-sided | 16 (23.2%) | 1 | 0.71 | - | - | - |
| Left-sided | 53 (76.8%) | 1.11 (0.63–1.97) | - | - | - | |
| Number of Metastatic sites | ||||||
| 1 | 28 (50.9%) | 1 | 0.38 0.09 | 28 (50.9%) | 1 | 0.47 0.002 |
| 2 | 19 (34.5%) | 1.28 (0.74–2.22) | 19 (34.5%) | 1.27 (0.67–2.40) | ||
| >3 | 8 (14.5%) | 1.75 (0.91–3.37) | 8 (14.5%) | 2.94 (1.46–5.92) | ||
| KIR2DL1 | ||||||
| No | 6 (8.7%) | 1 | 0.736 | - | - | - |
| Yes | 63 (91.3%) | 1.16 (0.49–2.69) | - | - | - | |
| KIR2DL2 | ||||||
| No | 32 (46.4%) | 1 | 0.247 | - | - | - |
| Yes | 37 (53.6%) | 0.75 (0.46–1.22) | - | - | - | |
| KIR2DL3 | ||||||
| No | 7 (10.1%) | 1 | 0.598 | - | - | - |
| Yes | 62 (89.9%) | 1.24 (0.56–2.74) | - | - | - | |
| KIR2DL5 | ||||||
| No | 30 (43.5%) | 1 | 0.039 | - | - | - |
| Yes | 39 (56.5%) | 0.59 (0.35–0.97) | - | - | - | |
| KIR2DS1 | ||||||
| No | 32 (46.4%) | 1 | 0.893 | - | - | - |
| Yes | 37 (53.6%) | 0.97 (0.6–1.56) | - | - | - | |
| KIR2DS2 | ||||||
| No | 33 (47.8%) | 1 | 0.178 | - | - | - |
| Yes | 36 (52.2%) | 0.71 (0.44–1.17) | - | - | - | |
| KIR2DS3 | ||||||
| No | 39 (56.5%) | 1 | 0.136 | - | - | - |
| Yes | 29 (42%) | 0.69 (0.43–1.12) | - | - | - | |
| KIR2DS4 | ||||||
| No | 6 (8.7%) | 1 | 0.806 | - | - | - |
| Yes | 63 (91.3%) | 1.11 (0.48–2.59) | - | - | - | |
| KIR2DS5 | ||||||
| No | 46 (66.7%) | 1 | 0.512 | - | - | - |
| Yes | 23 (33.3%) | 0.84 (0.51–1.4) | - | - | - | |
| KIR3DL1 | ||||||
| No | 7 (10.1%) | 1 | 0.947 | - | - | - |
| Yes | 62 (89.9%) | 0.97 (0.44–2.14) | - | - | - | |
| KIR3DS1 | ||||||
| No | 42 (60.9%) | 1 | 0.969 | - | - | - |
| Yes | 27 (39.1%) | 1.01 (0.62–1.64) | - | - | - | |
| KIR2DP1 | ||||||
| No | 7 (10.1%) | 1 | 0.977 | - | - | - |
| Yes | 62 (89.9%) | 1.01 (0.46–2.22) | - | - | - | |
| - | - | - | ||||
| KIR3DS1–Bw4 | ||||||
| No | 11 (40.7%) | 1 | 0.618 | - | - | - |
| Yes | 16 (59.3%) | 0.82 (0.38–1.79) | - | - | - | |
| 3DS1–HLAB | ||||||
| Heterozygous (w4w6) | 20 (28.9%) | 1 | 0.598 | - | - | - |
| Homozygous (w4w4 or w6w6) | 7 (10.1%) | 1.28 (0.51–3.22) | - | - | - | |
| Genotype | ||||||
| AB | 27 (44.3%) | 1 | 27 (44.3%) | 1 | ||
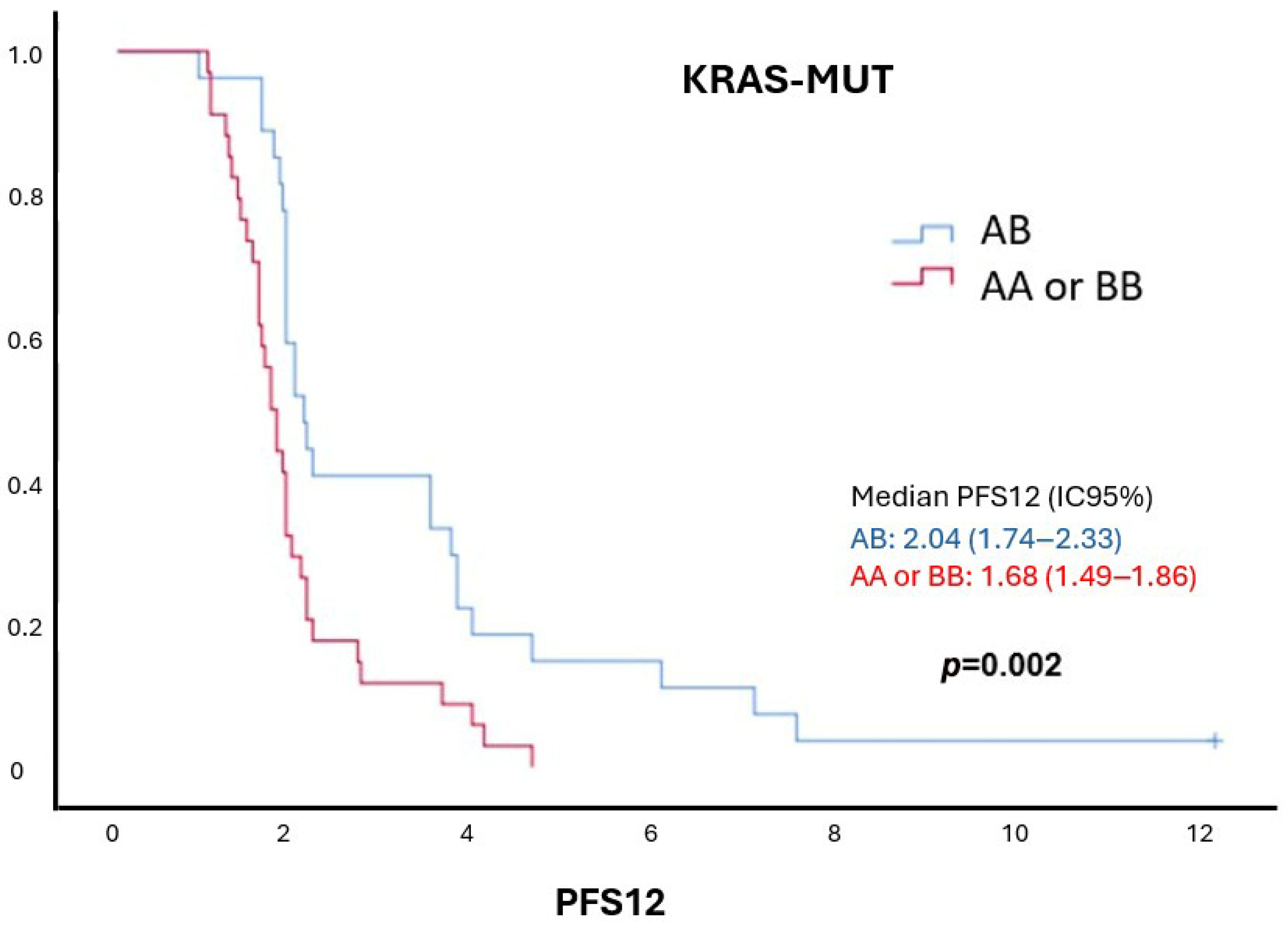
| AA | 16 (26.2%) | 2.27 (1.21–4.25) | 0.01 | 16 (26.2%) | 2.58 (1.31–5.10) | 0.006 |
| BB | 18 (29.5%) | 1.99 (1.12–3.55) | 0.02 | 18 (29.5%) | 1.93 (1.04–3.58) | 0.03 |
| Genotype | ||||||
| AB | 27 (44.3%) | 1 | 0.004 | - | 1 | 0.005 |
| AA or BB | 34 (55.7%) | 2.10 (1.28–3.47) | - | 2.16 (1.26–3.78) | ||
| Semi-haplotype | ||||||
| CENA/TELB or CENB/TELA | 26 (37.7%) | 1 | 0.03 | - | 1 | 0.04 |
| CENA/TELA | 16 (26.2%) | 2.31 (1.08–4.95) | - | 2.50 (1.02–6.14) | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gómez-Aguilera, M.; Manzanares-Martín, B.; Cebrián-Aranda, A.; Rodríguez-Ariza, A.; González-Fernández, R.; del Puerto-Nevado, L.; García-Foncillas, J.; Aranda, E. Immune Modulation Through KIR–HLA Interactions Influences Cetuximab Efficacy in Colorectal Cancer. Int. J. Mol. Sci. 2025, 26, 8062. https://doi.org/10.3390/ijms26168062
Gómez-Aguilera M, Manzanares-Martín B, Cebrián-Aranda A, Rodríguez-Ariza A, González-Fernández R, del Puerto-Nevado L, García-Foncillas J, Aranda E. Immune Modulation Through KIR–HLA Interactions Influences Cetuximab Efficacy in Colorectal Cancer. International Journal of Molecular Sciences. 2025; 26(16):8062. https://doi.org/10.3390/ijms26168062
Chicago/Turabian StyleGómez-Aguilera, María, Bárbara Manzanares-Martín, Arancha Cebrián-Aranda, Antonio Rodríguez-Ariza, Rafael González-Fernández, Laura del Puerto-Nevado, Jesús García-Foncillas, and Enrique Aranda. 2025. "Immune Modulation Through KIR–HLA Interactions Influences Cetuximab Efficacy in Colorectal Cancer" International Journal of Molecular Sciences 26, no. 16: 8062. https://doi.org/10.3390/ijms26168062
APA StyleGómez-Aguilera, M., Manzanares-Martín, B., Cebrián-Aranda, A., Rodríguez-Ariza, A., González-Fernández, R., del Puerto-Nevado, L., García-Foncillas, J., & Aranda, E. (2025). Immune Modulation Through KIR–HLA Interactions Influences Cetuximab Efficacy in Colorectal Cancer. International Journal of Molecular Sciences, 26(16), 8062. https://doi.org/10.3390/ijms26168062








