Genome-Wide mRNA and lncRNA Expression Profiling to Uncover Their Role in the Molecular Pathogenesis of Developmental Dysplasia of the Hip
Abstract
1. Introduction
2. Results
2.1. Mapping Efficiency
2.2. Differentially Expressed Genes in Hip Joint Capsules of DDH Patients
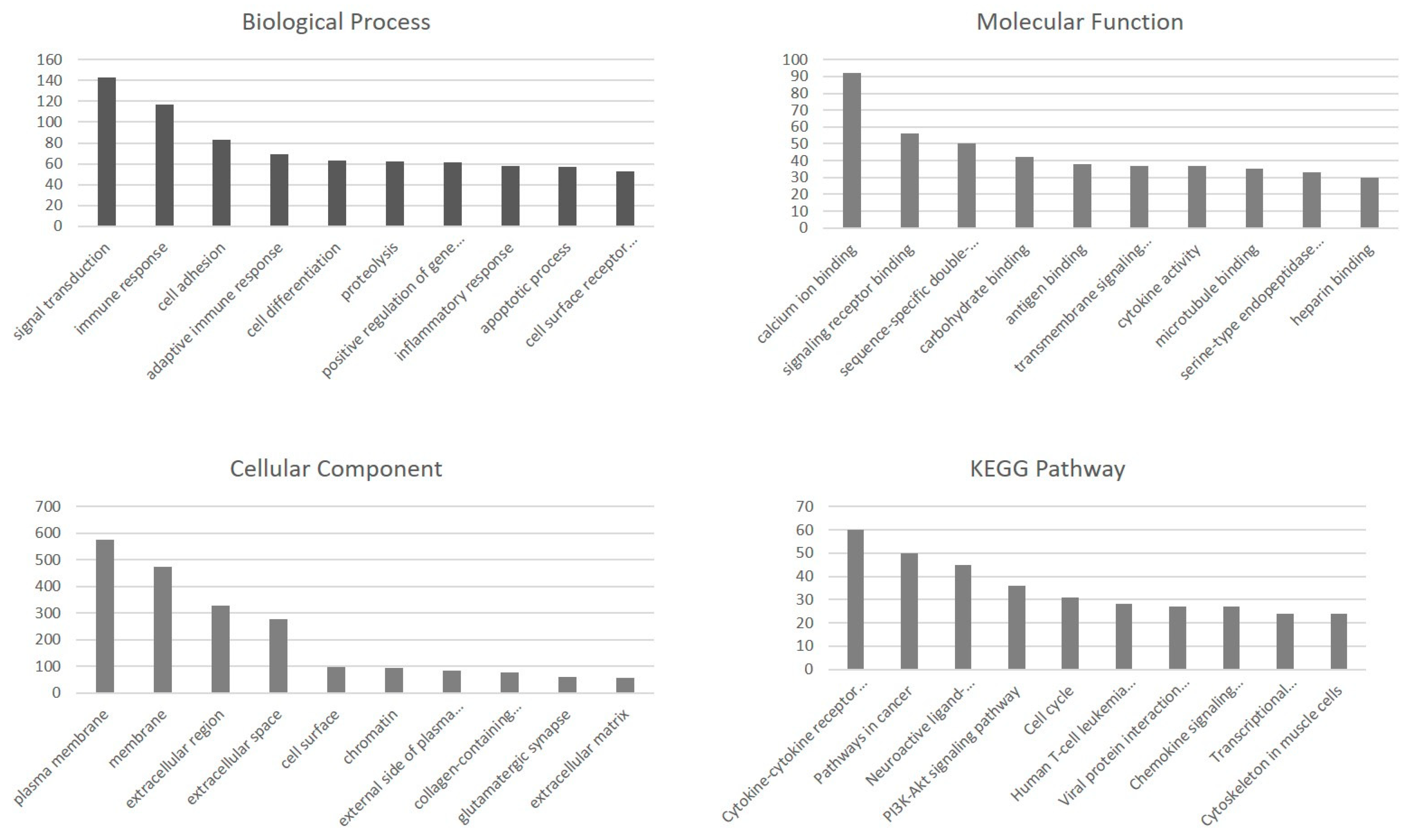
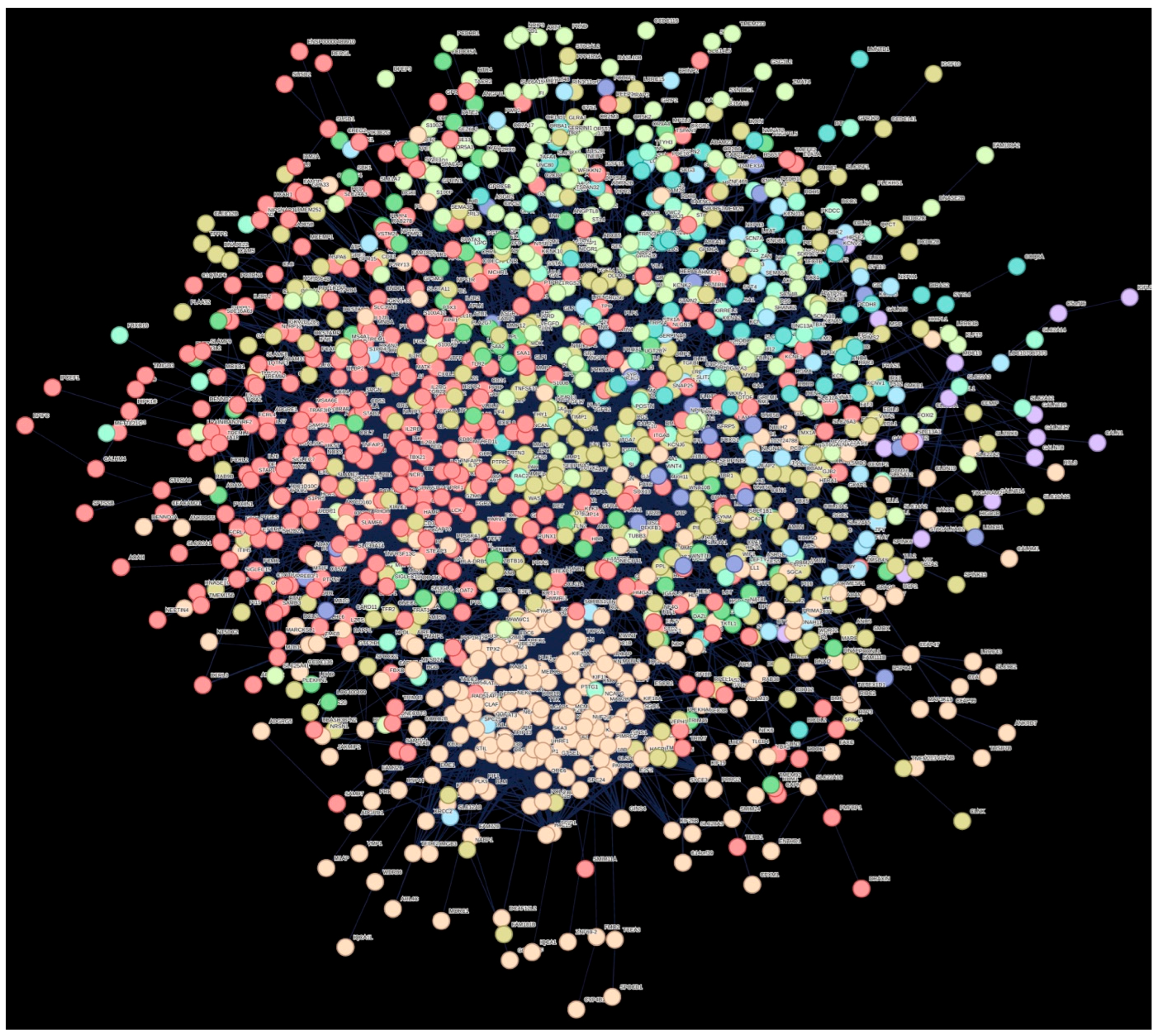
2.3. Functional Enrichment Analysis of DEGs
2.4. Identification of Long Non-Coding RNAs
3. Discussion
4. Materials and Methods
4.1. Data Acquisition
4.2. Transcript Data Analysis
4.3. Functional Enrichment Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| DDH | Developmental dysplasia of the hip |
| LncRNA | Long non-coding RNA |
| GO | Gene Ontology |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| DEGs | Differentially expressed genes |
| MF | Molecular function |
| BP | Biological process |
| CC | Cellular component |
| MMP13 | Matrix Metalloproteinase 13 |
| MMP3 | Matrix Metalloproteinase 3 |
| GDF5 | Growth Differentiation Factor 5 |
| DKK1 | Dickkopf WNT Signaling Pathway Inhibitor 1 |
| WIF1 | Wnt Inhibitor Factor 1 |
| MYOC | Myocilin |
| IRX | Iroquois Homeobox |
References
- Pollet, V.; Percy, V.; Prior, H.J. Relative risk and incidence for developmental dysplasia of the hip. J. Pediatr. 2017, 181, 202–207. [Google Scholar] [CrossRef]
- Loder, R.T.; Skopelja, E.N. The epidemiology and demographics of hip dysplasia. ISRN Orthop. 2011, 2011, 238607. [Google Scholar] [CrossRef] [PubMed]
- Kenanidis, E.; Gkekas, N.K.; Karasmani, A.; Anagnostis, P.; Christofilopoulos, P.; Tsiridis, E. Genetic predisposition to developmental dysplasia of the hip. J. Arthroplast. 2020, 35, 291–300.e1. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Jin, G.; Qian, K.; Zhang, C.; Zhi, W.; Yang, D.; Lu, Y.; Han, J. Comprehensive bioinformatics analysis of susceptibility genes for developmental dysplasia of the hip. Intractable Rare Dis. Res. 2022, 11, 70–80. [Google Scholar] [CrossRef] [PubMed]
- Jacobsen, K.K.; Laborie, L.B.; Kristiansen, H.; Schäfer, A.; Gundersen, T.; Zayats, T.; Rosendahl, K. Genetics of hip dysplasia—A systematic literature review. BMC Musculoskelet. Disord. 2024, 25, 762. [Google Scholar] [CrossRef]
- Wen, J.; Ping, H.; Kong, X.; Chai, W. Developmental dysplasia of the hip: A systematic review of susceptibility genes and epigenetics. Gene 2023, 853, 147067. [Google Scholar] [CrossRef]
- Cheng, B.; Jia, Y.; Wen, Y.; Hou, W.; Xu, K.; Liang, C.; Cheng, S.; Liu, L.; Chu, X.; Ye, J.; et al. Integrative analysis of microRNA and mRNA sequencing data identifies novel candidate genes and pathways for developmental dysplasia of hip. Cartilage 2021, 13, 1618S–1626S. [Google Scholar] [CrossRef]
- Harsanyi, S.; Zamborsky, R.; Kokavec, M.; Danisovic, L. Genetics of developmental dysplasia of the hip. Eur. J. Med. Genet. 2020, 63, 103990. [Google Scholar] [CrossRef]
- Gkiatas, I.; Boptsi, A.; Tserga, D.; Gelalis, I.; Kosmas, D.; Pakos, E. Developmental dysplasia of the hip: A systematic literature review of the genes related with its occurrence. EFORT Open Rev. 2019, 4, 595–601. [Google Scholar] [CrossRef]
- Li, C.; Peng, Z.; Zhou, Y.; Su, Y.; Bu, P.; Meng, X.; Li, B.; Xu, Y. Comprehensive analysis of pathological changes in hip joint capsule of patients with developmental dysplasia of the hip. Bone Jt. Res. 2021, 10, 558–570. [Google Scholar] [CrossRef]
- Liu, J.; Bao, Y.; Fan, J.; Chen, W.; Shu, Q. Microstructure changes and miRNA–mRNA network in a developmental dysplasia of the hip rat model. iScience 2024, 27, 109449. [Google Scholar] [CrossRef]
- Chen, L.-L.; Carmichael, G.G. Long noncoding RNAs in mammalian cells: What, where, and why? Wiley Interdiscip. Rev. RNA 2010, 1, 2–21. [Google Scholar] [CrossRef]
- Mishra, S.; Verma, S.S.; Rai, V.; Awasthee, N.; Chava, S.; Hui, K.M.; Kumar, A.P.; Challagundla, K.B.; Sethi, G.; Gupta, S.C. Long non-coding RNAs are emerging targets of phytochemicals for cancer and other chronic diseases. Cell Mol. Life Sci. 2019, 76, 1947–1966. [Google Scholar] [CrossRef]
- Uszczynska-Ratajczak, B.; Lagarde, J.; Frankish, A.; Guigó, R.; Johnson, R. Towards a complete map of the human long non-coding RNA transcriptome. Nat. Rev. Genet. 2018, 19, 535–548. [Google Scholar] [CrossRef]
- Fang, S.; Zhang, L.; Guo, J.; Niu, Y.; Wu, Y.; Li, H.; Zhao, L.; Li, X.; Teng, X.; Sun, X.; et al. NONCODEV5: A comprehensive annotation database for long non-coding RNAs. Nucleic Acids Res. 2018, 46, D308–D314. [Google Scholar] [CrossRef]
- Chen, G.; Wang, Z.; Wang, D.; Qiu, C.; Liu, M.; Chen, X.; Zhang, Q.; Yan, G.; Cui, Q. LncRNADisease: A database for long-non-coding RNA-associated diseases. Nucleic Acids Res. 2013, 41, D983–D986. [Google Scholar] [CrossRef]
- Zhang, G.; Wu, Y.; Xu, D.; Yan, X. Long noncoding RNA UFC1 promotes proliferation of chondrocyte in osteoarthritis by acting as a sponge for miR-34a. DNA Cell Biol. 2016, 35, 691–695. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Jia, L.; Li, X.; Guo, R.; Huang, Y.; Zheng, Y.; Li, W. Long noncoding RNAs: New players in the osteogenic differentiation of bone marrow- and adipose-derived mesenchymal stem cells. Stem Cell Rev. Rep. 2018, 14, 297–308. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Hossain, M.S.; Liu, D. Involvement of the long noncoding RNA H19 in osteogenic differentiation and bone regeneration. Stem Cell Res. Ther. 2021, 12, 74. [Google Scholar] [CrossRef]
- Ning, B.; Jin, R.; Wang, D.; Sun, J. The H19/let-7 feedback loop contributes to developmental dysplasia and dislocation of the hip. Physiol. Res. 2019, 68, 275–284. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.-L.; Zuo, B.; Li, D.; Zhu, J.-F.; Xiao, F.; Zhang, X.-L.; Chen, X.-D. The long noncoding RNA H19 attenuates force-driven cartilage degeneration via miR-483-5p/Dusp5. Biochem. Biophys. Res. Commun. 2020, 529, 210–217. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Durinck, S.; Spellman, P.T.; Birney, E.; Huber, W. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat. Protoc. 2009, 4, 1184–1191. [Google Scholar] [CrossRef]
- Sherman, B.T.; Hao, M.; Qiu, J.; Jiao, X.; Baseler, M.W.; Lane, H.C.; Imamichi, T.; Chang, W. DAVID: A web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res. 2022, 50, W216–W221. [Google Scholar] [CrossRef]
- Ning, B.; Sun, J.; Yuan, Y.; Yao, J.; Wang, P.; Ma, R. Early articular cartilage degeneration in a developmental dislocation of the hip model results from activation of β-catenin. Int. J. Clin. Exp. Pathol. 2014, 7, 1369–1378. [Google Scholar] [PubMed]
- Feng, W.-J.; Wang, H.; Shen, C.; Zhu, J.-F.; Chen, X.-D. Severe cartilage degeneration in patients with developmental dysplasia of the hip. IUBMB Life 2017, 69, 179–187. [Google Scholar] [CrossRef]
- Freemont, A.J.; Hampson, V.; Tilman, R.; Goupille, P.; Taiwo, Y.; Hoyland, J.A. Gene expression of matrix metalloproteinases 1, 3, and 9 by chondrocytes in osteoarthritic human knee articular cartilage is zone and grade specific. Ann. Rheum. Dis. 1997, 56, 542–549. [Google Scholar] [CrossRef]
- Dai, J.; Shi, D.; Zhu, P.; Qin, J.; Ni, H.; Xu, Y.; Yao, C.; Zhu, L.; Zhu, H.; Zhao, B.; et al. Association of a single nucleotide polymorphism in growth differentiate factor 5 with congenital dysplasia of the hip: A case-control study. Arthritis Res. Ther. 2008, 10, R126. [Google Scholar] [CrossRef]
- Enochson, L.; Stenberg, J.; Brittberg, M.; Lindahl, A. GDF5 reduces MMP13 expression in human chondrocytes via DKK1 mediated canonical Wnt signaling inhibition. Osteoarthr. Cartil. 2014, 22, 566–577. [Google Scholar] [CrossRef]
- Weng, L.-H.; Ko, J.-Y.; Wang, C.-J.; Sun, Y.-C.; Wang, F.-S. Dkk-1 promotes angiogenic responses and cartilage matrix proteinase secretion in synovial fibroblasts from osteoarthritic joints. Arthritis Rheum. 2012, 64, 3267–3277. [Google Scholar] [CrossRef]
- Weng, L.-H.; Wang, C.-J.; Ko, J.-Y.; Sun, Y.-C.; Wang, F.-S. Control of Dkk-1 ameliorates chondrocyte apoptosis, cartilage destruction, and subchondral bone deterioration in osteoarthritic knees. Arthritis Rheum. 2010, 62, 1393–1402. [Google Scholar] [CrossRef]
- Sun, Y.; You, Y.; Dai, K.; Zhang, J.; Yan, M.; Zhang, Y. Genetic variant of WIF1 gene is functionally associated with developmental dysplasia of the hip in Han Chinese population. Sci. Rep. 2019, 9, 285. [Google Scholar] [CrossRef]
- Church, V.L.; Francis-West, P. Wnt signalling during limb development. Int. J. Dev. Biol. 2002, 46, 927–936. [Google Scholar] [PubMed]
- Chijimatsu, R.; Saito, T. Mechanisms of synovial joint and articular cartilage development. Cell Mol. Life Sci. 2019, 76, 3939–3952. [Google Scholar] [CrossRef] [PubMed]
- Zayzafoon, M. Calcium/calmodulin signaling controls osteoblast growth and differentiation. J. Cell Biochem. 2006, 97, 56–70. [Google Scholar] [CrossRef] [PubMed]
- Kajiya, H. Calcium signaling in osteoclast differentiation and bone resorption. Adv. Exp. Med. Biol. 2012, 740, 917–932. [Google Scholar]
- Duncan, R.L.; Akanbi, K.A.; Farach-Carson, M.C. Calcium signals and calcium channels in osteoblastic cells. Semin. Nephrol. 1998, 18, 178–190. [Google Scholar]
- Wu, M.; Wu, S.; Chen, W.; Li, Y.P. The roles and regulatory mechanisms of TGF-β and BMP signaling in bone and cartilage development, homeostasis and disease. Cell Res. 2024, 34, 101–123. [Google Scholar] [CrossRef]
- Kwon, H.-S.; Lee, H.-S.; Ji, Y.; Rubin, J.S.; Tomarev, S.I. Myocilin is a modulator of Wnt signaling. Mol. Cell Biol. 2009, 29, 2139–2154. [Google Scholar] [CrossRef] [PubMed]
- Atienzar-Aroca, R.; Aroca-Aguilar, J.-D.; Alexandre-Moreno, S.; Ferre-Fernández, J.-J.; Bonet-Fernández, J.-M.; Cabañero-Varela, M.-J.; Escribano, J. Knockout of myoc Provides Evidence for the Role of Myocilin in Zebrafish Sex Determination Associated with Wnt Signalling Downregulation. Biology 2021, 10, 188. [Google Scholar] [CrossRef] [PubMed]
- Catela, C.; Assimacopoulos, S.; Chen, Y.; Tsioras, K.; Feng, W.; Kratsios, P. The Iroquois (Iro/Irx) homeobox genes are conserved Hox targets involved in motor neuron development. iScience 2025, 28, 112210. [Google Scholar] [CrossRef] [PubMed]
- Janssens, S.; Denayer, T.; Deroo, T.; Van Roy, F.; Vleminckx, K. Direct control of Hoxd1 and Irx3 expression by Wnt/beta-catenin signaling during anteroposterior patterning of the neural axis in Xenopus. Int. J. Dev. Biol. 2010, 54, 1435–1442. [Google Scholar] [CrossRef]
- Liao, J.; Chen, B.; Zhu, Z.; Du, C.; Gao, S.; Zhao, G.; Zhao, P.; Wang, Y.; Wang, A.; Schwartz, Z.; et al. Long noncoding RNA (lncRNA) H19: An essential developmental regulator with expanding roles in cancer, stem cell differentiation, and metabolic diseases. Genes Dis. 2023, 10, 1351–1366. [Google Scholar] [CrossRef]
- Bassani, S.; Chrast, J.; Ambrosini, G.; Voisin, N.; Schütz, F.; Brusco, A.; Sirchia, F.; Turban, L.; Schubert, S.; Jamra, R.A.; et al. Variant-specific pathophysiological mechanisms of AFF3 differently influence transcriptome profiles. Genome Med. 2024, 16, 72. [Google Scholar] [CrossRef]
- Chen, J.-W.; Wang, Y.-X.; Gao, R.-R.; Ma, L.-Y.; Zhong, J.; Yang, J.-X.; Deng, Z.-H.; Li, Y.-Y.; Li, X.-L.; Shu, Y.-H.; et al. CDK14 regulates the development and repair of lung. Cell Death Discov. 2025, 11, 12. [Google Scholar] [CrossRef]


| ID | Treatment | Mapping Rate (%) | Read Number (2 × 150 bp) |
|---|---|---|---|
| SRR14055716 | DDH | 80.74 | 24,847,026 |
| SRR14055717 | DDH | 84.91 | 33,533,704 |
| SRR14055718 | DDH | 80.85 | 23,836,127 |
| SRR14055719 | DDH | 98.25 | 27,104,541 |
| SRR14055720 | DDH | 87.71 | 24,866,901 |
| SRR14055721 | DDH | 94.21 | 30,621,536 |
| SRR14055722 | Control | 85.35 | 23,421,990 |
| SRR14055723 | Control | 93.55 | 23,402,707 |
| SRR14055724 | Control | 83.99 | 25,453,109 |
| SRR14055725 | Control | 83.48 | 20,408,199 |
| SRR14055726 | Control | 91.22 | 25,169,198 |
| SRR14055727 | Control | 90.25 | 21,844,255 |
| ID | log2 Fold Change | p-Value | Regulation in DDH |
|---|---|---|---|
| ENSG00000034971 | −9.1 | 1.82 × 10−33 | Up |
| ENSG00000309097 | −7.6 | 0.011025698 | Up |
| ENSG00000256513 | −7 | 2.82 × 10−6 | Up |
| ENSG00000170549 | −6.5 | 7.46 × 10−9 | Up |
| ENSG00000250421 | −6.5 | 2.88 × 10−7 | Up |
| ENSG00000279096 | −6.4 | 1.67 × 10−5 | Up |
| ENSG00000306524 | −6.2 | 5.87 × 10−8 | Up |
| ENSG00000168447 | −5.9 | 1.51 × 10−14 | Up |
| ENSG00000240654 | −5.6 | 4.60 × 10−7 | Up |
| ENSG00000160097 | −5.5 | 3.54 × 10−9 | Up |
| ENSG00000286415 | −5.4 | 0.000715357 | Up |
| ENSG00000305982 | −5.4 | 0.007306598 | Up |
| ENSG00000305491 | −5.4 | 0.010014926 | Up |
| ENSG00000181408 | −5.3 | 2.18 × 10−9 | Up |
| ENSG00000179915 | −5.3 | 6.20 × 10−9 | Up |
| ENSG00000156076 | −5.3 | 1.73 × 10−5 | Up |
| ENSG00000271239 | −5.3 | 0.015421818 | Up |
| ENSG00000142973 | −5.2 | 7.44 × 10−12 | Up |
| ENSG00000214866 | −5.2 | 2.19 × 10−5 | Up |
| ENSG00000118733 | −5.2 | 0.000432547 | Up |
| ENSG00000274833 | −5.2 | 0.002264803 | Up |
| ENSG00000265962 | −5.2 | 0.004283014 | Up |
| ENSG00000301580 | −5.1 | 5.39 × 10−6 | Up |
| ENSG00000188803 | −5.1 | 2.66 × 10−5 | Up |
| ENSG00000305239 | −5.1 | 0.000191428 | Up |
| ENSG00000229236 | 6.5 | 1.94 × 10−6 | Down |
| ENSG00000187689 | 6.5 | 4.20 × 10−6 | Down |
| ENSG00000149968 | 6.6 | 1.96 × 10−14 | Down |
| ENSG00000266604 | 6.7 | 1.93 × 10−5 | Down |
| ENSG00000116147 | 6.7 | 0.002750078 | Down |
| ENSG00000211934 | 6.7 | 0.025736049 | Down |
| ENSG00000231131 | 6.8 | 9.52 × 10−11 | Down |
| ENSG00000012817 | 6.8 | 2.32 × 10−6 | Down |
| ENSG00000211660 | 6.8 | 0.023562957 | Down |
| ENSG00000249306 | 6.9 | 9.30 × 10−17 | Down |
| ENSG00000260197 | 6.9 | 1.98 × 10−7 | Down |
| ENSG00000229876 | 7 | 1.05 × 10−6 | Down |
| ENSG00000183878 | 7 | 4.61 × 10−6 | Down |
| ENSG00000114374 | 7.1 | 4.07 × 10−7 | Down |
| ENSG00000067048 | 7.2 | 2.85 × 10−14 | Down |
| ENSG00000126545 | 7.2 | 5.33 × 10−9 | Down |
| ENSG00000129824 | 7.3 | 3.60 × 10−8 | Down |
| ENSG00000231535 | 7.4 | 2.40 × 10−8 | Down |
| ENSG00000211598 | 7.5 | 4.07 × 10−7 | Down |
| ENSG00000165246 | 7.5 | 1.11 × 10−6 | Down |
| ENSG00000266995 | 7.7 | 6.79 × 10−14 | Down |
| ENSG00000291033 | 7.7 | 7.11 × 10−14 | Down |
| ENSG00000067646 | 7.9 | 1.88 × 10−13 | Down |
| ENSG00000137745 | 8.7 | 5.16 × 10−26 | Down |
| ENSG00000198692 | 9.4 | 8.86 × 10−7 | Down |
| Cluster Number | Cluster Color | Gene Count | Primary Description |
|---|---|---|---|
| 1 | Red | 339 | Immune response |
| 2 | Brown | 266 | Mitotic cell cycle process |
| 3 | Dark goldenrod | 255 | Extracellular matrix |
| 4 | Green-yellow | 133 | - |
| 5 | Green 2 | 65 | - |
| 6 | Green | 56 | - |
| 7 | Blue | 53 | - |
| 8 | Light sky blue | 41 | Postsynaptic cell membrane and protein–protein interactions at synapses |
| 9 | Medium blue | 25 | ncRNAs involved in Wnt signaling in hepatocellular carcinoma, and regulation of FZD by ubiquitination |
| 10 | Purple | 16 | O-linked glycosylation of mucins |
| ID | BaseMean | Log2 Fold Change | lfcSE | Stat | p-Value | padj | Regulation in DDH |
|---|---|---|---|---|---|---|---|
| ENSG00000309097 | 20.0553182 | −7.6 | 3.00449956 | −2.5418833 | 0.0110257 | 0.12197728 | Up |
| ENSG00000256513 | 12.8646141 | −7 | 1.49363454 | −4.6834569 | 2.82 × 10−6 | 0.00036368 | Up |
| ENSG00000250421 | 13.1509542 | −6.5 | 1.2755177 | −5.1312126 | 2.88 × 10−7 | 6.37 × 10−5 | Up |
| ENSG00000306524 | 7.35762862 | −6.2 | 1.14212948 | −5.4227401 | 5.87 × 10−8 | 1.76 × 10−5 | Up |
| ENSG00000286415 | 4.37306511 | −5.4 | 1.6074669 | −3.3836247 | 0.00071536 | 0.02130454 | Up |
| ENSG00000305982 | 4.31991575 | −5.4 | 2.0210034 | −2.6825374 | 0.0073066 | 0.09516642 | Up |
| ENSG00000305491 | 4.18734128 | −5.4 | 2.08974346 | −2.5753135 | 0.01001493 | 0.1148278 | Up |
| ENSG00000271239 | 12.0861402 | −5.3 | 2.17536921 | −2.4223187 | 0.01542182 | 0.1463899 | Up |
| ENSG00000274833 | 3.70182773 | −5.2 | 1.70130402 | −3.0531135 | 0.0022648 | 0.04671496 | Up |
| ENSG00000265962 | 3.6288218 | −5.2 | 1.81060089 | −2.8565286 | 0.00428301 | 0.07013193 | Up |
| ENSG00000301580 | 6.83679945 | −5.1 | 1.11596197 | −4.5490905 | 5.39 × 10−6 | 0.00057765 | Up |
| ENSG00000305239 | 6.90491206 | −5.1 | 1.36677843 | −3.7300681 | 0.00019143 | 0.00839207 | Up |
| ENSG00000233845 | 3.34256322 | −5.1 | 1.40746917 | −3.5916966 | 0.00032853 | 0.01242405 | Up |
| ENSG00000300502 | 3.29931021 | −5 | 1.71918577 | −2.9309225 | 0.00337957 | 0.06050065 | Up |
| ENSG00000295404 | 12.0694117 | −5 | 2.17474927 | −2.2856073 | 0.02227725 | 0.18085266 | Up |
| ENSG00000307068 | 4.26292628 | −4.9 | 1.76580602 | −2.7621872 | 0.00574155 | 0.08279327 | Up |
| ENSG00000226562 | 2.83969294 | −4.8 | 2.15712697 | −2.2323878 | 0.02558934 | 0.19291473 | Up |
| ENSG00000267653 | 18.6393025 | −4.6 | 1.18355532 | −3.8857992 | 0.00010199 | 0.00535485 | Up |
| ENSG00000298690 | 2.40410301 | −4.6 | 1.31131045 | −3.4816176 | 0.0004984 | 0.01688615 | Up |
| ENSG00000304163 | 3.60829109 | −4.6 | 1.63774551 | −2.8282687 | 0.00468005 | 0.07329761 | Up |
| ENSG00000300437 | 2.48886314 | −4.6 | 1.7087183 | −2.7103252 | 0.00672173 | 0.09071955 | Up |
| ENSG00000293757 | 2.45471648 | −4.6 | 2.2150069 | −2.0801695 | 0.03750999 | 0.23607729 | Up |
| ENSG00000285649 | 2.24907601 | −4.5 | 1.49009445 | −3.006602 | 0.00264185 | NA | Up |
| ENSG00000276831 | 2.26891435 | −4.5 | 2.15967333 | −2.084345 | 0.03712879 | NA | Up |
| ENSG00000224239 | 3.14919595 | −4.4 | 1.21015928 | −3.6694311 | 0.00024309 | 0.00996896 | Up |
| ENSG00000296719 | 2.07942702 | −4.4 | 1.29583942 | −3.3770964 | 0.00073255 | NA | Up |
| ENSG00000295613 | 2.11341551 | −4.4 | 1.77626226 | −2.4722183 | 0.01342775 | NA | Up |
| ENSG00000298074 | 43.2261772 | −4.3 | 0.68673232 | −6.227569 | 4.74 × 10−10 | 3.71 × 10−7 | Up |
| ENSG00000274478 | 3.94265842 | −4.3 | 1.38362988 | −3.0753408 | 0.00210262 | 0.04461646 | Up |
| ENSG00000227487 | 4.06682931 | −4.3 | 1.58140449 | −2.721663 | 0.00649543 | 0.08932244 | Up |
| ENSG00000231132 | 1.92513856 | −4.3 | 1.86805998 | −2.2793749 | 0.02264479 | NA | Up |
| ENSG00000296003 | 1.94909901 | −4.3 | 1.89990295 | −2.2485052 | 0.02454399 | NA | Up |
| ENSG00000298193 | 1.92079493 | −4.3 | 1.96128058 | −2.16822 | 0.03014195 | NA | Up |
| ENSG00000285936 | 1.97756865 | −4.3 | 2.1797068 | −1.9693765 | 0.04890988 | NA | Up |
| ENSG00000300255 | 1.91034707 | −4.2 | 1.34504573 | −3.1580307 | 0.00158839 | NA | Up |
| ENSG00000286468 | 1.92446954 | −4.2 | 1.69334955 | −2.5096469 | 0.0120852 | NA | Up |
| ENSG00000297456 | 1.79753544 | −4.2 | 1.97057075 | −2.1102525 | 0.03483661 | NA | Up |
| ENSG00000264007 | 2.74415058 | −4.2 | 2.02575856 | −2.0950347 | 0.0361679 | 0.23135695 | Up |
| ENSG00000231419 | 10.5586343 | −4.1 | 0.95511704 | −4.2721184 | 1.94 × 10−5 | 0.00153673 | Up |
| ENSG00000297199 | 9.18829618 | −4.1 | 1.06625967 | −3.8898188 | 0.00010032 | 0.00528289 | Up |
| ENSG00000305011 | 2.48231308 | −4.1 | 1.18927314 | −3.4505029 | 0.00055954 | 0.01805428 | Up |
| ENSG00000229495 | 1.74392169 | −4.1 | 1.5299145 | −2.6866744 | 0.00721673 | NA | Up |
| ENSG00000275358 | 3.47818747 | −4.1 | 1.70141215 | −2.4009139 | 0.01635418 | 0.15200984 | Up |
| ENSG00000300949 | 1.67487121 | −4.1 | 1.84983805 | −2.1963248 | 0.0280687 | NA | Up |
| ENSG00000286411 | 2.505807 | −4.1 | 1.89232231 | −2.1742414 | 0.02968701 | 0.20751407 | Up |
| ENSG00000285686 | 3.5641727 | −4.1 | 1.91437549 | −2.1534663 | 0.03128205 | 0.21402918 | Up |
| ENSG00000286818 | 6.03882055 | −4 | 1.25554434 | −3.2155005 | 0.00130217 | 0.0325075 | Up |
| ENSG00000294628 | 1.600431 | −4 | 1.54122597 | −2.5909005 | 0.00957252 | NA | Up |
| ENSG00000300397 | 3.36778179 | −4 | 1.71935962 | −2.3439341 | 0.01908155 | 0.1658802 | Up |
| ENSG00000234944 | 3.24048221 | −4 | 1.72676428 | −2.2928222 | 0.02185824 | 0.1789809 | Up |
| ENSG00000204971 | 3.44993353 | 5.4 | 1.39793787 | 3.84619167 | 0.00011997 | 0.00603911 | Down |
| ENSG00000298268 | 3.46510039 | 5.4 | 1.48640556 | 3.60463808 | 0.00031259 | 0.01198425 | Down |
| ENSG00000258183 | 4.83347254 | 5.4 | 1.56660674 | 3.41522649 | 0.00063729 | 0.01970541 | Down |
| ENSG00000303389 | 67.4491212 | 5.4 | 1.59960987 | 3.39016959 | 0.00069849 | 0.02101712 | Down |
| ENSG00000249667 | 3.57232375 | 5.4 | 1.78879945 | 3.02570312 | 0.00248056 | 0.04947514 | Down |
| ENSG00000249993 | 3.51827941 | 5.4 | 1.81714131 | 2.96381834 | 0.00303848 | 0.05650863 | Down |
| ENSG00000310062 | 3.48578139 | 5.4 | 1.8301654 | 2.93439046 | 0.00334204 | 0.06010549 | Down |
| ENSG00000304111 | 3.54345937 | 5.4 | 1.85545923 | 2.90663633 | 0.00365338 | 0.06364457 | Down |
| ENSG00000308399 | 3.52372208 | 5.4 | 2.06650839 | 2.60844294 | 0.00909552 | 0.10839416 | Down |
| ENSG00000290670 | 3.52399192 | 5.4 | 2.08902577 | 2.57982723 | 0.00988498 | 0.1137742 | Down |
| ENSG00000300640 | 3.53914029 | 5.4 | 2.12685373 | 2.53647022 | 0.01119763 | 0.12259215 | Down |
| ENSG00000306164 | 3.45792879 | 5.4 | 2.15200616 | 2.49173312 | 0.01271215 | 0.13155437 | Down |
| ENSG00000250822 | 3.60090773 | 5.4 | 2.48073063 | 2.18592409 | 0.02882116 | 0.20492453 | Down |
| ENSG00000290840 | 5.17888445 | 5.5 | 1.50803576 | 3.61858637 | 0.00029622 | 0.01151552 | Down |
| ENSG00000300947 | 3.78576114 | 5.5 | 1.63901592 | 3.34744641 | 0.0008156 | 0.02356484 | Down |
| ENSG00000289707 | 3.82574307 | 5.5 | 1.73352772 | 3.18330533 | 0.00145604 | 0.03503209 | Down |
| ENSG00000300565 | 3.73979054 | 5.5 | 1.7681675 | 3.09858123 | 0.0019445 | 0.04225892 | Down |
| ENSG00000250348 | 3.81618728 | 5.5 | 2.45002286 | 2.2518587 | 0.0243312 | 0.1891298 | Down |
| ENSG00000230838 | 194.501423 | 5.6 | 0.70577156 | 7.98793817 | 1.37 × 10−15 | 3.81 × 10−12 | Down |
| ENSG00000298768 | 22.8322869 | 5.6 | 0.95737791 | 5.85599323 | 4.74 × 10−9 | 2.68 × 10−6 | Down |
| ENSG00000224099 | 4.19313483 | 5.6 | 1.77249083 | 3.18113846 | 0.00146698 | 0.03521975 | Down |
| ENSG00000308513 | 5.8501961 | 5.6 | 1.88810388 | 2.97828392 | 0.00289867 | 0.05482292 | Down |
| ENSG00000295056 | 4.32612076 | 5.7 | 1.36349558 | 4.18252197 | 2.88 × 10−5 | 0.00207142 | Down |
| ENSG00000259937 | 8.83425397 | 5.7 | 1.44977142 | 3.95282857 | 7.72 × 10−5 | 0.00441933 | Down |
| ENSG00000288015 | 4.27678936 | 5.7 | 1.46219699 | 3.87282695 | 0.00010758 | 0.00555274 | Down |
| ENSG00000308731 | 4.33753956 | 5.7 | 1.55232652 | 3.66561513 | 0.00024675 | 0.01005271 | Down |
| ENSG00000288049 | 4.24690662 | 5.7 | 1.69421782 | 3.34957101 | 0.00080937 | 0.02348628 | Down |
| ENSG00000253554 | 4.33106158 | 5.7 | 1.7507366 | 3.24846626 | 0.00116029 | 0.03006132 | Down |
| ENSG00000232596 | 6.40357577 | 5.8 | 1.51402082 | 3.79913474 | 0.0001452 | 0.00686811 | Down |
| ENSG00000298049 | 4.64546188 | 5.8 | 1.77170343 | 3.26603666 | 0.00109064 | 0.02881119 | Down |
| ENSG00000294222 | 17.6914436 | 5.9 | 1.47771016 | 4.01898815 | 5.84 × 10−5 | 0.00358626 | Down |
| ENSG00000293442 | 155.369799 | 5.9 | 1.48639466 | 3.93570269 | 8.30 × 10−5 | 0.00466193 | Down |
| ENSG00000264985 | 5.19724286 | 5.9 | 1.56786443 | 3.79214101 | 0.00014935 | 0.00698876 | Down |
| ENSG00000294508 | 5.07296798 | 5.9 | 1.75683438 | 3.36730502 | 0.00075907 | 0.02236649 | Down |
| ENSG00000251670 | 5.27532038 | 6 | 1.25670454 | 4.7454957 | 2.08 × 10−6 | 0.00029154 | Down |
| ENSG00000233854 | 5.74368619 | 6.1 | 1.30938943 | 4.65113408 | 3.30 × 10−6 | 0.0004047 | Down |
| ENSG00000286028 | 5.90442962 | 6.1 | 1.45659643 | 4.21037416 | 2.55 × 10−5 | 0.00189082 | Down |
| ENSG00000298839 | 32.2989896 | 6.2 | 1.03885417 | 5.94096616 | 2.83 × 10−9 | 1.80 × 10−6 | Down |
| ENSG00000293047 | 12.2817787 | 6.2 | 1.11489762 | 5.57238667 | 2.51 × 10−8 | 8.72 × 10−6 | Down |
| ENSG00000249343 | 8.40798677 | 6.2 | 1.55287593 | 3.96157443 | 7.45 × 10−5 | 0.00429032 | Down |
| ENSG00000176728 | 57.9891864 | 6.5 | 1.11309151 | 5.79756785 | 6.73 × 10−9 | 3.12 × 10−6 | Down |
| ENSG00000271216 | 15.2594445 | 6.5 | 1.36837552 | 4.77529373 | 1.79 × 10−6 | 0.0002611 | Down |
| ENSG00000229236 | 10.3349528 | 6.5 | 1.35769107 | 4.75996551 | 1.94 × 10−6 | 0.00027647 | Down |
| ENSG00000266604 | 8.70411474 | 6.7 | 1.56681229 | 4.27280749 | 1.93 × 10−5 | 0.00153597 | Down |
| ENSG00000231131 | 18.9261187 | 6.8 | 1.05759833 | 6.47436337 | 9.52 × 10−11 | 8.31 × 10−8 | Down |
| ENSG00000249306 | 151.620051 | 6.9 | 0.82875406 | 8.3133538 | 9.30 × 10−17 | 4.06 × 10−13 | Down |
| ENSG00000260197 | 19.7348345 | 6.9 | 1.32944921 | 5.20130859 | 1.98 × 10−7 | 4.72 × 10−5 | Down |
| ENSG00000229876 | 10.5213785 | 7 | 1.42708418 | 4.88236894 | 1.05 × 10−6 | 0.00017407 | Down |
| ENSG00000231535 | 27.0797506 | 7.4 | 1.32158065 | 5.58007893 | 2.40 × 10−8 | 8.44 × 10−6 | Down |
| ENSG00000291033 | 276.125148 | 7.7 | 1.03227535 | 7.48581379 | 7.11 × 10−14 | 1.21 × 10−10 | Down |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kaya, İ.; Türktaş, M.; Yaş, S.; Bircan, R. Genome-Wide mRNA and lncRNA Expression Profiling to Uncover Their Role in the Molecular Pathogenesis of Developmental Dysplasia of the Hip. Int. J. Mol. Sci. 2025, 26, 8058. https://doi.org/10.3390/ijms26168058
Kaya İ, Türktaş M, Yaş S, Bircan R. Genome-Wide mRNA and lncRNA Expression Profiling to Uncover Their Role in the Molecular Pathogenesis of Developmental Dysplasia of the Hip. International Journal of Molecular Sciences. 2025; 26(16):8058. https://doi.org/10.3390/ijms26168058
Chicago/Turabian StyleKaya, İbrahim, Mine Türktaş, Semih Yaş, and Resul Bircan. 2025. "Genome-Wide mRNA and lncRNA Expression Profiling to Uncover Their Role in the Molecular Pathogenesis of Developmental Dysplasia of the Hip" International Journal of Molecular Sciences 26, no. 16: 8058. https://doi.org/10.3390/ijms26168058
APA StyleKaya, İ., Türktaş, M., Yaş, S., & Bircan, R. (2025). Genome-Wide mRNA and lncRNA Expression Profiling to Uncover Their Role in the Molecular Pathogenesis of Developmental Dysplasia of the Hip. International Journal of Molecular Sciences, 26(16), 8058. https://doi.org/10.3390/ijms26168058






