A Single DNA Binding Site of DprA Dimer Is Required to Facilitate RecA Filament Nucleation
Abstract
1. Introduction
2. Results and Discussion
2.1. DprA Significantly Enhances Recombinational Exchange Frequency (FRE)
2.2. DprABsu Promotes SSBEco Displacement from ssDNA
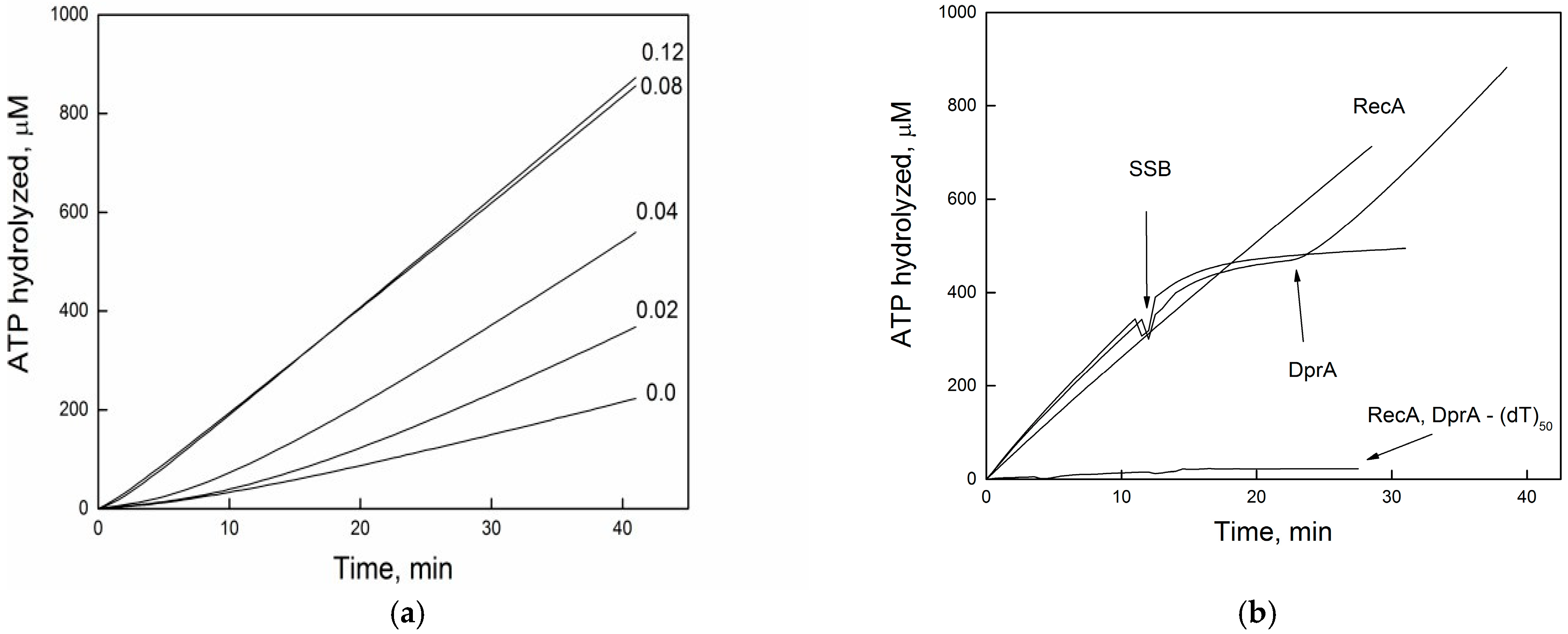
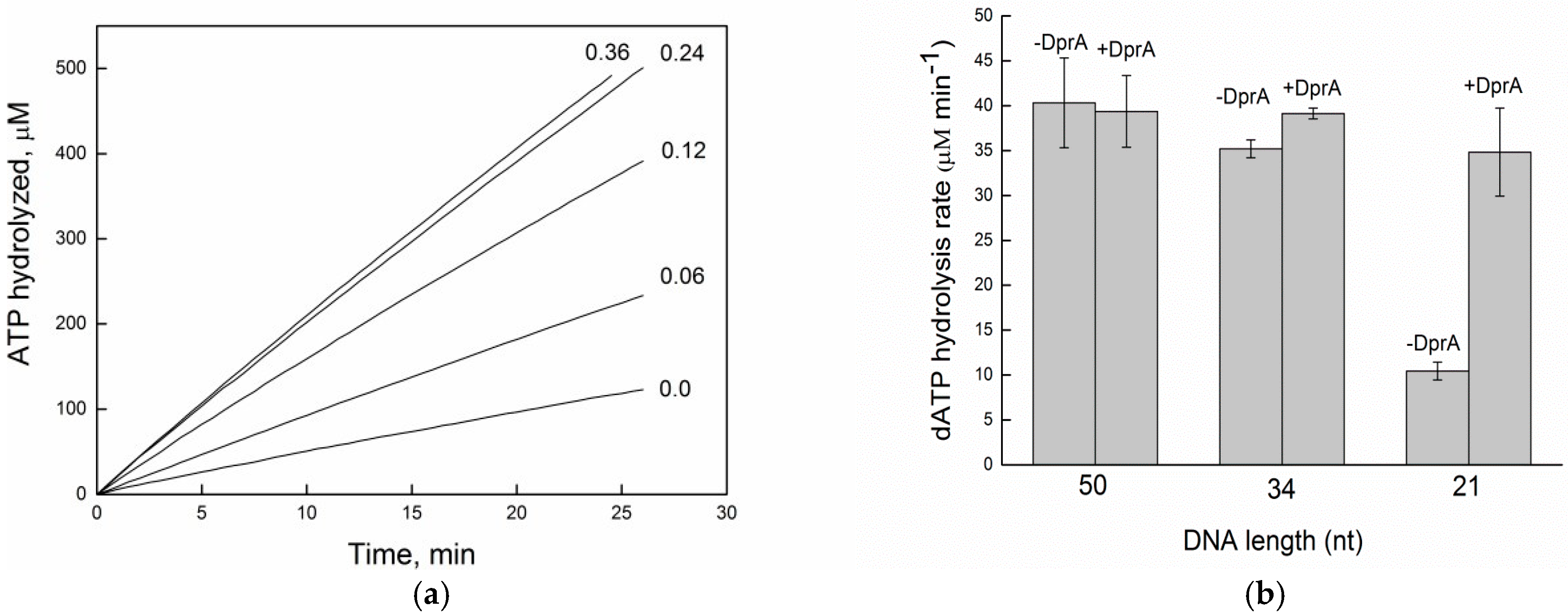
2.3. DprABsu Facilitates RecA Nucleation onto Short (dTn) Oligos
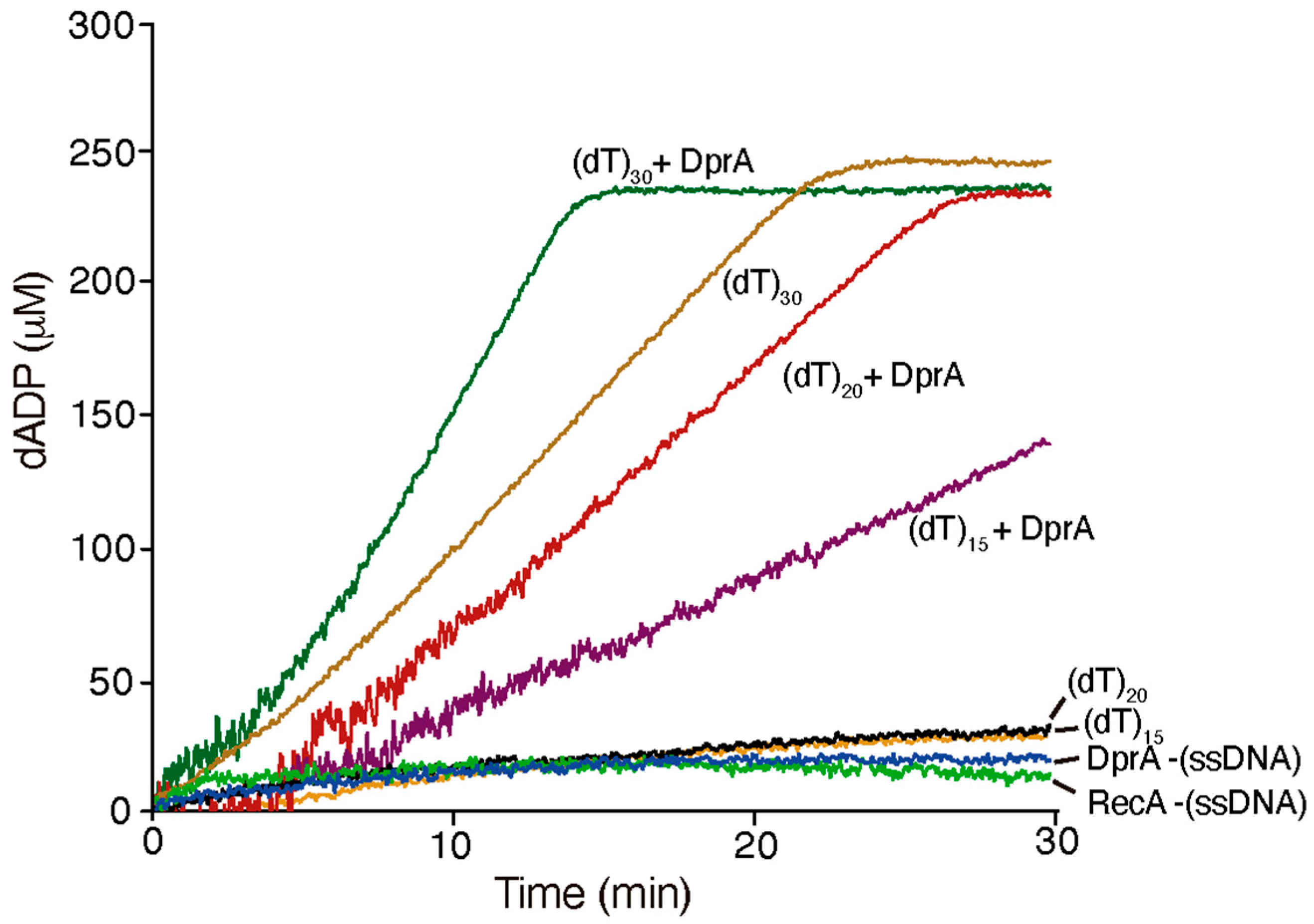
2.4. DprABsu Promotes Dynamic RecABsu·dATP Filaments on Short ssDNA
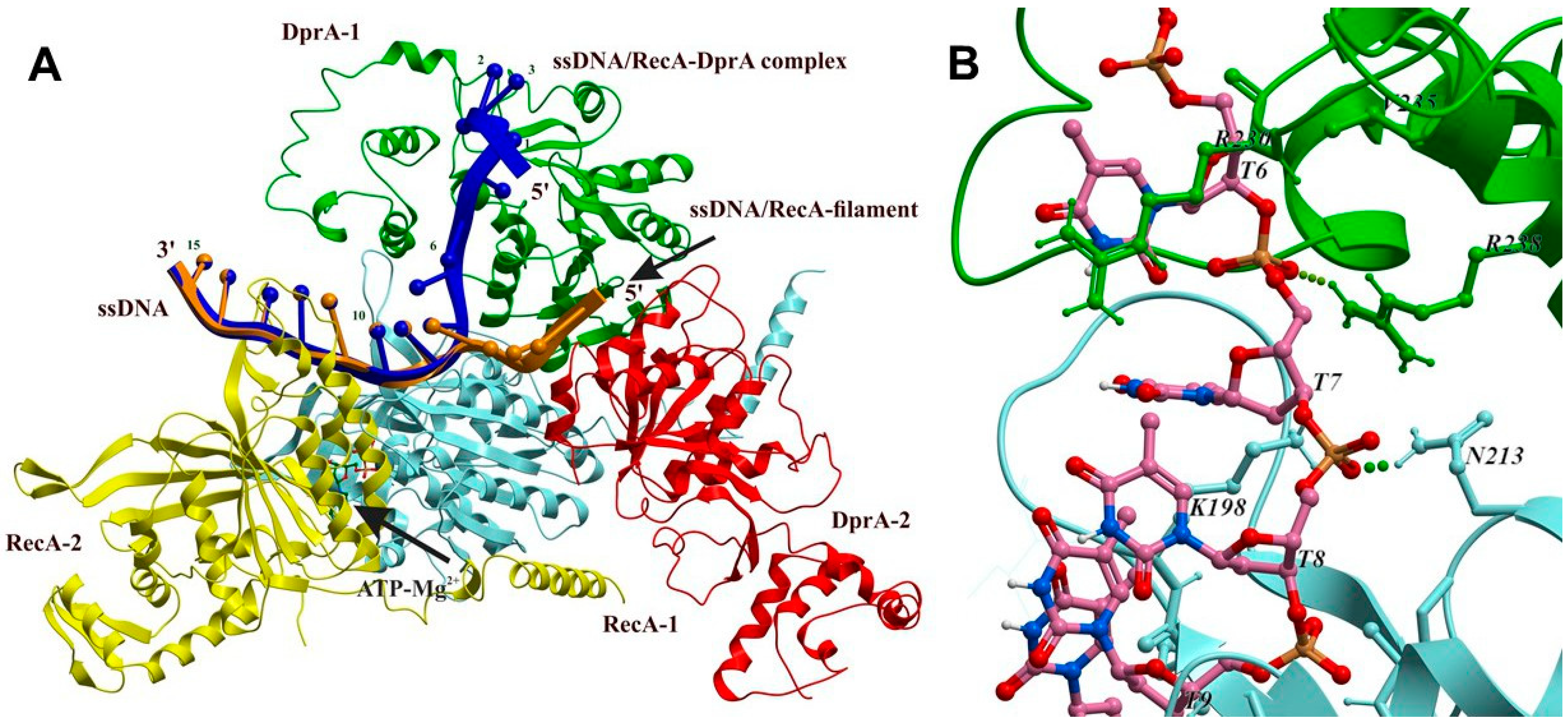
2.5. Structural Model of the RecA-DprA-ssDNA Complex
3. Materials and Methods
3.1. Strains and Plasmids
3.2. Proteins
3.3. (d)ATP Hydrolysis Assays
3.4. Conjugation
3.5. Reconstruction of the Structure of the RecA-ssDNA-DprABsu Complex
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| FRE | frequency of recombination exchanges per DNA unit length |
| SSB | single-stranded DNA binding protein |
| ssDNA | single-strand DNA |
| poly(dT) | extended polymers of deoxythymidine |
| nt | nucleotide(s) |
| EMSA | electrophoretic mobility shift assay |
References
- Kramer, N.; Hahn, J.; Dubnau, D. Multiple interactions among the competence proteins of Bacillus subtilis. Mol. Microbiol. 2007, 65, 454–464. [Google Scholar] [CrossRef] [PubMed]
- Mortier-Barriere, I.; Velten, M.; Dupaigne, P.; Mirouze, N.; Pietrement, O.; McGovern, S.; Fichant, G.; Martin, B.; Noirot, P.; Le Cam, E.; et al. A key presynaptic role in transformation for a widespread bacterial protein: DprA conveys incoming ssDNA to RecA. Cell 2007, 130, 824–836. [Google Scholar] [CrossRef] [PubMed]
- Kidane, D.; Ayora, S.; Sweasy, J.B.; Graumann, P.L.; Alonso, J.C. The cell pole: The site of cross talk between the DNA uptake and genetic recombination machinery. Crit. Rev. Biochem. Mol. Biol. 2012, 47, 531–555. [Google Scholar] [CrossRef]
- Quevillon-Cheruel, S.; Campo, N.; Mirouze, N.; Mortier-Barriere, I.; Brooks, M.A.; Boudes, M.; Durand, D.; Soulet, A.L.; Lisboa, J.; Noirot, P.; et al. Structure-function analysis of pneumococcal DprA protein reveals that dimerization is crucial for loading RecA recombinase onto DNA during transformation. Proc. Natl. Acad. Sci. USA 2012, 109, E2466–E2475. [Google Scholar] [CrossRef]
- Yadav, T.; Carrasco, B.; Hejna, J.; Suzuki, Y.; Takeyasu, K.; Alonso, J.C. Bacillus subtilis DprA recruits RecA onto single-stranded DNA and mediates annealing of complementary strands coated by SsbB and SsbA. J. Biol. Chem. 2013, 288, 22437–22450. [Google Scholar] [CrossRef]
- Sharma, D.K.; Misra, H.S.; Soni, I.; Rajpurohit, Y.S. Characterization of DNA Processing Protein A (DprA) of the Radiation-Resistant Bacterium Deinococcus radiodurans. Microbiol. Spectr. 2022, 10, e0347022. [Google Scholar] [CrossRef]
- Yadav, T.; Carrasco, B.; Serrano, E.; Alonso, J.C. Roles of Bacillus subtilis DprA and SsbA in RecA-mediated genetic recombination. J. Biol. Chem. 2014, 289, 27640–27652. [Google Scholar] [CrossRef]
- Le, S.; Serrano, E.; Kawamura, R.; Carrasco, B.; Yan, J.; Alonso, J.C. Bacillus subtilis RecA with DprA-SsbA antagonizes RecX function during natural transformation. Nucleic Acids Res. 2017, 45, 8873–8885. [Google Scholar] [CrossRef]
- Mirouze, N.; Berge, M.A.; Soulet, A.-L.; Mortier-Barriere, I.; Quentin, Y.; Fichant, G.; Granadel, C.; Noirot-Gros, M.-F.; Noirot, P.; Polard, P. Direct involvement of DprA, the transformation-dedicated RecA loader, in the shut-off of pneumococcal competence. Proc. Natl. Acad. Sci. USA 2013, 110, E1035–E1044. [Google Scholar] [CrossRef]
- Johnston, C.; Mortier-Barriere, I.; Khemici, V.; Polard, P. Fine-tuning cellular levels of DprA ensures transformant fitness in the human pathogen Streptococcus pneumoniae. Mol. Microbiol. 2018, 109, 663–675. [Google Scholar] [CrossRef]
- Brito, I.L. Examining horizontal gene transfer in microbial communities. Nat. Rev. Microbiol. 2021, 19, 442–453. [Google Scholar] [CrossRef]
- Smeets, L.C.; Becker, S.C.; Barcak, G.J.; Vandenbroucke-Grauls, C.M.; Bitter, W.; Goosen, N. Functional characterization of the competence protein DprA/Smf in Escherichia coli. FEMS Microbiol. Lett. 2006, 263, 223–228. [Google Scholar] [CrossRef]
- Lisboa, J.; Andreani, J.; Sanchez, D.; Boudes, M.; Collinet, B.; Liger, D.; van Tilbeurgh, H.; Guerois, R.; Quevillon-Cheruel, S. Molecular determinants of the DprA-RecA interaction for nucleation on ssDNA. Nucleic Acids Res. 2014, 42, 7395–7408. [Google Scholar] [CrossRef]
- Wang, W.; Ding, J.; Zhang, Y.; Hu, Y.; Wang, D.C. Structural insights into the unique single-stranded DNA-binding mode of Helicobacter pylori DprA. Nucleic Acids Res. 2014, 42, 3478–3491. [Google Scholar] [CrossRef] [PubMed]
- Lawley, T.D.; Gordon, G.S.; Wright, A.; Taylor, D.E. Bacterial conjugative transfer: Visualization of successful mating pairs and plasmid establishment in live Escherichia coli. Mol. Microbiol. 2002, 44, 947–956. [Google Scholar] [CrossRef] [PubMed]
- Lanzov, V.A.; Bakhlanova, I.V.; Clark, A.J. Conjugational hyperrecombination achieved by derepressing the LexA regulon, altering the properties of RecA protein and inactivating mismatch repair in Escherichia coli K-12. Genetics 2003, 163, 1243–1254. [Google Scholar] [CrossRef] [PubMed]
- Lloyd, R.G. Hyper-recombination in Escherichia coli K-12 mutants constitutive for protein X synthesis. J. Bacteriol. 1978, 134, 929–935. [Google Scholar] [CrossRef]
- Bakhlanova, I.V.; Dudkina, A.V.; Wood, E.A.; Lanzov, V.A.; Cox, M.M.; Baitin, D.M. DNA Metabolism in Balance: Rapid Loss of a RecA-Based Hyperrec Phenotype. PLoS ONE 2016, 11, e0154137. [Google Scholar] [CrossRef]
- Arenson, T.A.; Tsodikov, O.V.; Cox, M.M. Quantitative analysis of the kinetics of end-dependent disassembly of RecA filaments from ssDNA. J. Mol. Biol. 1999, 288, 391–401. [Google Scholar] [CrossRef]
- Bork, J.M.; Cox, M.M.; Inman, R.B. RecA protein filaments disassemble in the 5′ to 3′ direction on single-stranded DNA. J. Biol. Chem. 2001, 276, 45740–45743. [Google Scholar] [CrossRef]
- Hovland, E.; Beyene, G.T.; Frye, S.A.; Homberset, H.; Balasingham, S.V.; Gomez-Munoz, M.; Derrick, J.P.; Tonjum, T.; Ambur, O.H. DprA from Neisseria meningitidis: Properties and role in natural competence for transformation. Microbiology 2017, 163, 1016–1029. [Google Scholar] [CrossRef] [PubMed]
- Brenner, S.L.; Mitchell, R.S.; Morrical, S.W.; Neuendorf, S.K.; Schutte, B.C.; Cox, M.M. recA protein-promoted ATP hydrolysis occurs throughout recA nucleoprotein filaments. J. Biol. Chem. 1987, 262, 4011–4016. [Google Scholar] [CrossRef]
- Sussman, R.; Sharma, S.K.; Kuzirian, A. Catalytic activities of recA protein are dependent on the lattice length of the single-strand DNA ligand. Cell Cycle 2008, 7, 89–95. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Galletto, R.; Amitani, I.; Baskin, R.J.; Kowalczykowski, S.C. Direct observation of individual RecA filaments assembling on single DNA molecules. Nature 2006, 443, 875–878. [Google Scholar] [CrossRef] [PubMed]
- Bell, J.C.; Plank, J.L.; Dombrowski, C.C.; Kowalczykowski, S.C. Direct imaging of RecA nucleation and growth on single molecules of SSB-coated ssDNA. Nature 2012, 491, 274–278. [Google Scholar] [CrossRef]
- Menetski, J.P.; Kowalczykowski, S.C. Enhancement of Escherichia coli RecA protein enzymatic function by dATP. Biochemistry 1989, 28, 5871–5881. [Google Scholar] [CrossRef]
- Shan, Q.; Cox, M.M. RecA protein dynamics in the interior of RecA nucleoprotein filaments. J. Mol. Biol. 1996, 257, 756–774. [Google Scholar] [CrossRef][Green Version]
- Chen, Z.; Yang, H.; Pavletich, N.P. Mechanism of homologous recombination from the RecA-ssDNA/dsDNA structures. Nature 2008, 453, 489–494. [Google Scholar] [CrossRef]
- Sharma, D.K.; Misra, H.S.; Bihani, S.C.; Rajpurohit, Y.S. Biochemical Properties and Roles of DprA Protein in Bacterial Natural Transformation, Virulence, and Pilin Variation. J. Bacteriol. 2023, 205, e0046522. [Google Scholar] [CrossRef]
- Csonka, L.N.; Clark, A.J. Construction of an Hfr strain useful for transferring recA mutations between Escherichia coli strains. J. Bacteriol. 1980, 143, 529–530. [Google Scholar] [CrossRef]
- Cox, M.M.; McEntee, K.; Lehman, I.R. A simple and rapid procedure for the large scale purification of the recA protein of Escherichia coli. J. Biol. Chem. 1981, 256, 4676–4678. [Google Scholar] [CrossRef] [PubMed]
- Manfredi, C.; Carrasco, B.; Ayora, S.; Alonso, J.C. Bacillus subtilis RecO nucleates RecA onto SsbA-coated single-stranded DNA. J. Biol. Chem. 2008, 283, 24837–24847. [Google Scholar] [CrossRef] [PubMed]
- Drees, J.C.; Lusetti, S.L.; Chitteni-Pattu, S.; Inman, R.B.; Cox, M.M. A RecA filament capping mechanism for RecX protein. Mol. Cell 2004, 15, 789–798. [Google Scholar] [CrossRef] [PubMed]
- Lohman, T.M.; Green, J.M.; Beyer, R.S. Large-scale overproduction and rapid purification of the Escherichia coli ssb gene product. Expression of the ssb gene under lambda PL control. Biochemistry 1986, 25, 21–25. [Google Scholar] [CrossRef]
- Morrical, S.W.; Lee, J.; Cox, M.M. Continuous association of Escherichia coli single-stranded DNA binding protein with stable complexes of recA protein and single-stranded DNA. Biochemistry 1986, 25, 1482–1494. [Google Scholar] [CrossRef]
- Lindsley, J.E.; Cox, M.M. Assembly and disassembly of RecA protein filaments occur at opposite filament ends. Relationship to DNA strand exchange. J. Biol. Chem. 1990, 265, 9043–9054. [Google Scholar] [CrossRef]
- Abagyan, R.; Totrov, M. Biased probability Monte Carlo conformational searches and electrostatic calculations for peptides and proteins. J. Mol. Biol. 1994, 235, 983–1002. [Google Scholar] [CrossRef]
- Arnautova, Y.A.; Jagielska, A.; Scheraga, H.A. A new force field (ECEPP-05) for peptides, proteins, and organic molecules. J. Phys. Chem. B 2006, 110, 5025–5044. [Google Scholar] [CrossRef]
- Baitin, D.M.; Bakhlanova, I.V.; Chervyakova, D.V.; Kil, Y.V.; Lanzov, V.A.; Cox, M.M. Two RecA protein types that mediate different modes of hyperrecombination. J. Bacteriol. 2008, 190, 3036–3045. [Google Scholar] [CrossRef][Green Version]
- Bakhlanova, I.V.; Dudkina, A.V.; Baitin, D.M.; Knight, K.L.; Cox, M.M.; Lanzov, V.A. Modulating cellular recombination potential through alterations in RecA structure and regulation. Mol. Microbiol. 2010, 78, 1523–1538. [Google Scholar] [CrossRef]
- Yakimov, A.; Pobegalov, G.; Bakhlanova, I.; Khodorkovskii, M.; Petukhov, M.; Baitin, D. Blocking the RecA activity and SOS-response in bacteria with a short alpha-helical peptide. Nucleic Acids Res. 2017, 45, 9788–9796. [Google Scholar] [CrossRef]
- Yakimov, A.; Bakhlanova, I.; Baitin, D. Targeting evolution of antibiotic resistance by SOS response inhibition. Comput. Struct. Biotechnol. J. 2021, 19, 777–783. [Google Scholar] [CrossRef]
| Expression Plasmids | Linkage (μ) Between Selected thr+ and Unselected leu+ Markers | Yield of thr+ Str R Recombinants (% to Donors) | FRE | ΔFRE |
|---|---|---|---|---|
| (a) | ||||
| ABpT7, ΔdprApET21 | 0.93 ± 0.025 (600) * | 1.6% | 4.5 ± 1.0 | 1.0 |
| ABpT7, ΔdprApDprA | 0.84 ± 0.032 (600) | 0.71% | 11.7 ± 1.5 | 2.6 |
| ABpT7, ΔdprApDprABsu | 0.69 ± 0.021 (400) | 0.22% | 28.9 ± 5.3 | 6.4 |
| (b) | ||||
| ABpT7, ΔdprApET21 | 0.949 ± 0.024 (600) * | 1.7% | 3.2 ± 0.6 | 1.0 |
| ABΔdprA, pET21 | 0.94 ± 0.018 (600) | 3.1% | 3.6 ± 0.5 | 1.1 |
| ABpT7, ΔdprApDprA | 0.59 ± 0.025 (800) | 0.2% | 55.5 ± 6.3 | 17.3 |
| ABpT7, ΔdprApDprABsu | 0.56 ± 0.036 (600) | 0.02% | 61.2 ± 8.5 | 19.1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bakhlanova, I.; Carrasco, B.; Alekseev, A.; Yakunina, M.; Morozova, N.; Khodorkovskii, M.; Petukhov, M.; Baitin, D. A Single DNA Binding Site of DprA Dimer Is Required to Facilitate RecA Filament Nucleation. Int. J. Mol. Sci. 2025, 26, 7873. https://doi.org/10.3390/ijms26167873
Bakhlanova I, Carrasco B, Alekseev A, Yakunina M, Morozova N, Khodorkovskii M, Petukhov M, Baitin D. A Single DNA Binding Site of DprA Dimer Is Required to Facilitate RecA Filament Nucleation. International Journal of Molecular Sciences. 2025; 26(16):7873. https://doi.org/10.3390/ijms26167873
Chicago/Turabian StyleBakhlanova, Irina, Begoña Carrasco, Aleksandr Alekseev, Maria Yakunina, Natalia Morozova, Mikhail Khodorkovskii, Michael Petukhov, and Dmitry Baitin. 2025. "A Single DNA Binding Site of DprA Dimer Is Required to Facilitate RecA Filament Nucleation" International Journal of Molecular Sciences 26, no. 16: 7873. https://doi.org/10.3390/ijms26167873
APA StyleBakhlanova, I., Carrasco, B., Alekseev, A., Yakunina, M., Morozova, N., Khodorkovskii, M., Petukhov, M., & Baitin, D. (2025). A Single DNA Binding Site of DprA Dimer Is Required to Facilitate RecA Filament Nucleation. International Journal of Molecular Sciences, 26(16), 7873. https://doi.org/10.3390/ijms26167873






