Beyond the Microscope: Integrating Liquid Biopsies into the Molecular Pathology Era of Endometrial Cancer
Abstract
1. Introduction
2. Endometrial Carcinoma in the Molecular Era: Diagnostic Refinements and Emerging Roles for Pathologists
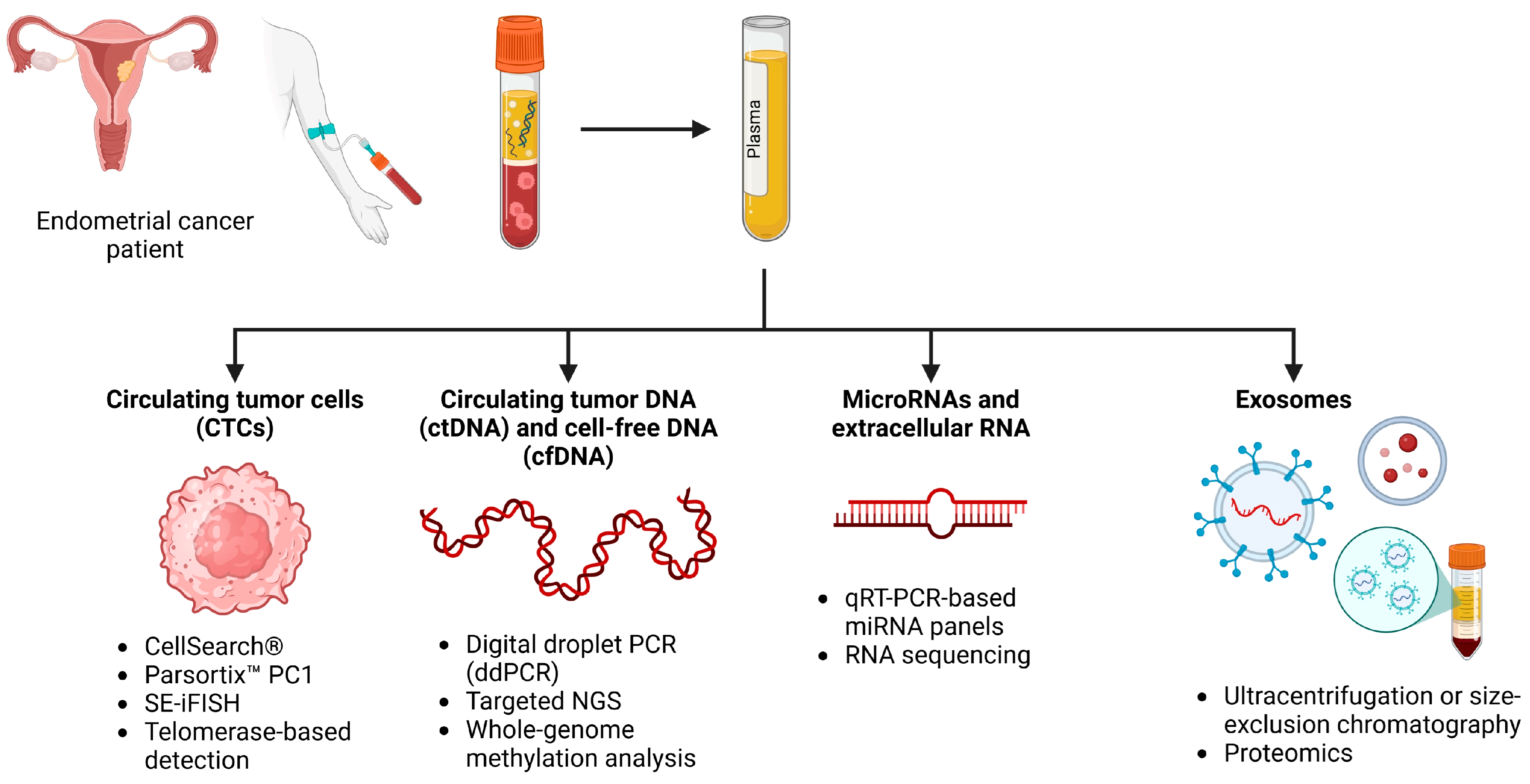
3. Liquid Biopsies: Technologies and Biomarkers
3.1. Circulating Tumor Cells (CTCs)
- CellSearch®, FDA-approved, utilizes immunomagnetic separation (nanoparticles) targeting EpCAM followed by cytokeratin (CK 8, 18, 19; to confirm epithelial origin) and DAPI staining (to confirm intact cells), with CD45 (to exclude leukocytes) [65]. However, it fails to detect CTCs that undergo EMT, which frequently occurs in high-grade EC.
- Parsortix™ PC1 is a microfluidics-based platform that isolates CTCs based on physical characteristics such as size and deformability, independent of epithelial markers [66]. It uses whole blood, and it is usually loaded via a microfluidic cassette into a Parsortix instrument. This method has an advantage over the CellSearch® method due to its lack of dependence on CTCs having to express epithelial markers such as EpCAM. The system is relatively slow, which may make it difficult to work with in the clinical setting, but it would detect CTCs including those that may have lost epithelial markers during EMT.
- SE-iFISH (Subtraction Enrichment and Immunostaining-FISH) combines the removal of leukocytes and erythrocytes with immunostaining and fluorescence in situ hybridization to detect CTCs. It allows for the identification of CTCs regardless of EpCAM expression and provides insights into chromosomal abnormalities. While still under investigation, its ability to detect a diverse range of CTCs makes it a promising tool in EC research [64].
- Telomerase-specific adenovirus-mediated fluorescence detection uses telomerase specific and replication selective adenovirus. In rapidly diving cells such as cancer cells, the activity of telomerase in DNA is highly elevated, which allows us to identify a wide range of tumors without having to do CTCs enrichment, which means concentrating the DNA to a higher density. The virus expresses a green fluorescent protein (GFO) that allows for the direct visualization of the detected tumor cell under a fluorescence microscope [67]. This method is very time-consuming and requires a lot of training for technicians due to its complexity, especially when analyzing larger volumes of body fluids. Lack of standardized clinical validation cutoffs limits its usefulness in clinical practice.
3.2. Circulating Tumor DNA (ctDNA) and Cell-Free DNA (cfDNA)
3.3. Circulating Cell-Free RNA (cfRNA)
3.4. Extracellular RNA and miRNAs
- qRT-PCR-based miRNA panels from plasma or uterine fluid.
- RNA sequencing of tumor-educated platelets (TEPs): A novel approach where platelets modulate their RNA content upon interaction with tumor cells and show promise in detecting EC with more than 95% accuracy [54].
3.5. Exosomes
- Ultracentrifugation or size-exclusion chromatography for exosome isolation.
- Proteomics (e.g., Liquid Chromatography–Parallel Reaction Monitoring [LC-PRM]) for profiling exosome-derived protein biomarkers such as MMP9 and PKM.
4. Mutation Panels and Methylation Profiling
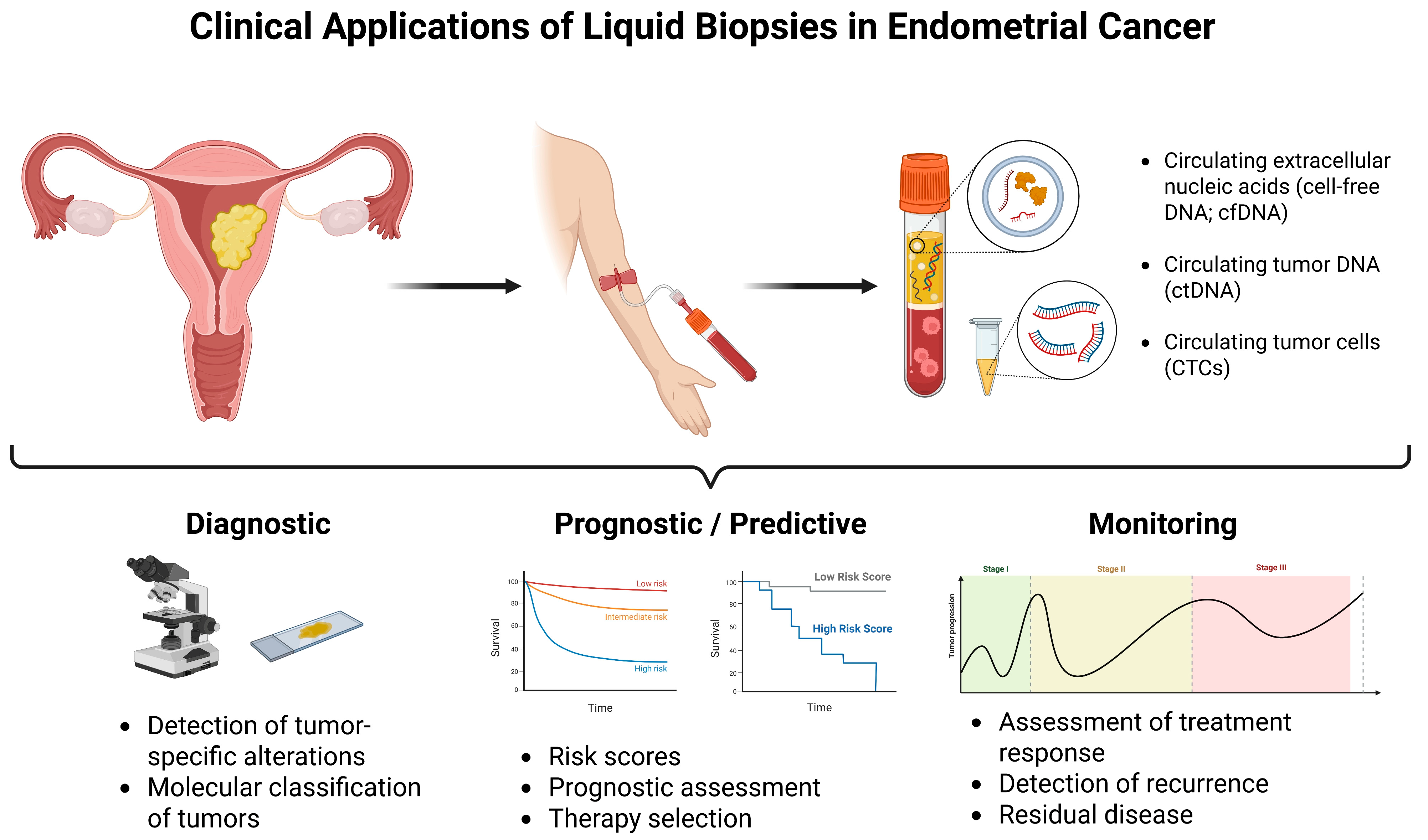
5. Clinical Applications of Liquid Biopsy in Endometrial Cancer
5.1. Potential Predictive Biomarkers Detectable via Liquid Biopsy
5.2. Application of Liquid Biopsies to Cervical Cytology Samples for Endometrial Cancer Detection
5.3. Liquid Biopsies in Clinical Trials
6. Challenges, Innovations, and Future Directions
6.1. Integrating Liquid Biopsies with Traditional Surgical Pathology
6.2. Overcoming Current Limitations
6.3. The “Needle in a Haystack” Problem and the Challenge of False Negatives
6.4. Equity, Histologic Subgroups, and Generalizability
6.5. Innovations in Liquid Biopsy Technologies
6.6. Artificial Intelligence
6.7. Personalized Oncology and Molecular Tailoring
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Constantine, G.D.; Kessler, G.; Graham, S.; Goldstein, S.R. Increased Incidence of Endometrial Cancer Following the Women’s Health Initiative: An Assessment of Risk Factors. J. Women’s Health 2019, 28, 237–243. [Google Scholar] [CrossRef]
- Siegel, R.L.; Kratzer, T.B.; Giaquinto, A.N.; Sung, H.; Jemal, A. Cancer statistics, 2025. CA A Cancer J. Clin. 2025, 75, 10–45. [Google Scholar] [CrossRef]
- National Cancer Institute. Cancer Stat Facts: Uterine Cancer. Available online: https://seer.cancer.gov/statfacts/html/corp.html?utm_source=chatgpt.com (accessed on 13 August 2025).
- Rodriguez, V.E.; Tanjasiri, S.P.; Ro, A.; Hoyt, M.A.; Bristow, R.E.; LeBrón, A.M.W. Trends in endometrial cancer incidence in the United States by race/ethnicity and age of onset from 2000 to 2019. Am. J. Epidemiol. 2025, 194, 103–113. [Google Scholar] [CrossRef]
- Howlader, N.; Noone, A.-M.; Krapcho, M.; Garshell, J.; Neyman, N.; Altekruse, S.; Kosary, C.; Yu, M.; Ruhl, J.; Tatalovich, Z. SEER cancer statistics review, 1975–2010. Bethesda MD Natl. Cancer Inst. 2013, 21, 12. [Google Scholar]
- Jemal, A.; Ward, E.M.; Johnson, C.J.; Cronin, K.A.; Ma, J.; Ryerson, B.; Mariotto, A.; Lake, A.J.; Wilson, R.; Sherman, R.L.; et al. Annual Report to the Nation on the Status of Cancer, 1975-2014, Featuring Survival. J. Natl. Cancer Inst. 2017, 109, djx030. [Google Scholar] [CrossRef] [PubMed]
- Bolivar, A.M.; Luthra, R.; Mehrotra, M.; Chen, W.; Barkoh, B.A.; Hu, P.; Zhang, W.; Broaddus, R.R. Targeted next-generation sequencing of endometrial cancer and matched circulating tumor DNA: Identification of plasma-based, tumor-associated mutations in early stage patients. Mod. Pathol. 2019, 32, 405–414. [Google Scholar] [CrossRef]
- Muinelo-Romay, L.; Casas-Arozamena, C.; Abal, M. Liquid Biopsy in Endometrial Cancer: New Opportunities for Personalized Oncology. Int. J. Mol. Sci. 2018, 19, 2311. [Google Scholar] [CrossRef]
- Ashley, C.W.; Selenica, P.; Patel, J.; Wu, M.; Nincevic, J.; Lakhman, Y.; Zhou, Q.; Shah, R.H.; Berger, M.F.; Da Cruz Paula, A.; et al. High-Sensitivity Mutation Analysis of Cell-Free DNA for Disease Monitoring in Endometrial Cancer. Clin. Cancer Res. 2023, 29, 410–421. [Google Scholar] [CrossRef]
- Kodada, D.; Hyblova, M.; Krumpolec, P.; Janostiakova, N.; Barath, P.; Grendar, M.; Blandova, G.; Petrovic, O.; Janega, P.; Repiska, V.; et al. The Potential of Liquid Biopsy in Detection of Endometrial Cancer Biomarkers: A Pilot Study. Int. J. Mol. Sci. 2023, 24, 7811. [Google Scholar] [CrossRef]
- Pereira, E.; Camacho-Vanegas, O.; Anand, S.; Sebra, R.; Catalina Camacho, S.; Garnar-Wortzel, L.; Nair, N.; Moshier, E.; Wooten, M.; Uzilov, A.; et al. Personalized Circulating Tumor DNA Biomarkers Dynamically Predict Treatment Response and Survival In Gynecologic Cancers. PLoS ONE 2015, 10, e0145754. [Google Scholar] [CrossRef]
- Zheng, W. Molecular Classification of Endometrial Cancer and the 2023 FIGO Staging: Exploring the Challenges and Opportunities for Pathologists. Cancers 2023, 15, 4101. [Google Scholar] [CrossRef]
- Yang, Y.; Wu, S.F.; Bao, W. Molecular subtypes of endometrial cancer: Implications for adjuvant treatment strategies. Int. J. Gynecol. Obstet. 2024, 164, 436–459. [Google Scholar] [CrossRef]
- Bradley, C.; Casey, C.; Daniel, S.; Saketh, G. Endometrial cancer: Molecular classification and future treatments. BMJ Med. 2022, 1, e000152. [Google Scholar] [CrossRef]
- Asami, Y.; Kobayashi Kato, M.; Hiranuma, K.; Matsuda, M.; Shimada, Y.; Ishikawa, M.; Koyama, T.; Komatsu, M.; Hamamoto, R.; Nagashima, M.; et al. Utility of molecular subtypes and genetic alterations for evaluating clinical outcomes in 1029 patients with endometrial cancer. Br. J. Cancer 2023, 128, 1582–1591. [Google Scholar] [CrossRef]
- RAINBO Research Consortium. Refining adjuvant treatment in endometrial cancer based on molecular features: The RAINBO clinical trial program. Int. J. Gynecol. Cancer 2023, 33, 109–117. [Google Scholar] [CrossRef]
- Matias-Guiu, X.; Oliva, E.; McCluggage, W.G.; Nucci, M.R.; Longacre, T.A. Tumours of the uterine corpus. In WHO Classification of Tumours Editorial Board. Female Genital Tumours [Internet], 5th ed.; Oliva, E., Matias-Guiu, X., Eds.; International Agency for Research on Cancer: Lyon, France, 2020; Volume 4. [Google Scholar]
- Piñeiro-Pérez, R.; Abal, M.; Muinelo-Romay, L. Liquid Biopsy for Monitoring EC Patients: Towards Personalized Treatment. Cancers 2022, 14, 1405. [Google Scholar] [CrossRef]
- Turashvili, G.; Karnezis, A.N.; Crothers, B.A.; Krishnamurti, U.G.; McCluggage, G.; Rabban, J.; Soslow, R. Protocol for the Examination of Specimens from Patients with Carcinoma of the Endometrium; College of American Pathologists: Northfield, IL, USA, 2024. [Google Scholar]
- Bokhman, J.V. Two pathogenetic types of endometrial carcinoma. Gynecol. Oncol. 1983, 15, 10–17. [Google Scholar] [CrossRef]
- Kandoth, C.; Schultz, N.; Cherniack, A.D.; Akbani, R.; Liu, Y.; Shen, H.; Robertson, A.G.; Pashtan, I.; Shen, R.; Benz, C.C.; et al. Integrated genomic characterization of endometrial carcinoma. Nature 2013, 497, 67–73. [Google Scholar] [CrossRef]
- Talhouk, A.; McConechy, M.K.; Leung, S.; Li-Chang, H.H.; Kwon, J.S.; Melnyk, N.; Yang, W.; Senz, J.; Boyd, N.; Karnezis, A.N.; et al. A clinically applicable molecular-based classification for endometrial cancers. Br. J. Cancer 2015, 113, 299–310. [Google Scholar] [CrossRef]
- Stelloo, E.; Nout, R.A.; Osse, E.M.; Jürgenliemk-Schulz, I.J.; Jobsen, J.J.; Lutgens, L.C.; van der Steen-Banasik, E.M.; Nijman, H.W.; Putter, H.; Bosse, T.; et al. Improved Risk Assessment by Integrating Molecular and Clinicopathological Factors in Early-stage Endometrial Cancer-Combined Analysis of the PORTEC Cohorts. Clin. Cancer Res. 2016, 22, 4215–4224. [Google Scholar] [CrossRef]
- McAlpine, J.; Leon-Castillo, A.; Bosse, T. The rise of a novel classification system for endometrial carcinoma; integration of molecular subclasses. J. Pathol. 2018, 244, 538–549. [Google Scholar] [CrossRef]
- Momeni-Boroujeni, A.; Dahoud, W.; Vanderbilt, C.M.; Chiang, S.; Murali, R.; Rios-Doria, E.V.; Alektiar, K.M.; Aghajanian, C.; Abu-Rustum, N.R.; Ladanyi, M.; et al. Clinicopathologic and Genomic Analysis of TP53-Mutated Endometrial Carcinomas. Clin. Cancer Res. 2021, 27, 2613–2623. [Google Scholar] [CrossRef]
- DeLair, D.F.; Burke, K.A.; Selenica, P.; Lim, R.S.; Scott, S.N.; Middha, S.; Mohanty, A.S.; Cheng, D.T.; Berger, M.F.; Soslow, R.A.; et al. The genetic landscape of endometrial clear cell carcinomas. J. Pathol. 2017, 243, 230–241. [Google Scholar] [CrossRef]
- Kuhn, E.; Ayhan, A.; Bahadirli-Talbott, A.; Zhao, C.; Shih Ie, M. Molecular characterization of undifferentiated carcinoma associated with endometrioid carcinoma. Am. J. Surg. Pathol. 2014, 38, 660–665. [Google Scholar] [CrossRef]
- Tung, H.-J.; Wu, R.-C.; Lin, C.-Y.; Lai, C.-H. Rare Subtype of Endometrial Cancer: Undifferentiated/Dedifferentiated Endometrial Carcinoma, from Genetic Aspects to Clinical Practice. Int. J. Mol. Sci. 2022, 23, 3794. [Google Scholar] [CrossRef]
- Coatham, M.; Li, X.; Karnezis, A.N.; Hoang, L.N.; Tessier-Cloutier, B.; Meng, B.; Soslow, R.A.; Blake Gilks, C.; Huntsman, D.G.; Stewart, C.J.; et al. Concurrent ARID1A and ARID1B inactivation in endometrial and ovarian dedifferentiated carcinomas. Mod. Pathol. 2016, 29, 1586–1593. [Google Scholar] [CrossRef]
- Köbel, M.; Hoang, L.N.; Tessier-Cloutier, B.; Meng, B.; Soslow, R.A.; Stewart, C.J.R.; Lee, C.H. Undifferentiated Endometrial Carcinomas Show Frequent Loss of Core Switch/Sucrose Nonfermentable Complex Proteins. Am. J. Surg. Pathol. 2018, 42, 76–83. [Google Scholar] [CrossRef] [PubMed]
- Ramalingam, P.; Croce, S.; McCluggage, W.G. Loss of expression of SMARCA4 (BRG1), SMARCA2 (BRM) and SMARCB1 (INI1) in undifferentiated carcinoma of the endometrium is not uncommon and is not always associated with rhabdoid morphology. Histopathology 2017, 70, 359–366. [Google Scholar] [CrossRef]
- Strehl, J.D.; Wachter, D.L.; Fiedler, J.; Heimerl, E.; Beckmann, M.W.; Hartmann, A.; Agaimy, A. Pattern of SMARCB1 (INI1) and SMARCA4 (BRG1) in poorly differentiated endometrioid adenocarcinoma of the uterus: Analysis of a series with emphasis on a novel SMARCA4-deficient dedifferentiated rhabdoid variant. Ann. Diagn. Pathol. 2015, 19, 198–202. [Google Scholar] [CrossRef] [PubMed]
- Karnezis, A.N.; Hoang, L.N.; Coatham, M.; Ravn, S.; Almadani, N.; Tessier-Cloutier, B.; Irving, J.; Meng, B.; Li, X.; Chow, C.; et al. Loss of switch/sucrose non-fermenting complex protein expression is associated with dedifferentiation in endometrial carcinomas. Mod. Pathol. 2016, 29, 302–314. [Google Scholar] [CrossRef]
- Stewart, C.J.; Crook, M.L. SWI/SNF complex deficiency and mismatch repair protein expression in undifferentiated and dedifferentiated endometrial carcinoma. Pathology 2015, 47, 439–445. [Google Scholar] [CrossRef]
- Castilla, M.; Moreno-Bueno, G.; Romero-Pérez, L.; Van De Vijver, K.; Biscuola, M.; López-García, M.; Prat, J.; Matías-Guiu, X.; Cano, A.; Oliva, E.; et al. Micro-RNA signature of the epithelial-mesenchymal transition in endometrial carcinosarcoma. J. Pathol. 2011, 223, 72–80. [Google Scholar] [CrossRef] [PubMed]
- Romero-Pérez, L.; Castilla, M.; López-García, M.; Díaz-Martín, J.; Biscuola, M.; Ramiro-Fuentes, S.; Oliva, E.; Matias-Guiu, X.; Prat, J.; Cano, A.; et al. Molecular events in endometrial carcinosarcomas and the role of high mobility group AT-hook 2 in endometrial carcinogenesis. Hum. Pathol. 2013, 44, 244–254. [Google Scholar] [CrossRef]
- Leskela, S.; Pérez-Mies, B.; Rosa-Rosa, J.M.; Cristobal, E.; Biscuola, M.; Palacios-Berraquero, M.L.; Ong, S.; Matias-Guiu Guia, X.; Palacios, J. Molecular Basis of Tumor Heterogeneity in Endometrial Carcinosarcoma. Cancers 2019, 11, 964. [Google Scholar] [CrossRef]
- Clarke, M.A.; Devesa, S.S.; Harvey, S.V.; Wentzensen, N. Hysterectomy-Corrected Uterine Corpus Cancer Incidence Trends and Differences in Relative Survival Reveal Racial Disparities and Rising Rates of Nonendometrioid Cancers. J. Clin. Oncol. 2019, 37, 1895–1908. [Google Scholar] [CrossRef]
- Piulats, J.M.; Guerra, E.; Gil-Martín, M.; Roman-Canal, B.; Gatius, S.; Sanz-Pamplona, R.; Velasco, A.; Vidal, A.; Matias-Guiu, X. Molecular approaches for classifying endometrial carcinoma. Gynecol. Oncol. 2017, 145, 200–207. [Google Scholar] [CrossRef]
- Eggink, F.A.; Van Gool, I.C.; Leary, A.; Pollock, P.M.; Crosbie, E.J.; Mileshkin, L.; Jordanova, E.S.; Adam, J.; Freeman-Mills, L.; Church, D.N.; et al. Immunological profiling of molecularly classified high-risk endometrial cancers identifies POLE-mutant and microsatellite unstable carcinomas as candidates for checkpoint inhibition. Oncoimmunology 2017, 6, e1264565. [Google Scholar] [CrossRef]
- Bakhsh, S.; Kinloch, M.; Hoang, L.N.; Soslow, R.A.; Köbel, M.; Lee, C.H.; McAlpine, J.N.; McConechy, M.K.; Gilks, C.B. Histopathological features of endometrial carcinomas associated with POLE mutations: Implications for decisions about adjuvant therapy. Histopathology 2016, 68, 916–924. [Google Scholar] [CrossRef]
- Hussein, Y.R.; Weigelt, B.; Levine, D.A.; Schoolmeester, J.K.; Dao, L.N.; Balzer, B.L.; Liles, G.; Karlan, B.; Köbel, M.; Lee, C.H.; et al. Clinicopathological analysis of endometrial carcinomas harboring somatic POLE exonuclease domain mutations. Mod. Pathol. 2015, 28, 505–514. [Google Scholar] [CrossRef]
- Mehnert, J.M.; Panda, A.; Zhong, H.; Hirshfield, K.; Damare, S.; Lane, K.; Sokol, L.; Stein, M.N.; Rodriguez-Rodriquez, L.; Kaufman, H.L.; et al. Immune activation and response to pembrolizumab in POLE-mutant endometrial cancer. J. Clin. Investig. 2016, 126, 2334–2340. [Google Scholar] [CrossRef]
- Mirkovic, J.; McFarland, M.; Garcia, E.; Sholl, L.M.; Lindeman, N.; MacConaill, L.; Dong, F.; Hirsch, M.; Nucci, M.R.; Quick, C.M.; et al. Targeted Genomic Profiling Reveals Recurrent KRAS Mutations in Mesonephric-like Adenocarcinomas of the Female Genital Tract. Am. J. Surg. Pathol. 2018, 42, 227–233. [Google Scholar] [CrossRef]
- Na, K.; Kim, H.S. Clinicopathologic and Molecular Characteristics of Mesonephric Adenocarcinoma Arising From the Uterine Body. Am. J. Surg. Pathol. 2019, 43, 12–25. [Google Scholar] [CrossRef]
- Euscher, E.D.; Bassett, R.; Duose, D.Y.; Lan, C.; Wistuba, I.; Ramondetta, L.; Ramalingam, P.; Malpica, A. Mesonephric-like Carcinoma of the Endometrium: A Subset of Endometrial Carcinoma With an Aggressive Behavior. Am. J. Surg. Pathol. 2020, 44, 429–443. [Google Scholar] [CrossRef] [PubMed]
- Koontz, J.I.; Soreng, A.L.; Nucci, M.; Kuo, F.C.; Pauwels, P.; van Den Berghe, H.; Dal Cin, P.; Fletcher, J.A.; Sklar, J. Frequent fusion of the JAZF1 and JJAZ1 genes in endometrial stromal tumors. Proc. Natl. Acad. Sci. USA 2001, 98, 6348–6353. [Google Scholar] [CrossRef]
- Chiang, S.; Lee, C.H.; Stewart, C.J.R.; Oliva, E.; Hoang, L.N.; Ali, R.H.; Hensley, M.L.; Arias-Stella, J.A., 3rd; Frosina, D.; Jungbluth, A.A.; et al. BCOR is a robust diagnostic immunohistochemical marker of genetically diverse high-grade endometrial stromal sarcoma, including tumors exhibiting variant morphology. Mod. Pathol. 2017, 30, 1251–1261. [Google Scholar] [CrossRef] [PubMed]
- Cotzia, P.; Benayed, R.; Mullaney, K.; Oliva, E.; Felix, A.; Ferreira, J.; Soslow, R.A.; Antonescu, C.R.; Ladanyi, M.; Chiang, S. Undifferentiated Uterine Sarcomas Represent Under-Recognized High-grade Endometrial Stromal Sarcomas. Am. J. Surg. Pathol. 2019, 43, 662–669. [Google Scholar] [CrossRef]
- Esposito, G.; D’Angelo, G.; De Falco, L.; Evangelista, E.; Savarese, G.; Fico, A.; Cinque, F.; Giampaolino, P.; Di Spiezio Sardo, A.; Bifulco, G.; et al. The Application of Liquid Biopsy for the Development and Validation of a Non-Invasive Screening and Diagnosis Test for Endometrial Premalignant and Malignant Lesions: A Prospective Innovative Pilot Study. Cancers 2025, 17, 1078. [Google Scholar] [CrossRef]
- Francini, S.; Duraes, M.; Rathat, G.; Macioce, V.; Mollevi, C.; Pages, L.; Ferrer, C.; Cayrefourcq, L.; Alix-Panabières, C. Circulating Tumor Cell Detection by Liquid Biopsy during Early-Stage Endometrial Cancer Surgery: A Pilot Study. Biomolecules 2023, 13, 428. [Google Scholar] [CrossRef]
- Shen, Y.; Shi, R.; Zhao, R.; Wang, H. Clinical application of liquid biopsy in endometrial carcinoma. Med. Oncol. 2023, 40, 92. [Google Scholar] [CrossRef]
- Alix-Panabières, C.; Pantel, K. Liquid Biopsy to Catch the Epigenetic Changes in Endometrial Cancer. Clin. Chem. 2022, 68, 745–747. [Google Scholar] [CrossRef] [PubMed]
- Łukasiewicz, M.; Pastuszak, K.; Łapińska-Szumczyk, S.; Różański, R.; Veld, S.; Bieńkowski, M.; Stokowy, T.; Ratajska, M.; Best, M.G.; Würdinger, T.; et al. Diagnostic Accuracy of Liquid Biopsy in Endometrial Cancer. Cancers 2021, 13, 5731. [Google Scholar] [CrossRef]
- Casas-Arozamena, C.; Díaz, E.; Moiola, C.P.; Alonso-Alconada, L.; Ferreirós, A.; Abalo, A.; Gil, C.L.; Oltra, S.S.; de Santiago, J.; Cabrera, S.; et al. Genomic Profiling of Uterine Aspirates and cfDNA as an Integrative Liquid Biopsy Strategy in Endometrial Cancer. J. Clin. Med. 2020, 9, 585. [Google Scholar] [CrossRef]
- Herrero, C.; de la Fuente, A.; Casas-Arozamena, C.; Sebastian, V.; Prieto, M.; Arruebo, M.; Abalo, A.; Colás, E.; Moreno-Bueno, G.; Gil-Moreno, A.; et al. Extracellular Vesicles-Based Biomarkers Represent a Promising Liquid Biopsy in Endometrial Cancer. Cancers 2019, 11, 2000. [Google Scholar] [CrossRef]
- Santoro, A.; Angelico, G.; Travaglino, A.; Inzani, F.; Arciuolo, D.; Valente, M.; D’Alessandris, N.; Scaglione, G.; Fiorentino, V.; Raffone, A.; et al. New Pathological and Clinical Insights in Endometrial Cancer in View of the Updated ESGO/ESTRO/ESP Guidelines. Cancers 2021, 13, 2623. [Google Scholar] [CrossRef]
- Santoro, A.; Angelico, G.; Travaglino, A.; Raffone, A.; Arciuolo, D.; D’Alessandris, N.; Inzani, F.; Zannoni, G.F. Clinico-pathological significance of TCGA classification and SWI/SNF proteins expression in undifferentiated/dedifferentiated endometrial carcinoma: A possible prognostic risk stratification. Gynecol. Oncol. 2021, 161, 629–635. [Google Scholar] [CrossRef] [PubMed]
- Cardoso, F.; Paluch-Shimon, S.; Senkus, E.; Curigliano, G.; Aapro, M.S.; André, F.; Barrios, C.H.; Bergh, J.; Bhattacharyya, G.S.; Biganzoli, L.; et al. 5th ESO-ESMO international consensus guidelines for advanced breast cancer (ABC 5). Ann. Oncol. 2020, 31, 1623–1649. [Google Scholar] [CrossRef]
- Koh, W.J.; Abu-Rustum, N.R.; Bean, S.; Bradley, K.; Campos, S.M.; Cho, K.R.; Chon, H.S.; Chu, C.; Cohn, D.; Crispens, M.A.; et al. Uterine Neoplasms, Version 1.2018, NCCN Clinical Practice Guidelines in Oncology. J. Natl. Compr. Cancer Netw. 2018, 16, 170–199. [Google Scholar] [CrossRef]
- Martinelli, C.; Ercoli, A.; Vizzielli, G.; Burk, S.R.; Cuomo, M.; Satasiya, V.; Kacem, H.; Braccia, S.; Mazzarotti, G.; Miriello, I.; et al. Liquid biopsy in gynecological cancers: A translational framework from molecular insights to precision oncology and clinical practice. J. Exp. Clin. Cancer Res. 2025, 44, 140. [Google Scholar] [CrossRef]
- de Jager, V.D.; Giacomini, P.; Fairley, J.A.; Toledo, R.A.; Patton, S.J.; Joosse, S.A.; Koch, C.; Deans, Z.C.; Agelaki, S.; Andersen, C.L.; et al. Reporting of molecular test results from cell-free DNA analyses: Expert consensus recommendations from the 2023 European Liquid Biopsy Society ctDNA Workshop. eBioMedicine 2025, 114, 105636. [Google Scholar] [CrossRef] [PubMed]
- Sholl, L.M.; Oxnard, G.R.; Paweletz, C.P. Traditional Diagnostics versus Disruptive Technology: The Role of the Pathologist in the Era of Liquid Biopsy. Cancer Res. 2020, 80, 3197–3199. [Google Scholar] [CrossRef] [PubMed]
- Lin, P.P. Integrated EpCAM-independent subtraction enrichment and iFISH strategies to detect and classify disseminated and circulating tumors cells. Clin. Transl. Med. 2015, 4, 38. [Google Scholar] [CrossRef]
- Andree, K.C.; van Dalum, G.; Terstappen, L.W. Challenges in circulating tumor cell detection by the CellSearch system. Mol. Oncol. 2016, 10, 395–407. [Google Scholar] [CrossRef] [PubMed]
- Rushton, A.J.; Nteliopoulos, G.; Shaw, J.A.; Coombes, R.C. A Review of Circulating Tumour Cell Enrichment Technologies. Cancers 2021, 13, 970. [Google Scholar] [CrossRef] [PubMed]
- Kojima, T.; Hashimoto, Y.; Watanabe, Y.; Kagawa, S.; Uno, F.; Kuroda, S.; Tazawa, H.; Kyo, S.; Mizuguchi, H.; Urata, Y.; et al. A simple biological imaging system for detecting viable human circulating tumor cells. J. Clin. Investig. 2009, 119, 3172–3181. [Google Scholar] [CrossRef]
- Beinse, G.; Borghese, B.; Métairie, M.; Just, P.-A.; Poulet, G.; Garinet, S.; Parfait, B.; Didelot, A.; Bourreau, C.; Agueeff, N.; et al. Highly Specific Droplet-Digital PCR Detection of Universally Methylated Circulating Tumor DNA in Endometrial Carcinoma. Clin. Chem. 2022, 68, 782–793. [Google Scholar] [CrossRef]
- Wever, B.M.M.; van den Helder, R.; van Splunter, A.P.; van Gent, M.D.J.M.; Kasius, J.C.; Trum, J.W.; Verhoeve, H.R.; van Baal, W.M.; Hulbert, A.; Verhoef, L.; et al. DNA methylation testing for endometrial cancer detection in urine, cervicovaginal self-samples and cervical scrapes. Int. J. Cancer 2023, 153, 341–351. [Google Scholar] [CrossRef]
- Li, M.; Xia, Z.; Wang, R.; Xi, M.; Hou, M. Unveiling DNA methylation: Early diagnosis, risk assessment, and therapy for endometrial cancer. Front. Oncol. 2024, 14, 1455255. [Google Scholar] [CrossRef]
- Schreiberhuber, L.; Herzog, C.; Vavourakis, C.D.; Redl, E.; Kastner, C.; Jones, A.; Evans, I.; Zikan, M.; Cibula, D.; Widschwendter, P.; et al. The WID-qEC test: Performance in a hospital-based cohort and feasibility to detect endometrial and cervical cancers. Int. J. Cancer 2023, 152, 1269–1274. [Google Scholar] [CrossRef] [PubMed]
- Nesselbush, M.C.; Luca, B.A.; Jeon, Y.-J.; Jabara, I.; Meador, C.B.; Garofalo, A.; Binkley, M.S.; Hui, A.B.; van ‘t Erve, I.; Xu, N.; et al. An ultrasensitive method for detection of cell-free RNA. Nature 2025, 641, 759–768. [Google Scholar] [CrossRef]
- Wang, J.; Huang, J.; Hu, Y.; Guo, Q.; Zhang, S.; Tian, J.; Niu, Y.; Ji, L.; Xu, Y.; Tang, P.; et al. Terminal modifications independent cell-free RNA sequencing enables sensitive early cancer detection and classification. Nat. Commun. 2024, 15, 156. [Google Scholar] [CrossRef]
- Larson, M.H.; Pan, W.; Kim, H.J.; Mauntz, R.E.; Stuart, S.M.; Pimentel, M.; Zhou, Y.; Knudsgaard, P.; Demas, V.; Aravanis, A.M.; et al. A comprehensive characterization of the cell-free transcriptome reveals tissue- and subtype-specific biomarkers for cancer detection. Nat. Commun. 2021, 12, 2357. [Google Scholar] [CrossRef]
- Bao, P.; Wang, T.; Liu, X.; Xing, S.; Ruan, H.; Ma, H.; Tao, Y.; Zhan, Q.; Belmonte-Reche, E.; Qin, L.; et al. Peak analysis of cell-free RNA finds recurrently protected narrow regions with clinical potential. Genome Biol. 2025, 26, 119. [Google Scholar] [CrossRef]
- Ju, C.-W.; Lyu, R.; Li, H.; Wei, J.; Parra Vitela, A.J.; Dougherty, U.; Kwesi, A.; Luna, A.; Zhu, X.; Shen, S.; et al. Modifications of microbiome-derived cell-free RNA in plasma discriminates colorectal cancer samples. Nat. Biotechnol. 2025. [Google Scholar] [CrossRef] [PubMed]
- Peddu, V.; Hill, A.; Maroli, S.L.V.; Mattingly, C.; Gardner, J.M.V.; Miga, K.H.; Fitzgerald, R.C.; Kim, D.H. RNA liquid biopsy via nanopore sequencing for novel biomarker discovery and cancer early detection. bioRxiv 2025. [Google Scholar] [CrossRef]
- Ling, H.; Fabbri, M.; Calin, G.A. MicroRNAs and other non-coding RNAs as targets for anticancer drug development. Nat. Rev. Drug Discov. 2013, 12, 847–865. [Google Scholar] [CrossRef]
- Zhang, C.; Sun, C.; Zhao, Y.; Wang, Q.; Guo, J.; Ye, B.; Yu, G. Overview of MicroRNAs as Diagnostic and Prognostic Biomarkers for High-Incidence Cancers in 2021. Int. J. Mol. Sci. 2022, 23, 11389. [Google Scholar] [CrossRef]
- Xiao, J.; Sluijter, J.P.G. Extracellular vesicles in cardiovascular homeostasis and disease: Potential role in diagnosis and therapy. Nat. Rev. Cardiol. 2025. [Google Scholar] [CrossRef]
- Su, J.; Song, Y.; Zhu, Z.; Huang, X.; Fan, J.; Qiao, J.; Mao, F. Cell–cell communication: New insights and clinical implications. Signal Transduct. Target. Ther. 2024, 9, 196. [Google Scholar] [CrossRef] [PubMed]
- Yao, Y.; Shi, L.; Zhu, X. Four differentially expressed exosomal miRNAs as prognostic biomarkers and therapy targets in endometrial cancer: Bioinformatic analysis. Medicine 2023, 102, e34998. [Google Scholar] [CrossRef] [PubMed]
- Sykaras, A.G.; Christofidis, K.; Politi, E.; Theocharis, S. Exosomes Endometrial Cancer: A Biomark. Treasure Trove? Cancers 2022, 14, 1733. [Google Scholar] [CrossRef] [PubMed]
- Bogani, G.; Liu, M.C.; Dowdy, S.C.; Cliby, W.A.; Kerr, S.E.; Kalli, K.R.; Kipp, B.R.; Halling, K.C.; Campion, M.B.; Mariani, A. Detection of circulating tumor cells in high-risk endometrial cancer. Anticancer. Res. 2015, 35, 683–687. [Google Scholar]
- Shintani, D.; Hihara, T.; Ogasawara, A.; Sato, S.; Yabuno, A.; Tai, K.; Fujiwara, K.; Watanabe, K.; Hasegawa, K. Tumor-related mutations in cell-free DNA in pre-operative plasma as a prognostic indicator of recurrence in endometrial cancer. Int. J. Gynecol. Cancer 2020, 30, 1340–1346. [Google Scholar] [CrossRef]
- Moss, E.L.; Gorsia, D.N.; Collins, A.; Sandhu, P.; Foreman, N.; Gore, A.; Wood, J.; Kent, C.; Silcock, L.; Guttery, D.S. Utility of Circulating Tumor DNA for Detection and Monitoring of Endometrial Cancer Recurrence and Progression. Cancers 2020, 12, 2231. [Google Scholar] [CrossRef] [PubMed]
- Bellone, S.; McNamara, B.; Mutlu, L.; Demirkiran, C.; Hartwich, T.M.P.; Harold, J.; Yang-Hartwich, Y.; Siegel, E.R.; Santin, A.D. Monitoring Treatment Response, Early Recurrence, and Survival in Uterine Serous Carcinoma and Carcinosarcoma Patients Using Personalized Circulating Tumor DNA Biomarkers. Int. J. Mol. Sci. 2023, 24, 8873. [Google Scholar] [CrossRef]
- Casas-Arozamena, C.; Vilar, A.; Cueva, J.; Arias, E.; Sampayo, V.; Diaz, E.; Oltra, S.S.; Moiola, C.P.; Cabrera, S.; Cortegoso, A.; et al. Role of cfDNA and ctDNA to improve the risk stratification and the disease follow-up in patients with endometrial cancer: Towards the clinical application. J. Exp. Clin. Cancer Res. 2024, 43, 264. [Google Scholar] [CrossRef]
- Rao, Q.; Zhou, H.; Weng, W.; He, Y.; Tang, W.; Chen, X.; Wu, X.; Bao, H.; Lin, Z.; Zhang, B. Early detection, clinicopathological subtyping and prognosis prediction for patients with endometrial cancer using fragmentoimcs-based liquid-biopsy assay. J. Clin. Oncol. 2024, 42, e17584. [Google Scholar] [CrossRef]
- Feng, W.; Jia, N.; Jiao, H.; Chen, J.; Chen, Y.; Zhang, Y.; Zhu, M.; Zhu, C.; Shen, L.; Long, W. Circulating tumor DNA as a prognostic marker in high-risk endometrial cancer. J. Transl. Med. 2021, 19, 51. [Google Scholar] [CrossRef]
- Vizza, E.; Corrado, G.; De Angeli, M.; Carosi, M.; Mancini, E.; Baiocco, E.; Chiofalo, B.; Patrizi, L.; Zampa, A.; Piaggio, G.; et al. Serum DNA integrity index as a potential molecular biomarker in endometrial cancer. J. Exp. Clin. Cancer Res. 2018, 37, 16. [Google Scholar] [CrossRef]
- Abbink, K.; Zusterzeel, P.L.; Geurts-Moespot, A.J.; Herwaarden, A.E.V.; Pijnenborg, J.M.; Sweep, F.C.; Massuger, L.F. HE4 is superior to CA125 in the detection of recurrent disease in high-risk endometrial cancer patients. Tumour Biol. 2018, 40, 1010428318757103. [Google Scholar] [CrossRef] [PubMed]
- Cicchillitti, L.; Corrado, G.; De Angeli, M.; Mancini, E.; Baiocco, E.; Patrizi, L.; Zampa, A.; Merola, R.; Martayan, A.; Conti, L.; et al. Circulating cell-free DNA content as blood based biomarker in endometrial cancer. Oncotarget 2017, 8, 115230–115243. [Google Scholar] [CrossRef] [PubMed]
- Lemech, C.R.; Ensell, L.; Paterson, J.C.; Eminowicz, G.; Lowe, H.; Arora, R.; Arkenau, H.T.; Widschwendter, M.; MacDonald, N.; Olaitan, A.; et al. Enumeration and Molecular Characterisation of Circulating Tumour Cells in Endometrial Cancer. Oncology 2016, 91, 48–54. [Google Scholar] [CrossRef]
- Tsukamoto, O.; Miura, K.; Mishima, H.; Abe, S.; Kaneuchi, M.; Higashijima, A.; Miura, S.; Kinoshita, A.; Yoshiura, K.; Masuzaki, H. Identification of endometrioid endometrial carcinoma-associated microRNAs in tissue and plasma. Gynecol. Oncol. 2014, 132, 715–721. [Google Scholar] [CrossRef]
- Dobrzycka, B.; Terlikowski, S.J.; Mazurek, A.; Kowalczuk, O.; Niklinska, W.; Chyczewski, L.; Kulikowski, M. Circulating free DNA, p53 antibody and mutations of KRAS gene in endometrial cancer. Int. J. Cancer 2010, 127, 612–621. [Google Scholar] [CrossRef] [PubMed]
- Li, L.M.; Zhu, Y.X.; Zhong, Y.; Su, T.; Fan, X.M.; Xi, Q.; Li, M.Y.; Fu, J.; Tan, H.; Liu, S. Human epididymis protein 4 in endometrial cancer: A meta-analysis. Clin. Chim. Acta 2018, 482, 215–223. [Google Scholar] [CrossRef]
- Bignotti, E.; Ragnoli, M.; Zanotti, L.; Calza, S.; Falchetti, M.; Lonardi, S.; Bergamelli, S.; Bandiera, E.; Tassi, R.A.; Romani, C.; et al. Diagnostic and prognostic impact of serum HE4 detection in endometrial carcinoma patients. Br. J. Cancer 2011, 104, 1418–1425. [Google Scholar] [CrossRef]
- Zhang, Y.; Qu, X.; Qu, P.P. Value of circulating tumor cells positive for thyroid transcription factor-1 (TTF-1) to predict recurrence and survival rates for endometrial carcinoma. J. Buon. 2016, 21, 1491–1495. [Google Scholar]
- Ni, T.; Sun, X.; Shan, B.; Wang, J.; Liu, Y.; Gu, S.L.; Wang, Y.D. Detection of circulating tumour cells may add value in endometrial cancer management. Eur. J. Obs. Gynecol. Reprod. Biol. 2016, 207, 1–4. [Google Scholar] [CrossRef]
- Zhang, Y.; Tian, L. Advances and challenges in the use of liquid biopsy in gynaecological oncology. Heliyon 2024, 10, e39148. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Yang, J.-X.; Yu, M. Evaluation of Cell-free DNA from Papanicolaou Smears and Peripheral Blood to Detect Endometrial Cancer. Front. Oncol. 2025, 15, 1570938. [Google Scholar] [CrossRef]
- Wang, Y.; Li, L.; Douville, C.; Cohen, J.D.; Yen, T.T.; Kinde, I.; Sundfelt, K.; Kjær, S.K.; Hruban, R.H.; Shih, I.M.; et al. Evaluation of liquid from the Papanicolaou test and other liquid biopsies for the detection of endometrial and ovarian cancers. Sci. Transl. Med. 2018, 10, eaap8793. [Google Scholar] [CrossRef] [PubMed]
- Latham, A.; Mueller, J.J.; Charalambous, K.; Liu, Y.L.; Shah, R.; Long, K.; Praiss, A.; Milli, L.; Borio, M.; Nagarajan, T. Feasibility Pap-Deriv. Ctdna Detect. Sporadic Lynch-Assoc. Endometrial Cancer. 2025, 43, 16. [Google Scholar] [CrossRef]
- Kaigorodova, E.; Zavaruev, I.; Chernyshova, A.; Grishchenko, M.Y. Prognostic significance of atypical/hybrid forms of EpCAM+ CD45+ cells in the blood of patients with endometrial cancer. Tumors Female Reprod. Syst. 2023, 19, 104–108. [Google Scholar] [CrossRef]
- Cheng, J.C.; Swarup, N.; Wong, D.T.W.; Chia, D. A review on the impact of single-stranded library preparation on plasma cell-free diversity for cancer detection. Front. Oncol. 2024, 14, 1332004. [Google Scholar] [CrossRef] [PubMed]
- Medina, J.E.; Dracopoli, N.C.; Bach, P.B.; Lau, A.; Scharpf, R.B.; Meijer, G.A.; Andersen, C.L.; Velculescu, V.E. Cell-free DNA approaches for cancer early detection and interception. J. Immunother. Cancer 2023, 11, e006013. [Google Scholar] [CrossRef]
- Bartolomucci, A.; Nobrega, M.; Ferrier, T.; Dickinson, K.; Kaorey, N.; Nadeau, A.; Castillo, A.; Burnier, J.V. Circulating tumor DNA to monitor treatment response in solid tumors and advance precision oncology. NPJ Precis. Oncol. 2025, 9, 84. [Google Scholar] [CrossRef]
- Hoermann, G. Clinical Significance of Clonal Hematopoiesis of Indeterminate Potential in Hematology and Cardiovascular Disease. Diagnostics 2022, 12, 1613. [Google Scholar] [CrossRef] [PubMed]
- Abbosh, C.; Swanton, C.; Birkbak, N.J. Clonal haematopoiesis: A source of biological noise in cell-free DNA analyses. Ann. Oncol. 2019, 30, 358–359. [Google Scholar] [CrossRef]
- Razavi, P.; Li, B.T.; Brown, D.N.; Jung, B.; Hubbell, E.; Shen, R.; Abida, W.; Juluru, K.; De Bruijn, I.; Hou, C.; et al. High-intensity sequencing reveals the sources of plasma circulating cell-free DNA variants. Nat. Med. 2019, 25, 1928–1937. [Google Scholar] [CrossRef]
- Berger, S.; Pachkov, M.; Arnold, P.; Omidi, S.; Kelley, N.; Salatino, S.; van Nimwegen, E. Crunch: Integrated processing and modeling of ChIP-seq data in terms of regulatory motifs. Genome Res. 2019, 29, 1164–1177. [Google Scholar] [CrossRef]
- Kwinten, K.J.J.; Lemain, V.A.; de Hullu, J.A.; Leenders, W.P.J.; Steenbeek, M.P.; van Altena, A.M.; Pijnenborg, J.M.A. Cervicovaginal specimen biomarkers for early detection of ovarian and endometrial cancer: A review. Cancer Med. 2024, 13, e70000. [Google Scholar] [CrossRef] [PubMed]
- O’Flynn, H.; Ryan, N.A.J.; Narine, N.; Shelton, D.; Rana, D.; Crosbie, E.J. Diagnostic accuracy of cytology for the detection of endometrial cancer in urine and vaginal samples. Nat. Commun. 2021, 12, 952. [Google Scholar] [CrossRef] [PubMed]
- Capasso, I.; Nero, C.; Anderson, G.; Del Re, M.; Perrone, E.; Fanfani, F.; Scambia, G.; Cucinella, G.; Mariani, A.; Choong, G.; et al. Circulating tumor DNA in endometrial cancer: Clinical significance and implications. Int. J. Gynecol. Cancer 2025, 35, 101656. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.S.; Cho, E.H.; Kim, B.; Hong, J.; Kim, Y.G.; Kim, Y.; Jang, J.H.; Lee, S.T.; Kong, S.Y.; Lee, W.; et al. Clinical Practice Guideline for Blood-based Circulating Tumor DNA Assays. Ann. Lab. Med. 2024, 44, 195–209. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Garcia, E.; Lesur, A.; Devis, L.; Cabrera, S.; Matias-Guiu, X.; Hirschfeld, M.; Asberger, J.; van Oostrum, J.; Casares de Cal, M.; Gómez-Tato, A.; et al. Targeted Proteomics Identifies Proteomic Signatures in Liquid Biopsies of the Endometrium to Diagnose Endometrial Cancer and Assist in the Prediction of the Optimal Surgical Treatment. Clin. Cancer Res. 2017, 23, 6458–6467. [Google Scholar] [CrossRef] [PubMed]
| Biomarker | Clinical Significance | Type of Sample | Cohort | Technology | References |
|---|---|---|---|---|---|
| ctDNA (PTEN, PIK3R1, KMT2C, etc.) | ctDNA mutations detected in 93% of EC or AEH patients; mutations correlated with higher grade and myometrial invasion; 65% concordance with tissue biopsy | Plasma | n = 63 | NGS-based ctDNA panel | Esposito et al. (2025) [50] |
| cfDNA/ctDNA monitoring | High levels of cfDNA and detectable ctDNA predict poor DFS and DSS; longitudinal monitoring identifies early recurrence | Plasma, urine aspirates | n = 198 | Targeted NGS (Oncomine™), ddPCR | Casas-Arozamena et al. (2024) [88] |
| Fragmentomics-based cfDNA | Early detection, clinicopathological subtyping (stage, grade, histology, MSI), and recurrence prediction in EC | Plasma | Training: 120 EC + 120 healthy; Testing: 62 EC + 62 healthy | Low-pass whole genome sequencing + machine learning (ensemble model) | Rao et al. (2024) [89] |
| ctDNA mutations (including DNMT3A, TP53, FGFR2) | ctDNA mutations were detected in 100% of patients; DNMT3A mutations were most frequent. Demonstrated feasibility of liquid biopsy for EC molecular profiling | Plasma | n = 21 | Targeted NGS | Kodada et al. (2023) [10] |
| CTCs | Detectable in ovarian vein during early-stage EC surgery; potential for recurrence risk stratification | Peripheral and ovarian vein blood | n = 10 | CellSearch® | Francini et al. (2023) [51] |
| Exosomal miRNAs (hsa-miR-17-3p, hsa-miR-99b-3p, hsa-miR-193a-5p, hsa-miR-320d) | Prognostic exosomal miRNAs predictive of poor survival in EC; identified as potential therapy targets | Plasma (Exosomal miRNA data from TCGA) | n = 566 | miRNA-seq + bioinformatic analysis (TCGA/NCBI + KM survival modeling) | Yao et al. (2023) [82] |
| DNA methylation (e.g., GYPC, ZSCAN12) | High sensitivity and specificity in detecting EC via self-sampling | Urine, self-sample, cervical scrape | n = 103 EC and 317 controls | qMSP | Wever et al. (2023) [69] |
| Methylated DNA (WID-qEC: ZSCAN12, GYPC) | AUC 0.99, sensitivity 100%, specificity 82.5%; validated in hospital cohort | Cervicovaginal self-sample | n = 330 | qPCR | Schreiberhuber et al. (2023) [71] |
| ctDNA (personalized SNV-based) | Monitoring treatment response, early recurrence, and survival in USC and carcinosarcoma | Plasma | n = 16 (14 USCs and 2 CSs) | ddPCR based on tumor-informed SSVs via NGS (Foundation Medicine) | Bellone et al. (2023) [87] |
| Universally methylated ctDNA (ZSCAN12, OXT) | Highly specific and sensitive detection of EC; potential for diagnosis and monitoring | Plasma | Retrospective: 78 EC tumors + 30 adjacent; Prospective pilot: 33 EC (stage I–IV); Controls: 55 non-cancer individuals | Methylation-specific droplet digital PCR (meth-ddPCR) | Beinse et al. (2022) [68] |
| ctDNA as prognostic marker | Detection of ctDNA postoperatively significantly associated with progression and decreased OS | Plasma | n = 9 | ddPCR | Feng et al. (2021) [90] |
| ctDNA (PIK3CA and KRAS mutations) | Presence of ctDNA in plasma correlated with advanced FIGO stage, non-endometrioid histology, LVSI, and poorer recurrence-free and overall survival | Plasma | n = 199 (68 had tumor mutations; 10 had matched ctDNA) | ddPCR (PIK3CA, KRAS) | Shintani et al. (2020) [85] |
| Uterine aspirate + cfDNA NGS | ctDNA present in 41.2% overall; enriched in high-risk subtypes | Uterine aspirates, plasma | n = 60 | NGS panel | Casas-Arozamena et al. (2020) [55] |
| ctDNA mutation burden | Rising ctDNA precedes radiographic or clinical recurrence by months; captures emerging MSI; dynamic real-time monitoring | Serial plasma | n = 13 | ddPCR + targeted NGS | Moss et al. (2020) [86] |
| cfDNA content & integrity index | Increased total cfDNA and Alu integrity ratio correlate with higher grade and LVSI, independent of hypertension or obesity | Serum | n = 60 | qPCR–Alu quantification | Vizza et al. (2018) [91] |
| Serum HE4 and CA125 | HE4 is more sensitive than CA-125 for detecting recurrence; elevated levels correlate with advanced stage, myometrial invasion, nodal metastases, and shorter survival | Serum | n = 174 | Enzyme immunoassay | Abbink et al. (2018) [92] |
| Total cfDNA and cfmtDNA | Elevated total cfDNA in higher-grade tumors | Serum | n = 59 (12 G1, 30 G2, 17 G3) | RT-qPCR | Cicchillitti et al. (2017) [93] |
| CTC enumeration (CellSearch®) | CTC count correlated with stathmin expression and advanced disease | Whole blood | n = 30 | CellSearch® + immunofluorescence | Lemech et al. (2016) [94] |
| Personalized ctDNA panels | Personalized ctDNA detected residual disease and recurrence around 6 months ahead of CA-125 and imaging; associated with shorter PFS/OS | Tissue, serum | n = 44 (17 EC cases) | Tumor-specific ddPCR | Pereira et al. (2015) [11] |
| miR-135b, miR-205, miR-30a-3p | miR-135b and miR-205 elevated in tissue/plasma; levels drop post-hysterectomy | Tissue/plasma | n = 24 | NGS + qRT-PCR | Tsukamoto et al. (2014) [95] |
| cfDNA, p53 autoantibody, KRAS mut | cfDNA/KRAS detected in 19% of Type II and 11.9% of higher-grade EC; autoantibodies in 20% | Plasma | n = 109 (87 Type I, 22 Type II) | PCR-RFLP | Dobrzycka et al. (2010) [96] |
| Identifier | Title | Aim(s) and Intervention(s) | Study Type | Start Date (Actual) | Study Completion (Estimated) | Enrollment (Estimated) | Status |
|---|---|---|---|---|---|---|---|
| NCT05504161 | Detection of Tumor DNA Through Cervical Smear and Liquid Biopsy in EC Patients and Evaluation of Prognostic and Predictive Values of Tumor DNA Assay | To compare the ctDNA mutation detection rate based on cervical swab and whole blood at the time of surgery | Observational | 30 December 2020 | December 2023 | 300 | Unknown |
| NCT06846775 | The Clinical Utility of DNA Methylation Testing in Patient-collected Urine and Vaginal Samples to Detect EC: a Case-control Study | Diagnostic Test: DNA-methylation testing of methylation markers CDO1, GHSR and ZIC1 for patient-collected vaginal samples and GHSR, CDH13 and SST for patient-collected urine samples | Observational [Patient Registry] | 15 April 2025 | 30 November 2027 | 120 | Recruiting |
| NCT04456972 | Reliability and Interest of Circulating Tumor DNA in ECs | To determine the concordance rate between molecular analysis of tumor tissue and that of ctDNA in patients with EC during treatment | Interventional | 19 June 2020 | 8 January 2022 | 44 | Completed |
| NCT06341855 | Exploring the Potential of ctDNA-MRD for Recurrence Surveillance and Prognostic Evaluation in High-risk EC | To explore the feasibility of ctDNA-MRD in monitoring recurrence and evaluating prognosis of high-risk endometrial carcinoma | Interventional | 25 January 2024 | 30 January 2026 | 100 | Recruiting |
| NCT05099978 | Asian Multicenter Prospective Study of ctDNA Sequencing (A-TRAIN) | NGS analysis will be performed on cfDNA extracted from peripheral blood samples of target patients to determine the types and incidences of genetic abnormalities | Observational | 1 November 2021 | 31 December 2024 | 506 | Active, not recruiting |
| NCT03744962 | MSI in Circulatory DNA of EC | to analyze the MSI in the circulatory tumor DNA and in the tumor tissue in the patients diagnosed with uterine EC | Observational | 10 November 2018 | 23 December 2020 | 100 | Unknown |
| NCT05366881 | cfDNA Assay Prospective Observational Validation for Early Cancer Detection and Minimal Residual Disease (CAMPERR) | To train and validate a genome-wide methylome enrichment platform to detect multiple cancer types and to differentiate amongst cancer types, including EC | Observational | 3 May 2022 | December 2026 | 7000 | Recruiting |
| NCT04651738 | Cell-free DNA Methylation for EC | To perform methylation testing of host DNA, namely, BHLHE22, CELF4, HAND2, and ZNF177, in the peripheral serum to discover the diagnostic and supervision roles of DNA methylation in EC | Interventional | 18 December 2020 | 1 January 2023 | 400 | Unknown |
| NCT05955079 | Circulating Tumor DNA Study in Patients With EC (ctDNA-endo) | To identify a population at risk of early recurrence after oncologic resection surgery of a primary uterine tumor based on the detection of ctDNA | Observational | 1 January 2021 | January 2026 | 130 | Recruiting |
| NCT06083779 | Early Detection of EC Using Plasma Cell-free DNA Fragmentomics | To enable non-invasive early detection of EC in high-risk populations through the establishment of a multimodal machine learning model using plasma cell-free DNA fragmentomics | Observational | 1 August 2023 | 30 April 2024 | 216 | Recruiting |
| NCT06028724 | A Study on the Prevalence of Clinically Useful Mutations in Solid Tumor Characterized by NGS Methods on Liquid Biopsy Analysis (POPCORN) (POPCORN) | To evaluate the real-world prevalence of clinically useful mutations in patients who are receiving therapy for advanced and locally advanced solid tumor through liquid biopsy, including EC | Observational | 26 May 2023 | 31 May 2030 | 782 | Recruiting |
| NCT05059444 | ORACLE: Observation of ResiduAl Cancer With Liquid Biopsy Evaluation (ORACLE) | To demonstrate the ability of a novel ctDNA assay developed by Guardant Health to detect recurrence in individuals treated for early-stage solid tumors, including EC | Prospective Cohort | 7 September 2021 | February 2028 | 1050 | Recruiting |
| NCT05051722 | Leveraging Methylated DNA Markers (MDMs) in the Detection of EC, Ovarian Cancer, and Cervical Cancer (ECHO) | To develop a pan-gynecologic cancer detection test using gynecologic (unique endometrial, cervical, and ovarian cancer) cancer-specific methylated DNA markers and high-risk human papilloma virus (HR-HPV) detected in vaginal fluid and/or plasma | Observational | 3 August 2021 | 30 December 2026 | 3110 | Recruiting |
| NCT05049538 | Determine the Utility of Liquid Biopsies and Tumor Molecular Profiling in Predicting Recurrence in ECs | To find out how well liquid biopsies work as a non-invasive alternative to other methods of finding cancer cells (such as a tissue biopsy) in patients with EC by comparing TP53, FBXW7 and other mutated genes in ctDNA samples obtained at several timepoints of disease progression/treatment | Observational | 18 June 2019 | 30 June 2028 | 1000 | Recruiting |
| NCT04817501 | Phenotypic Spectrum of CTCs in Tumors of the Female Reproductive System | To evaluate the level and molecular profiles of different CTC populations as markers for predicting the risk of developing hematogenous metastases and the effectiveness of treatment in patients with tumors of the female reproductive system including EC | Observational | 14 February 2014 | 1 December 2022 | 150 | Completed [105] |
| NCT03776630 | Exploring the Potential of Novel Biomarkers Based on Plasma miRNAs for a Better Management of Pelvic Gynecologic Tumors (GYNO-MIR) | To validate the 5-miR index assessed in plasma samples as a diagnostic marker to assess the risk of lymph node metastases | Interventional | 23 May 2019 | May 2027 | 363 | Active, not recruiting |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Perez, M.; Carvajal, L.L.; Wong, A.; Poppiti, R.; Ruiz-Cordero, R.; Castellano-Sánchez, A.A.; Bahmad, H.F. Beyond the Microscope: Integrating Liquid Biopsies into the Molecular Pathology Era of Endometrial Cancer. Int. J. Mol. Sci. 2025, 26, 7987. https://doi.org/10.3390/ijms26167987
Perez M, Carvajal LL, Wong A, Poppiti R, Ruiz-Cordero R, Castellano-Sánchez AA, Bahmad HF. Beyond the Microscope: Integrating Liquid Biopsies into the Molecular Pathology Era of Endometrial Cancer. International Journal of Molecular Sciences. 2025; 26(16):7987. https://doi.org/10.3390/ijms26167987
Chicago/Turabian StylePerez, Miguel, Luis Lorenzo Carvajal, Andres Wong, Robert Poppiti, Roberto Ruiz-Cordero, Amilcar A. Castellano-Sánchez, and Hisham F. Bahmad. 2025. "Beyond the Microscope: Integrating Liquid Biopsies into the Molecular Pathology Era of Endometrial Cancer" International Journal of Molecular Sciences 26, no. 16: 7987. https://doi.org/10.3390/ijms26167987
APA StylePerez, M., Carvajal, L. L., Wong, A., Poppiti, R., Ruiz-Cordero, R., Castellano-Sánchez, A. A., & Bahmad, H. F. (2025). Beyond the Microscope: Integrating Liquid Biopsies into the Molecular Pathology Era of Endometrial Cancer. International Journal of Molecular Sciences, 26(16), 7987. https://doi.org/10.3390/ijms26167987






