AI-Based Facial Phenotyping Supports a Shared Molecular Axis in PACS1-, PACS2-, and WDR37-Related Syndromes
Abstract
1. Introduction
2. Results
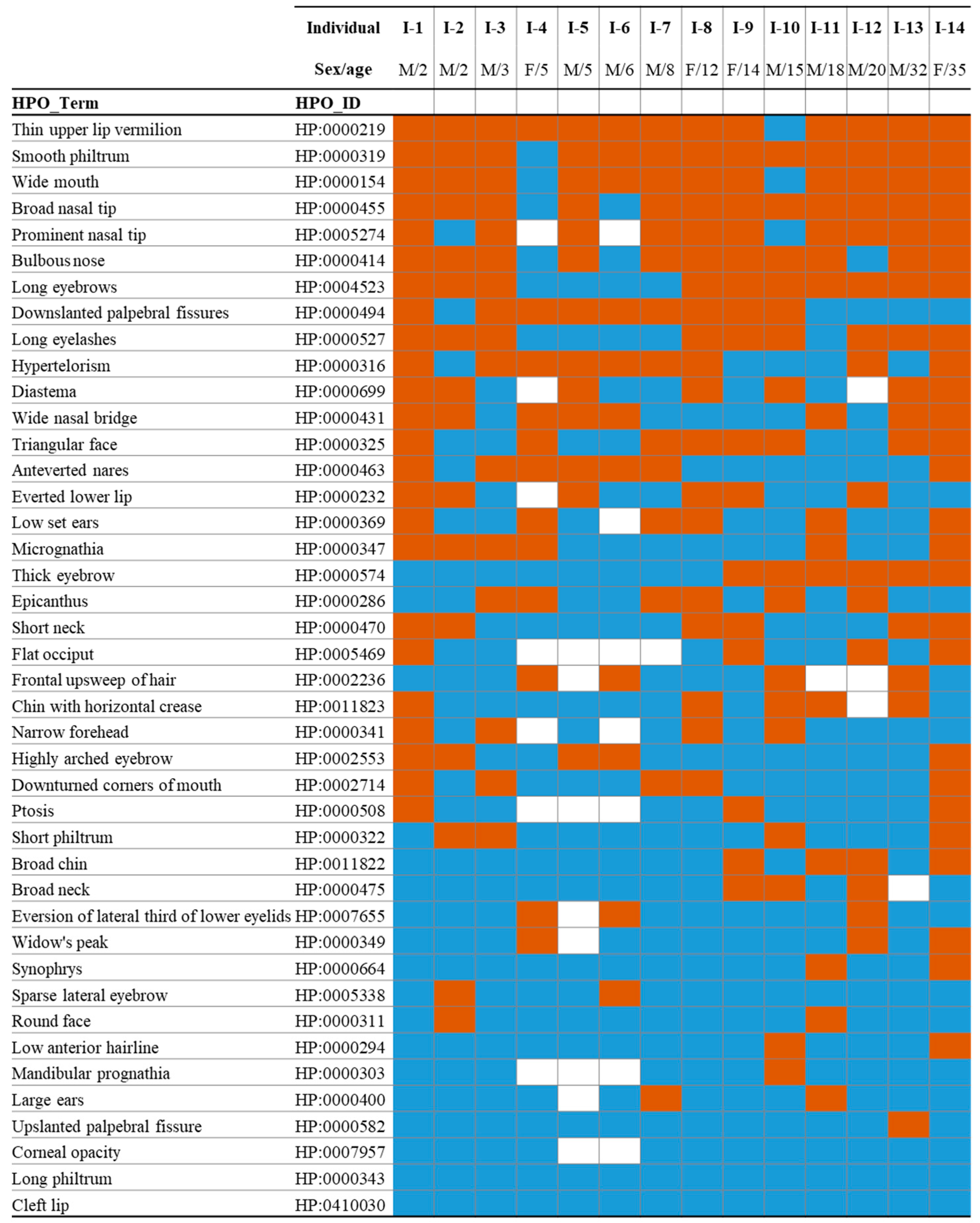
2.1. Dysmorphic Facial Phenotype in Individuals with Schuurs–Hoeijmakers Syndrome
2.2. Common Differential Syndromes of SHMS Based on Gestalt Analysis
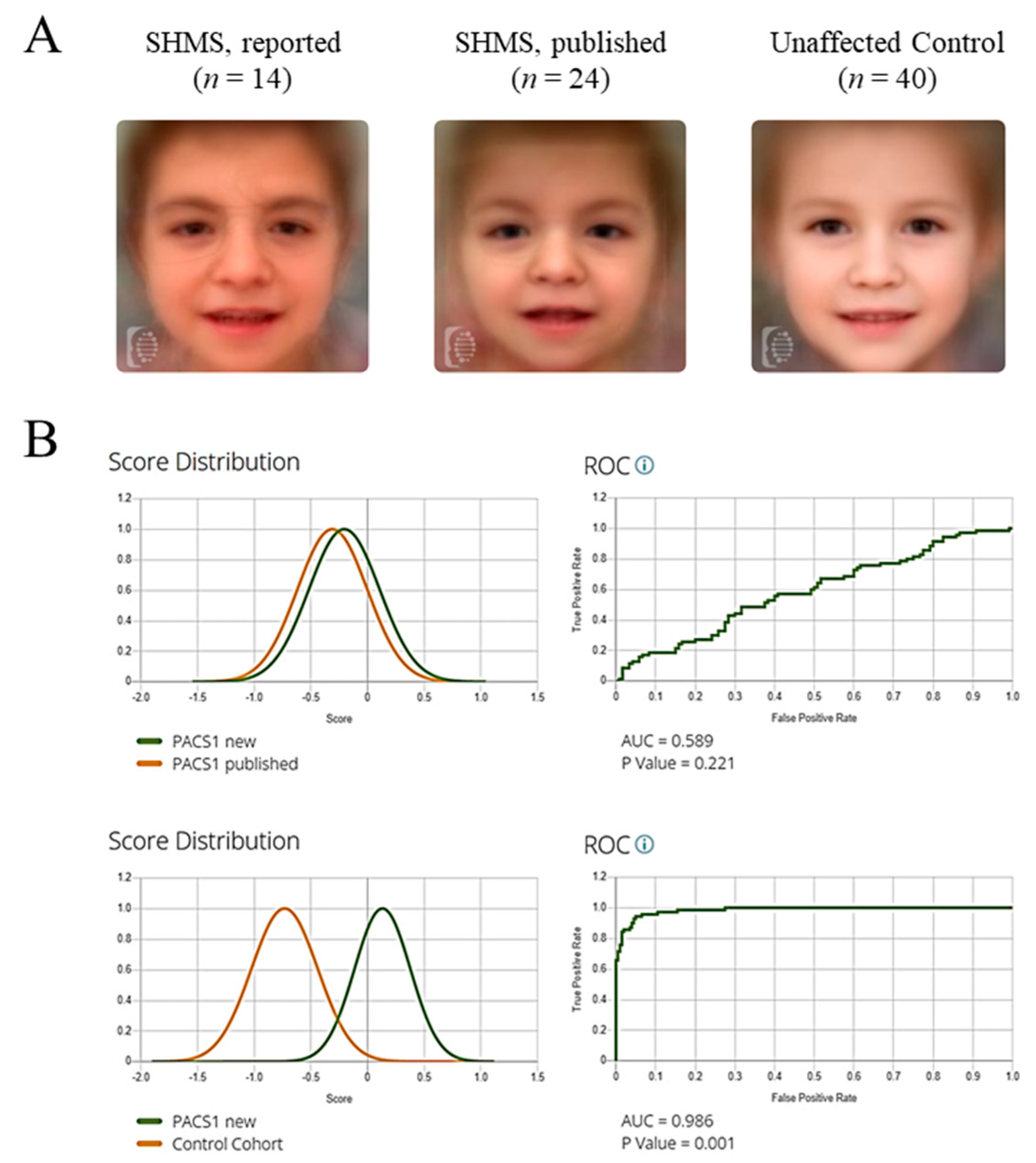
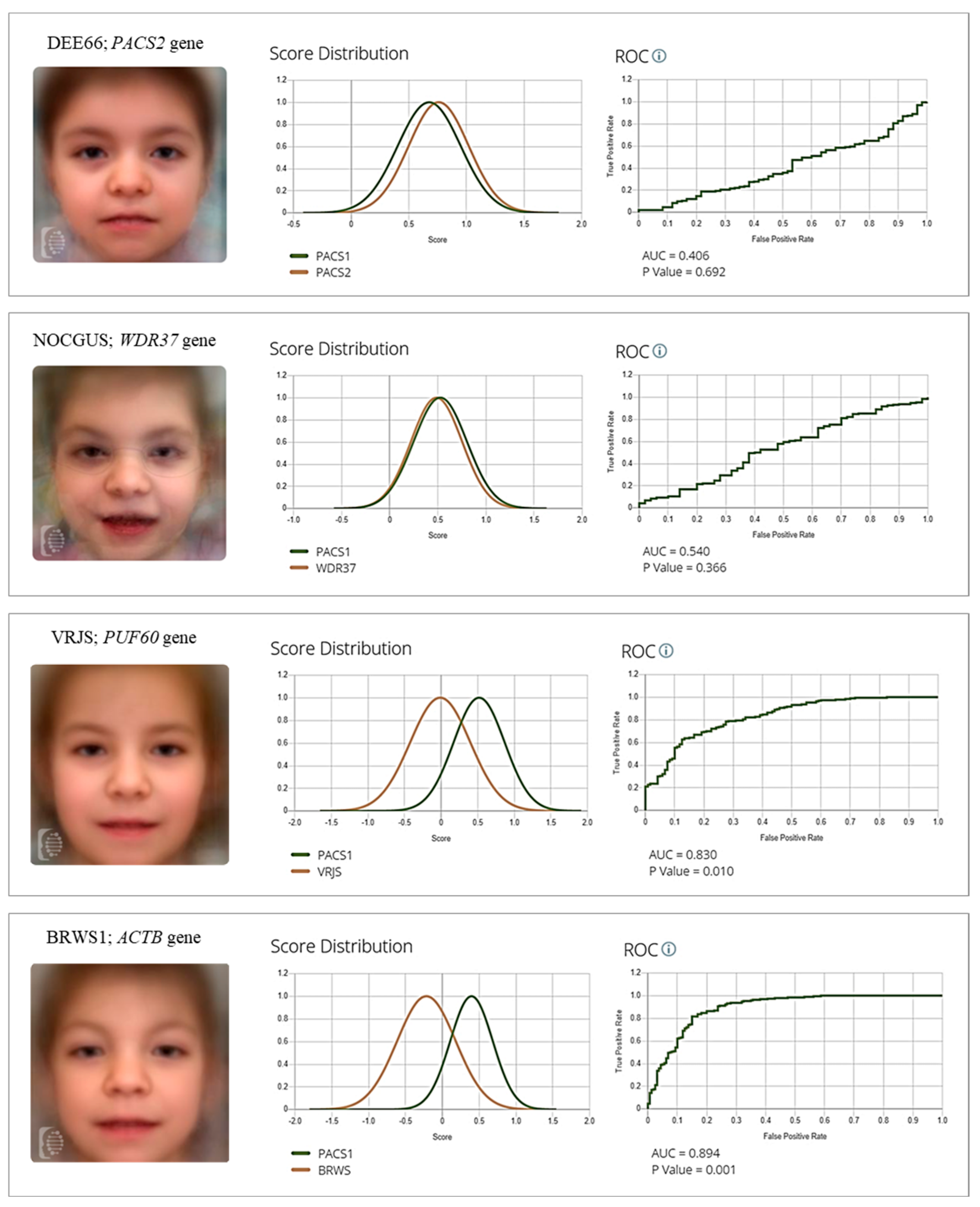
2.3. Quantification of Facial Gestalt Similarities Between Syndromes
3. Discussion
4. Materials and Methods
4.1. Patient Recruitment and Clinical Imaging
4.2. Clinical Facial Deep Phenotyping
4.3. Computational Facial Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AI | Artificial intelligence |
| BRWS1 | Baraitser–Winter syndrome 1 |
| CdLS | Cornelia de Lange syndrome |
| CEICA | Ethics and clinical research committee of the Government of Aragon |
| CNS | Central nervous system |
| DEE66 | Developmental and Epileptic Encephalopathy-66 |
| F2G | Face2Gene |
| HDAC | Histone deacetylase |
| HPO | Human phenotype ontology |
| NDD | Neurodevelopmental disorder |
| NOCGUS | Neuro-Oculo-Cardio-Genitourinary syndrome |
| OMIM | Online Mendelian Inheritance in Man |
| RGD | Rare genetic disorder |
| SHMS | Schuurs–Hoeijmakers syndrome |
| PPI | Protein–protein interaction |
| VRJS | Verheij syndrome |
References
- Wakap, S.N.; Lambert, D.M.; Olry, A.; Rodwell, C.; Gueydan, C.; Lanneau, V.; Murphy, D.; Le Cam, Y.; Rath, A. Estimating cumulative point prevalence of rare diseases: Analysis of the Orphanet database. Eur. J. Hum. Genet. 2020, 28, 165–173. [Google Scholar] [CrossRef]
- Online Mendelian Inheritance in Man, OMIM®. McKusick-Nathans Institute of Genetic Medicine, Johns Hopkins University (Baltimore, MD). Available online: https://omim.org (accessed on 22 July 2025).
- Monaco, L.; Zanello, G.; Baynam, G.; Jonker, A.H.; Julkowska, D.; Hartman, A.L.; O’cOnnor, D.; Wang, C.M.; Wong-Rieger, D.; Pearce, D.A. Research on rare diseases: Ten years of progress and challenges at IRDiRC. Nat. Rev. Drug. Discov. 2022, 21, 319–320. [Google Scholar] [CrossRef] [PubMed]
- Boycott, K.M.; Rath, A.; Chong, J.X.; Hartley, T.; Alkuraya, F.S.; Baynam, G.; Brookes, A.J.; Brudno, M.; Carracedo, A.; Dunnen, J.T.D.; et al. International Cooperation to Enable the Diagnosis of All Rare Genetic Diseases. Am. J. Hum. Genet. 2017, 100, 695–705. [Google Scholar] [CrossRef] [PubMed]
- Bamshad, M.J.; Nickerson, D.A.; Chong, J.X. Mendelian Gene Discovery: Fast and Furious with No End in Sight. Am. J. Hum. Genet. 2019, 105, 448–455. [Google Scholar] [CrossRef] [PubMed]
- McRae, J.F.; Clayton, S.; Fitzgerald, T.W.; Kaplanis, J.; Prigmore, E.; Rajan, D.; Sifrim, A.; Aitken, S.; Akawi, N.; Alvi, M.; et al. Prevalence and architecture of de novo mutations in developmental disorders. Nature 2017, 542, 433–438. [Google Scholar] [CrossRef]
- Sanders, S.J.; Sahin, M.; Hostyk, J.; Thurm, A.; Jacquemont, S.; Avillach, P.; Douard, E.; Martin, C.L.; Modi, M.E.; Moreno-De-Luca, A.; et al. A framework for the investigation of rare genetic disorders in neuropsychiatry. Nat. Med. 2019, 25, 1477–1487. [Google Scholar] [CrossRef]
- Lee, C.E.; Singleton, K.S.; Wallin, M.; Faundez, V. Rare Genetic Diseases: Nature’s Experiments on Human Development. iScience 2020, 23, 101123. [Google Scholar] [CrossRef]
- Barabási, A.L.; Gulbahce, N.; Loscalzo, J. Network medicine: A network-based approach to human disease. Nat. Rev. Genet. 2011, 12, 56–68. [Google Scholar] [CrossRef]
- Parenti, I.; Rabaneda, L.G.; Schoen, H.; Novarino, G. Neurodevelopmental Disorders: From Genetics to Functional Pathways. Trends Neurosci. 2020, 43, 608–621. [Google Scholar] [CrossRef]
- Buphamalai, P.; Kokotovic, T.; Nagy, V.; Menche, J. Network analysis reveals rare disease signatures across multiple levels of biological organization. Nat. Commun. 2021, 12, 6306. [Google Scholar] [CrossRef]
- Porras, A.R.; Rosenbaum, K.; Tor-Diez, C.; Summar, M.; Linguraru, M.G. Development and evaluation of a machine learning-based point-of-care screening tool for genetic syndromes in children: A multinational retrospective study. Lancet Digit. Health 2021, 3, e635–e643. [Google Scholar] [CrossRef]
- Hennocq, Q.; Willems, M.; Amiel, J.; Arpin, S.; Attie-Bitach, T.; Bongibault, T.; Bouygues, T.; Cormier-Daire, V.; Corre, P.; Dieterich, K.; et al. Next generation phenotyping for diagnosis and phenotype-genotype correlations in Kabuki syndrome. Sci. Rep. 2024, 14, 2330. [Google Scholar] [CrossRef]
- Ciancia, S.; Goedegebuure, W.J.; Grootjen, L.N.; Hokken-Koelega, A.C.S.; Kerkhof, G.F.; van der Kaay, D.C.M. Computer-aided facial analysis as a tool to identify patients with Silver-Russell syndrome and Prader-Willi syndrome. Eur. J. Pediatr. 2023, 182, 2607–2614. [Google Scholar] [CrossRef] [PubMed]
- Gurovich, Y.; Hanani, Y.; Bar, O.; Nadav, G.; Fleischer, N.; Gelbman, D.; Basel-Salmon, L.; Krawitz, P.M.; Kamphausen, S.B.; Zenker, M.; et al. Identifying facial phenotypes of genetic disorders using deep learning. Nat. Med. 2019, 25, 60–64. [Google Scholar] [CrossRef] [PubMed]
- Pantel, J.T.; Hajjir, N.; Danyel, M.; Elsner, J.; Abad-Perez, A.T.; Hansen, P.; Mundlos, S.; Spielmann, M.; Horn, D.; Ott, C.-E.; et al. Efficiency of Computer-Aided Facial Phenotyping (DeepGestalt) in Individuals with and Without a Genetic Syndrome: Diagnostic Accuracy Study. J. Med. Int. Res. 2020, 22, e19263. [Google Scholar] [CrossRef] [PubMed]
- Latorre-Pellicer, A.; Ascaso, Á.; Trujillano, L.; Gil-Salvador, M.; Arnedo, M.; Lucia-Campos, C.; Antoñanzas-Pérez, R.; Marcos-Alcalde, I.; Parenti, I.; Bueno-Lozano, G.; et al. Evaluating face2gene as a tool to identify cornelia de lange syndrome by facial phenotypes. Int. J. Mol. Sci. 2020, 21, 1042. [Google Scholar] [CrossRef]
- Hsieh, T.C.; Bar-Haim, A.; Moosa, S.; Ehmke, N.; Gripp, K.W.; Pantel, J.T.; Danyel, M.; Mensah, M.A.; Horn, D.; Rosnev, S.; et al. GestaltMatcher facilitates rare disease matching using facial phenotype descriptors. Nat. Genet. 2022, 54, 349–357. [Google Scholar] [CrossRef]
- Schuurs-Hoeijmakers, J.H.M.; Oh, E.C.; Vissers, L.E.L.M.; Swinkels, M.E.M.; Gilissen, C.; Willemsen, M.A.; Holvoet, M.; Steehouwer, M.; Veltman, J.A.; de Vries, B.B.; et al. Recurrent de novo mutations in PACS1 cause defective cranial-neural-crest migration and define a recognizable intellectual-disability syndrome. Am. J. Hum. Genet. 2012, 91, 1122–1127. [Google Scholar] [CrossRef]
- Crump, C.M.; Xiang, Y.; Thomas, L.; Gu, F.; Austin, C.; Tooze, S.A.; Thomas, G. PACS-1 binding to adaptors is required for acidic cluster motif-mediated protein traffic. EMBO J. 2001, 20, 2191–2201. [Google Scholar] [CrossRef]
- Mani, C.; Tripathi, K.; Luan, S.; Clark, D.W.; Andrews, J.F.; Vindigni, A.; Thomas, G.; Palle, K. The multifunctional protein PACS-1 is required for HDAC2- and HDAC3-dependent chromatin maturation and genomic stability. Oncogene 2020, 39, 2583–2596. [Google Scholar] [CrossRef]
- Villar-Pazos, S.; Thomas, L.; Yang, Y.; Chen, K.; Lyles, J.B.; Deitch, B.J.; Ochaba, J.; Ling, K.; Powers, B.; Gingras, S.; et al. Neural deficits in a mouse model of PACS1 syndrome are corrected with PACS1- or HDAC6-targeting therapy. Nat. Commun. 2023, 14, 6547. [Google Scholar] [CrossRef]
- Arnedo, M.; Ascaso, Á.; Latorre-Pellicer, A.; Lucia-Campos, C.; Gil-Salvador, M.; Ayerza-Casas, A.; Pablo, M.J.; Gómez-Puertas, P.; Ramos, F.J.; Bueno-Lozano, G.; et al. Molecular Basis of the Schuurs-Hoeijmakers Syndrome: What We Know about the Gene and the PACS-1 Protein and Novel Therapeutic Approaches. Int. J. Mol. Sci. 2022, 23, 9649. [Google Scholar] [CrossRef]
- Sakaguchi, Y.; Yoshihashi, H.; Uehara, T.; Miyama, S.; Kosaki, K.; Takenouchi, T. Coloboma may be a shared feature in a spectrum of disorders caused by mutations in the WDR37-PACS1-PACS2 axis. Am. J. Med. Genet. A 2021, 185, 884–888. [Google Scholar] [CrossRef]
- Sorokina, E.A.; Reis, L.M.; Thompson, S.; Agre, K.; Babovic-Vuksanovic, D.; Ellingson, M.S.; Hasadsri, L.; van Bever, Y.; Semina, E.V. WDR37 syndrome: Identification of a distinct new cluster of disease-associated variants and functional analyses of mutant proteins. Hum. Genet. 2021, 140, 1775–1789. [Google Scholar] [CrossRef] [PubMed]
- Byrd, D.T.; Han, Z.C.; Piggott, C.A.; Jin, Y. PACS-1 variant protein is aberrantly localized in Caenorhabditis elegans model of PACS1/PACS2 syndromes. Genetics 2024, 228, iyae118. [Google Scholar] [CrossRef] [PubMed]
- Schuurs-Hoeijmakers, J.H.M.; Landsverk, M.L.; Foulds, N.; Kukolich, M.K.; Gavrilova, R.H.; Greville-Heygate, S.; Hanson-Kahn, A.; Chitayat, D.; Glass, J.; Bernstein, J.A.; et al. Clinical delineation of the PACS1-related syndrome--Report on 19 patients. Am. J. Med. Genet. A 2016, 170, 670–675. [Google Scholar] [CrossRef]
- Gadzicki, D.; Döcker, D.; Schubach, M.; Menzel, M.; Schmorl, B.; Stellmer, F.; Biskup, S.; Bartholdi, D. Expanding the phenotype of a recurrent de novo variant in PACS1 causing intellectual disability. Clin. Genet. 2015, 88, 300–302. [Google Scholar] [CrossRef] [PubMed]
- Tenorio-Castaño, J.; Morte, B.; Nevado, J.; Martinez-Glez, V.; Santos-Simarro, F.; García-Miñaúr, S.; Palomares-Bralo, M.; Pacio-Míguez, M.; Gómez, B.; Arias, P.; et al. Schuurs-Hoeijmakers Syndrome (PACS1 Neurodevelopmental Disorder): Seven Novel Patients and a Review. Genes 2021, 12, 738. [Google Scholar] [CrossRef]
- Hoshino, Y.; Enokizono, T.; Imagawa, K.; Tanaka, R.; Suzuki, H.; Fukushima, H.; Arai, J.; Sumazaki, R.; Uehara, T.; Takenouchi, T.; et al. Schuurs-Hoeijmakers syndrome in two patients from Japan. Am. J. Med. Genet. A 2019, 179, 341–343. [Google Scholar] [CrossRef]
- Kurt Colak, F.; Eyerci, N.; Aytekin, C.; Eksioglu, A.S. Renpenning Syndrome in a Turkish Patient: de novo Variant c.607C>T in PACS1 and Hypogammaglobulinemia Phenotype. Mol. Syndromol. 2020, 11, 157–161. [Google Scholar] [CrossRef]
- Abdulqader, S.A.; Wli, W.A.; Qaryaqos, S.H. Schuurs-Hoeijmakers syndrome in a patient from Iraq-Kirkuk. Clin. Case Rep. 2021, 9, e04897. [Google Scholar] [CrossRef]
- Olson, H.E.; Jean-Marçais, N.; Yang, E.; Heron, D.; Tatton-Brown, K.; van der Zwaag, P.A.; Bijlsma, E.K.; Kamsteeg, E.J.; Backer, E.; Krock, B.L.; et al. A Recurrent De Novo PACS2 Heterozygous Missense Variant Causes Neonatal-Onset Developmental Epileptic Encephalopathy, Facial Dysmorphism, and Cerebellar Dysgenesis. Am. J. Hum. Genet. 2018, 102, 995–1007. [Google Scholar] [CrossRef]
- Cesaroni, E.; Matricardi, S.; Cappanera, S.; Marini, C. First reported case of an inherited PACS2 pathogenic variant with variable expression. Epileptic Disord. 2022, 24, 572–576. [Google Scholar] [CrossRef]
- Terrone, G.; Marchese, F.; Vari, M.S.; Severino, M.; Madia, F.; Amadori, E.; Del Giudice, E.; Romano, A.; Gennaro, E.; Zara, F.; et al. A further contribution to the delineation of epileptic phenotype in PACS2-related syndrome. Seizure 2020, 79, 53–55. [Google Scholar] [CrossRef]
- Sánchez-Soler, M.J.; Serrano-Antón, A.T.; López-González, V.; Guillén-Navarro, E. New case with the recurrent c.635G>A pathogenic variant in the PACS2 gene: Expanding the phenotype. Neurologia 2021, 36, 716–719. [Google Scholar] [CrossRef] [PubMed]
- Dentici, M.L.; Barresi, S.; Niceta, M.; Ciolfi, A.; Trivisano, M.; Bartuli, A.; Digilio, M.C.; Specchio, N.; Dallapiccola, B.; Tartaglia, M. Expanding the clinical spectrum associated with PACS2 mutations. Clin. Genet. 2019, 95, 525–531. [Google Scholar] [CrossRef] [PubMed]
- El Chehadeh, S.; Kerstjens-Frederikse, W.S.; Thevenon, J.; Kuentz, P.; Bruel, A.L.; Thauvin-Robinet, C.; Bensignor, C.; Dollfus, H.; Laugel, V.; Rivière, J.B.; et al. Dominant variants in the splicing factor PUF60 cause a recognizable syndrome with intellectual disability, heart defects and short stature. Eur. J. Hum. Genet. 2016, 25, 43–51. [Google Scholar] [CrossRef] [PubMed]
- Yamada, M.; Uehara, T.; Suzuki, H.; Takenouchi, T.; Kosaki, K. Protein elongation variant of PUF60: Milder phenotypic end of the Verheij syndrome. Am. J. Med. Genet. A 2020, 182, 2709–2714. [Google Scholar] [CrossRef]
- Low, K.J.; Ansari, M.; Abou Jamra, R.; Clarke, A.; El Chehadeh, S.; FitzPatrick, D.R.; Greenslade, M.; Henderson, A.; Hurst, J.; Keller, K.; et al. PUF60 variants cause a syndrome of ID, short stature, microcephaly, coloboma, craniofacial, cardiac, renal and spinal features. Eur. J. Hum. Genet. 2017, 25, 552–559. [Google Scholar] [CrossRef]
- Fennell, A.P.; Baxter, A.E.; Berkovic, S.F.; Ellaway, C.J.; Forwood, C.; Hildebrand, M.S.; Kumble, S.; McKeown, C.; Mowat, D.; Poke, G.; et al. The diverse pleiotropic effects of spliceosomal protein PUF60: A case series of Verheij syndrome. Am. J. Med. Genet. A 2022, 188, 3432–3447. [Google Scholar] [CrossRef]
- Hay, E.; Henderson, R.H.; Mansour, S.; Deshpande, C.; Jones, R.; Nutan, S.; Mankad, K.; Young, R.M.; Arno, G.; Moosajee, M.; et al. Expanding the phenotypic spectrum consequent upon de novo WDR37 missense variants. Clin. Genet. 2020, 98, 191–197. [Google Scholar] [CrossRef] [PubMed]
- Reis, L.M.; Sorokina, E.A.; Thompson, S.; Muheisen, S.; Velinov, M.; Zamora, C.; Aylsworth, A.S.; Semina, E.V. De Novo Missense Variants in WDR37 Cause a Severe Multisystemic Syndrome. Am. J. Hum. Genet. 2019, 105, 425–433. [Google Scholar] [CrossRef] [PubMed]
- Kanca, O.; Andrews, J.C.; Lee, P.T.; Patel, C.; Braddock, S.R.; Slavotinek, A.M.; Cohen, J.S.; Gubbels, C.S.; Aldinger, K.A.; Williams, J.; et al. De Novo Variants in WDR37 Are Associated with Epilepsy, Colobomas, Dysmorphism, Developmental Delay, Intellectual Disability, and Cerebellar Hypoplasia. Am. J. Hum. Genet. 2019, 105, 672–674, Erratum in Am. J. Hum. Genet. 2019, 105, 413–424. [Google Scholar] [CrossRef]
- Nie, K.; Huang, J.; Liu, L.; Lv, H.; Chen, D.; Fan, W. Identification of a De Novo Heterozygous Missense ACTB Variant in Baraitser-Winter Cerebrofrontofacial Syndrome. Front. Genet. 2022, 13, 828120. [Google Scholar] [CrossRef] [PubMed]
- Baumann, M.; Beaver, E.M.; Palomares-Bralo, M.; Santos-Simarro, F.; Holzer, P.; Povysil, G.; Müller, T.; Valovka, T.; Janecke, A.R. Further delineation of putative ACTB loss-of-function variants: A 4-patient series. Hum. Mutat. 2020, 41, 753–758. [Google Scholar] [CrossRef]
- Chacon-Camacho, O.F.; Barragán-Arévalo, T.; Villarroel, C.E.; Almanza-Monterrubio, M.; Zenteno, J.C. Previously undescribed phenotypic findings and novel ACTG1 gene pathogenic variants in Baraitser-Winter cerebrofrontofacial syndrome. Eur. J. Med. Genet. 2020, 63, 103877. [Google Scholar] [CrossRef]
- Hampshire, K.; Martin, P.M.; Carlston, C.; Slavotinek, A. Baraitser-Winter cerebrofrontofacial syndrome: Report of two adult siblings. Am. J. Med. Genet. A 2020, 182, 1923–1932. [Google Scholar] [CrossRef]
- Di Donato, N.; Kuechler, A.; Vergano, S.; Heinritz, W.; Bodurtha, J.; Merchant, S.R.; Breningstall, G.; Ladda, R.; Sell, S.; Altmüller, J.; et al. Update on the ACTG1-associated Baraitser-Winter cerebrofrontofacial syndrome. Am. J. Med. Genet. A 2016, 170, 2644–2651. [Google Scholar] [CrossRef]
- Verloes, A.; Di Donato, N.; Masliah-Planchon, J.; Jongmans, M.; Abdul-Raman, O.A.; Albrecht, B.; Allanson, J.; Brunner, H.; Bertola, D.; Chassaing, N.; et al. Baraitser-Winter cerebrofrontofacial syndrome: Delineation of the spectrum in 42 cases. Eur. J. Hum. Genet. 2015, 23, 292–301. [Google Scholar] [CrossRef]
- Gargano, M.A.; Matentzoglu, N.; Coleman, B.; Addo-Lartey, E.B.; Anagnostopoulos, A.V.; Anderton, J.; Avillach, P.; Bagley, A.M.; Bakštein, E.; Balhoff, J.P.; et al. The Human Phenotype Ontology in 2024: Phenotypes around the world. Nucleic Acids Res. 2024, 52, D1333–D1346. [Google Scholar] [CrossRef]
- Krawitz, P.M.; Lesmann, H.; Klinkhammer, H. The future role of facial image analysis in ACMG classification guidelines. Med. Genet. 2023, 35, 115–121. [Google Scholar] [CrossRef]
- Pagano, S.; Lopergolo, D.; De Falco, A.; Meossi, C.; Satolli, S.; Pasquariello, R.; Trovato, R.; Tessa, A.; Casalini, C.; Pfanner, L.; et al. Expanding the Clinical Spectrum Associated with the Recurrent Arg203Trp Variant in PACS1: An Italian Cohort Study. Genes 2025, 16, 227. [Google Scholar] [CrossRef]
- Latorre-Pellicer, A.; Trujillano, L.; del Rincón, J.; Peña-Marco, M.; Gil-Salvador, M.; Lucia-Campos, C.; Arnedo, M.; Puisac, B.; Ramos, F.J.; Ayerza-Casas, A.; et al. Heart Disease Characterization and Myocardial Strain Analysis in Patients with PACS1 Neurodevelopmental Disorder. J. Clin. Med. 2023, 12, 4052. [Google Scholar] [CrossRef]
- Reiter, A.M.V.; Pantel, J.T.; Danyel, M.; Horn, D.; Ott, C.E.; Mensah, M.A. Validation of 3 Computer-Aided Facial Phenotyping Tools (DeepGestalt, GestaltMatcher, and D-Score): Comparative Diagnostic Accuracy Study. J. Med. Internet Res. 2024, 26, e42904. [Google Scholar] [CrossRef] [PubMed]
- Thirunavukarasu, A.J.; Ting, D.S.J.; Elangovan, K.; Gutierrez, L.; Tan, T.F.; Ting, D.S.W. Large language models in medicine. Nat. Med. 2023, 29, 1930–1940. [Google Scholar] [CrossRef] [PubMed]
- Ao, G.; Chen, M.; Li, J.; Nie, H.; Zhang, L.; Chen, Z. Comparative analysis of large language models on rare disease identification. Orphanet J. Rare Dis. 2025, 20, 150. [Google Scholar] [CrossRef] [PubMed]
- BrainSpan: Atlas of the Developing Human Brain; Allen Institute for Brain Science. Available online: https://www.brainspan.org (accessed on 23 July 2025).
- Selleri, L.; Rijli, F.M. Shaping faces: Genetic and epigenetic control of craniofacial morphogenesis. Nat. Rev. Genet. 2023, 24, 610–626. [Google Scholar] [CrossRef]
- Thomas, G.; Aslan, J.E.; Thomas, L.; Shinde, P.; Shinde, U.; Simmen, T. Caught in the act-protein adaptation and the expanding roles of the PACS proteins in tissue homeostasis and disease. J. Cell Sci. 2017, 130, 1865–1876. [Google Scholar] [CrossRef]
- Blagoveshchenskaya, A.D.; Thomas, L.; Feliciangeli, S.F.; Hung, C.H.; Thomas, G. HIV-1 Nef downregulates MHC-I by a PACS-1- and PI3K-regulated ARF6 endocytic pathway. Cell 2002, 111, 853–866. [Google Scholar] [CrossRef]
- Simmen, T.; Aslan, J.E.; Blagoveshchenskaya, A.D.; Thomas, L.; Wan, L.; Xiang, Y.; Crump, C.M.; Hung, C.H.; Feliciangeli, S.F.; Thomas, G. PACS-2 controls endoplasmic reticulum-mitochondria communication and Bid-mediated apoptosis. EMBO J. 2005, 24, 717–729. [Google Scholar] [CrossRef]
- Köttgen, M.; Benzing, T.; Simmen, T.; Tauber, R.; Buchholz, B.; Feliciangeli, S.; Huber, T.B.; Schermer, B.; Kramer-Zucker, A.; Höpker, K.; et al. Trafficking of TRPP2 by PACS proteins represents a novel mechanism of ion channel regulation. EMBO J. 2005, 24, 705–716. [Google Scholar] [CrossRef]
- Rylaarsdam, L.; Rakotomamonjy, J.; Pope, E.; Guemez-Gamboa, A. iPSC-derived models of PACS1 syndrome reveal transcriptional and functional deficits in neuron activity. Nat. Commun. 2024, 15, 827. [Google Scholar] [CrossRef]
- Nair-Gill, E.; Bonora, M.; Zhong, X.; Liu, A.; Miranda, A.; Stewart, N.; Ludwig, S.; Russell, J.; Gallagher, T.; Pinton, P.; et al. Calcium flux control by Pacs1-Wdr37 promotes lymphocyte quiescence and lymphoproliferative diseases. EMBO J. 2021, 40, e104888. [Google Scholar] [CrossRef]
| Syndrome_id | OMIM_id | Gene | # Patients | Gestalt_Score (Prom) |
|---|---|---|---|---|
| DeepGestalt | ||||
| SHMS | 615009 | PACS1 | 37 | 0.558 |
| BRWS1 | 243310 | ACTB | 33 | 0.244 |
| NS1 | 163950 | PTPN11 | 33 | 0.297 |
| KABUK1 | 147920 | KMT2D | 28 | 0.223 |
| RSTS1 | 180849 | CREBBP | 25 | 0.213 |
| AS | 105830 | UBE3A | 24 | 0.226 |
| NF1 | 162200 | NF1 | 24 | 0.219 |
| CdLS | 122470 | NIPBL | 23 | 0.215 |
| CSS | 135900 | ARID1B | 22 | 0.211 |
| HPMRS1 | 239300 | PIGV | 21 | 0.250 |
| KBGS | 148050 | ANKRD11 | 20 | 0.195 |
| GestaltMatcher | ||||
| DEE66 | 618067 | PACS2 | 33 | 0.358 |
| DeepGestalt and GestaltMatcher | ||||
| SHMS | 615009 | PACS1 | 37 | 0.467 |
| DEE66 | 618067 | PACS2 | 30 | 0.366 |
| VRJS | 615583 | PUF60 | 26 | 0.357 |
| NOCGUS | 618652 | WDR37 | 23 | 0.363 |
| BRWS1 | 243310 | ACTB | 20 | 0.359 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
del Rincón, J.; Gil-Salvador, M.; Lucia-Campos, C.; Acero, L.; Trujillano, L.; Arnedo, M.; Pamplona, P.; Ayerza-Casas, A.; Puisac, B.; Ramos, F.J.; et al. AI-Based Facial Phenotyping Supports a Shared Molecular Axis in PACS1-, PACS2-, and WDR37-Related Syndromes. Int. J. Mol. Sci. 2025, 26, 7964. https://doi.org/10.3390/ijms26167964
del Rincón J, Gil-Salvador M, Lucia-Campos C, Acero L, Trujillano L, Arnedo M, Pamplona P, Ayerza-Casas A, Puisac B, Ramos FJ, et al. AI-Based Facial Phenotyping Supports a Shared Molecular Axis in PACS1-, PACS2-, and WDR37-Related Syndromes. International Journal of Molecular Sciences. 2025; 26(16):7964. https://doi.org/10.3390/ijms26167964
Chicago/Turabian Styledel Rincón, Julia, Marta Gil-Salvador, Cristina Lucia-Campos, Laura Acero, Laura Trujillano, María Arnedo, Pilar Pamplona, Ariadna Ayerza-Casas, Beatriz Puisac, Feliciano J. Ramos, and et al. 2025. "AI-Based Facial Phenotyping Supports a Shared Molecular Axis in PACS1-, PACS2-, and WDR37-Related Syndromes" International Journal of Molecular Sciences 26, no. 16: 7964. https://doi.org/10.3390/ijms26167964
APA Styledel Rincón, J., Gil-Salvador, M., Lucia-Campos, C., Acero, L., Trujillano, L., Arnedo, M., Pamplona, P., Ayerza-Casas, A., Puisac, B., Ramos, F. J., Pié, J., & Latorre-Pellicer, A. (2025). AI-Based Facial Phenotyping Supports a Shared Molecular Axis in PACS1-, PACS2-, and WDR37-Related Syndromes. International Journal of Molecular Sciences, 26(16), 7964. https://doi.org/10.3390/ijms26167964






