Mouse Model of STAT3 Mutation Resulting in Job’s Syndrome Diverges from Human Pathology
Abstract
1. Introduction
2. Results
2.1. Generation of G656_M660del Mice
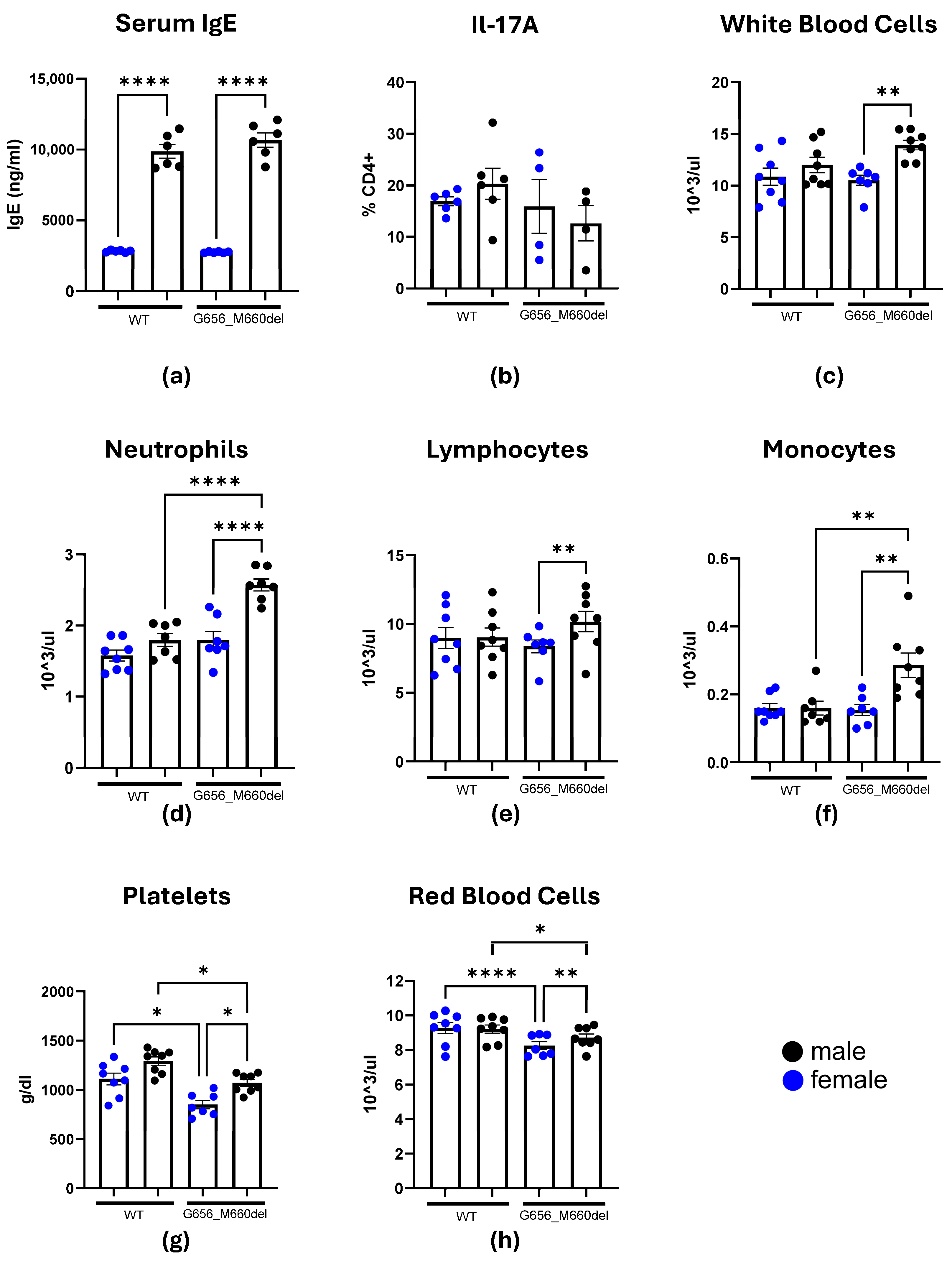
2.2. HIES Phenotype Investigation
2.3. Effects of the STAT3G656_M660del Deletion on Blood Cell Populations
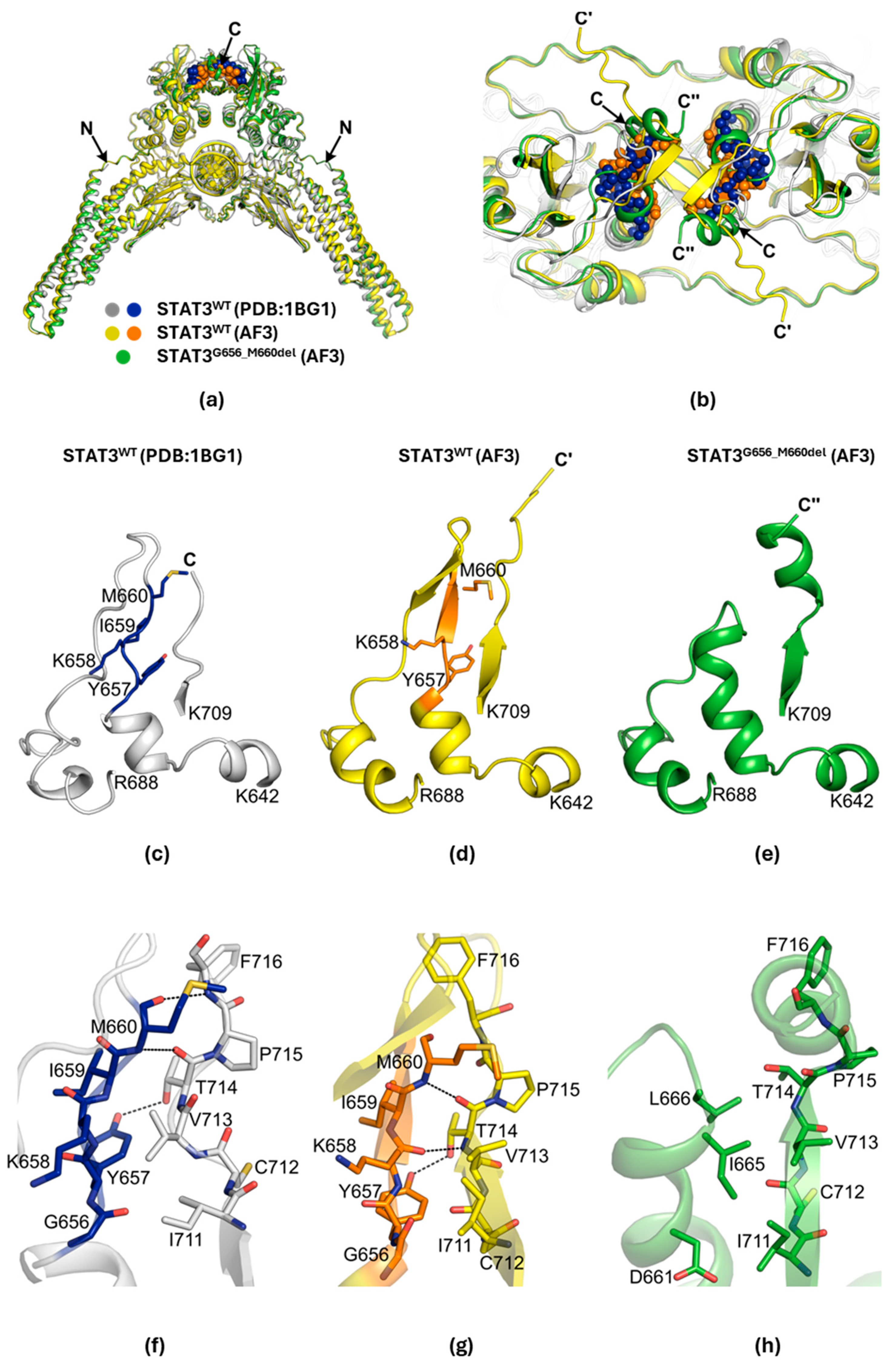
2.4. Structural Changes in the G656_M660del Mutant
3. Discussion
4. Materials and Methods
4.1. Mice
4.2. Serum IgE Measurement
4.3. Complete Blood Count and Flow Cytometry
4.4. Structural Analysis
4.5. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ACK Lysis Buffer | Ammonium–Chloride–Potassium Lysis Buffer |
| AD-HIES | Autosomal-Dominant Hyper IgE Syndrome |
| AF3 | AlphaFold3 |
| CBC | Complete Blood Count |
| SH2 Domain | Src Homology 2 Domain |
| STAT3 | Signal Transducer and Activator of Transcription 3 |
References
- Hillmer, E.J.; Zhang, H.; Li, H.S.; Watowich, S.S. STAT3 signaling in immunity. Cytokine Growth Factor Rev. 2016, 31, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Zou, S.; Tong, Q.; Liu, B.; Huang, W.; Tian, Y.; Fu, X. Targeting STAT3 in Cancer Immunotherapy. Mol. Cancer 2020, 19, 145–163. [Google Scholar] [CrossRef] [PubMed]
- Takeda, K.; Noguchi, K.; Shi, W.; Tanaka, T.; Matsumoto, M.; Yoshida, N.; Kishimoto, T.; Akira, S. Targeted disruption of the mouse Stat3 gene leads to early embryonic lethality. Proc. Natl. Acad. Sci. USA 1997, 94, 3801–3804. [Google Scholar] [CrossRef]
- Semenzato, G.; Calabretto, G.; Teramo, A.; Gasparini, V.R.; Rampazzo, E.; Barilà, G.; Zambello, R. The constitutive activation of STAT3 gene and its mutations are at the crossroad between LGL leukemia and autoimmune disorders. Blood Cancer J. 2024, 14, 13–20. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Zhang, Z.; Qu, X.; Zhu, X.; Zhao, L.; Wei, R.; Guo, Q.; Sun, L.; Yin, X.; Zhang, Y.; et al. Roles of STAT3 in leukemia (Review). Int. J. Oncol. 2018, 53, 7–20. [Google Scholar] [CrossRef]
- Vogel, T.P.; Milner, J.D.; Cooper, M.A. The Ying and Yang of STAT3 in Human Disease. J. Clin. Immunol. 2015, 35, 615–623. [Google Scholar] [CrossRef]
- Faletti, L.; Ehl, S.; Heeg, M. Germline STAT3 gain-of-function mutations in primary immunodeficiency: Impact on the cellular and clinical phenotype. Biomed. J. 2021, 44, 412–421. [Google Scholar] [CrossRef]
- Ott, N.; Faletti, L.; Heeg, M.; Andreani, V.; Grimbacher, B. JAKs and STATs from a Clinical Perspective: Loss-of-Function Mutations, Gain-of-Function Mutations, and Their Multidimensional Consequences. J. Clin. Immunol. 2023, 43, 1326–1359. [Google Scholar] [CrossRef]
- Lodi, L.; Faletti, L.E.; Maccari, M.E.; Consonni, F.; Groß, M.; Pagnini, I.; Ricci, S.; Heeg, M.; Simonini, G.; Azzari, C.; et al. STAT3-confusion-of-function: Beyond the loss and gain dualism. J. Allergy Clin. Immunol. 2022, 150, 1237–1241.e3. [Google Scholar] [CrossRef]
- Deng, Y.; Li, T.; Xie, X.; Xia, D.; Ding, L.; Xiang, H.; Ma, J.J.; Li, W. Hyper IgE syndrome associated with novel and recurrent STAT3 mutations: Two case reports. Medicine 2019, 98, e14003. [Google Scholar] [CrossRef]
- Grimbacher, B.; Holland, S.M.; Gallin, J.I.; Greenberg, F.; Hill, S.C.; Malech, H.L.; Miller, J.A.; O’Connell, A.C.; Puck, J.M. Hyper-IgE syndrome with recurrent infections—An autosomal dominant multisystem disorder. N. Engl. J. Med. 1999, 340, 692–702. [Google Scholar] [CrossRef]
- Woellner, C.; Gertz, E.M.; Schäffer, A.A.; Lagos, M.; Perro, M.; Glocker, E.O.; Pietrogrande, M.C.; Cossu, F.; Franco, J.L.; Matamoros, N.; et al. Mutations in STAT3 and diagnostic guidelines for hyper-IgE syndrome. J. Allergy Clin. Immunol. 2010, 125, 424–432.e8. [Google Scholar] [CrossRef]
- Criado, P.R.; Miot, H.A.; Ianhez, M. Eosinophilia and elevated IgE serum levels: A red flag: When your diagnosis is not a common atopic eczema or common allergy. Inflamm. Res. 2023, 72, 541–551. [Google Scholar] [CrossRef]
- Schmitt, E.G.; Toth, K.A.; Risma, S.I.; Kolicheski, A.; Saucier, N.; Berríos, R.J.F.; Greenberg, Z.J.; Leiding, J.W.; Bleesing, J.J.; Thatayatikom, A.; et al. A human STAT3 gain-of-function variant confers T cell dysregulation without predominant Treg dysfunction in mice. JCI Insight 2022, 7, e162695. [Google Scholar] [CrossRef] [PubMed]
- Siroux, V.; Curt, F.; Oryszczyn, M.P.; Maccario, J.; Kauffmann, F. Role of gender and hormone-related events on IgE, atopy, and eosinophils in the Epidemiological Study on the Genetics and Environment of Asthma, bronchial hyperresponsiveness and atopy. J. Allergy Clin. Immunol. 2004, 114, 491–498. [Google Scholar] [CrossRef]
- Abdullah, A.K.; el-Hazmi, M.A.; Uz-Zaman, A.; al-Nozha, M.; Lambourne, A. Serum IgE levels in adults with asthma. J. Asthma 1987, 24, 207–213. [Google Scholar] [CrossRef]
- Paula Couto, T.A.; Falsarella, N.; Mattos Cde, C.; Mattos, L.C. Total IgE plasma levels vary according to gender and age in Brazilian patients with allergic rhinitis. Clinics 2014, 69, 740–744. [Google Scholar] [CrossRef] [PubMed]
- Becker, S.; Groner, B.; Müller, C.W. Three-dimensional structure of the Stat3beta homodimer bound to DNA. Nature 1998, 394, 145–151. [Google Scholar] [CrossRef] [PubMed]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Abramson, J.; Adler, J.; Dunger, J.; Evans, R.; Green, T.; Pritzel, A.; Ronneberger, O.; Willmore, L.; Ballard, A.J.; Bambrick, J.; et al. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature 2024, 630, 493–500. [Google Scholar] [CrossRef]
- Yu, H.; Pardoll, D.; Jove, R. STATs in cancer inflammation and immunity: A leading role for STAT3. Nat. Rev. Cancer 2009, 9, 798–809. [Google Scholar] [CrossRef]
- Asano, T.; Khourieh, J.; Zhang, P.; Rapaport, F.; Spaan, A.N.; Li, J.; Lei, W.T.; Pelham, S.J.; Hum, D.; Chrabieh, M.; et al. Human STAT3 variants underlie autosomal dominant hyper-IgE syndrome by negative dominance. J. Exp. Med. 2021, 218, e20202592. [Google Scholar] [CrossRef]
- Carrabba, M.; Dellepiane, R.M.; Cortesi, M.; Baselli, L.A.; Soresina, A.; Cirillo, E.; Giardino, G.; Conti, F.; Dotta, L.; Finocchi, A.; et al. Long term longitudinal follow-up of an AD-HIES cohort: The impact of early diagnosis and enrollment to IPINet centers on the natural history of Job’s syndrome. Allergy Asthma Clin. Immunol. 2023, 19, 32–41. [Google Scholar] [CrossRef]
- Steward-Tharp, S.M.; Laurence, A.; Kanno, Y.; Kotlyar, A.; Villarino, A.V.; Sciume, G.; Kuchen, S.; Resch, W.; Wohlfert, E.A.; Jiang, K.; et al. A mouse model of HIES reveals pro- and anti-inflammatory functions of STAT3. Blood 2014, 123, 2978–2987. [Google Scholar] [CrossRef] [PubMed]
- Caetano, M.S.; Hassane, M.; Van, H.T.; Bugarin, E.; Cumpian, A.M.; McDowell, C.L.; Cavazos, C.G.; Zhang, H.; Deng, S.; Diao, L.; et al. Sex specific function of epithelial STAT3 signaling in pathogenesis of K-ras mutant lung cancer. Nat. Commun. 2018, 9, 4589–4599. [Google Scholar] [CrossRef]
- Wang, M.; Zhang, W.; Crisostomo, P.; Markel, T.; Meldrum, K.K.; Fu, X.Y.; Meldrum, D.R. Sex differences in endothelial STAT3 mediate sex differences in myocardial inflammation. Am. J. Physiol. Endocrinol. Metab. 2007, 293, E872–E877. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Mauracher, A.A.; Eekels, J.J.M.; Woytschak, J.; van Drogen, A.; Bosch, A.; Prader, S.; Felber, M.; Heeg, M.; Opitz, L.; Trück, J.; et al. Erythropoiesis defect observed in STAT3 GOF patients with severe anemia. J. Allergy Clin. Immunol. 2020, 145, 1297–1301. [Google Scholar] [CrossRef] [PubMed]
- Ciullini Mannurita, S.; Goda, R.; Schiavo, E.; Coniglio, M.L.; Azzali, A.; Fotzi, I.; Tondo, A.; Tintori, V.; Frenos, S.; Sanvito, M.C.; et al. Case Report: Signal Transducer and Activator of Transcription 3 Gain-of-Function and Spectrin Deficiency: A Life-Threatening Case of Severe Hemolytic Anemia. Front. Immunol. 2021, 11, 620046. [Google Scholar] [CrossRef]
- Kawakami, T.; Sekiguchi, N.; Kobayashi, J.; Imi, T.; Matsuda, K.; Yamane, T.; Nishina, S.; Senoo, Y.; Sakai, H.; Ito, T.; et al. Frequent STAT3 mutations in CD8+ T cells from patients with pure red cell aplasia. Blood Adv. 2018, 2, 2704–2712. [Google Scholar] [CrossRef]
- Zhang, N.; Sherwood, K.; Claggett, B.; Dhruva, S.; Sandhu, A.; Cheng, S.; Ebinger, J. Sex differences and the role of anemia in contrast-associated acute kidney injury after percutaneous coronary intervention. J. Invasive Cardiol. 2025, 37. online publication. [Google Scholar] [CrossRef]
- Xu, Z.; Xu, Y.J.; Hao, Y.N.; Ren, L.J.; Zhang, Z.B.; Xu, X.; Cao, B.Y.; Dai, K.S.; Zhu, L.; Fang, Q.; et al. A novel STAT3 inhibitor negatively modulates platelet activation and aggregation. Acta Pharmacol. Sin. 2017, 38, 651–659. [Google Scholar] [CrossRef]
- Lee, H.K.; Jankowski, J.; Liu, C.; Hennighausen, L. Disease-Associated Mutations of the STAT5B SH2 Domain Regulate Cytokine-Driven Enhancer Function and Mammary Development. J. Mammary Gland. Biol. Neoplasia 2025, 30, 7–26. [Google Scholar] [CrossRef]
- Lee, H.K.; Chen, J.; Philips, R.L.; Lee, S.G.; Feng, X.; Wu, Z.; Liu, C.; Schultz, A.B.; Dalzell, M.; Meggendorfer, M.; et al. STAT5B leukemic mutations, altering SH2 tyrosine 665, have opposing impacts on immune gene programs. Life Sci. Alliance 2025, 8, e202503222. [Google Scholar] [CrossRef] [PubMed]
- UniProt Consortium. UniProt: The Universal Protein Knowledgebase in 2025. Nucleic Acids Res. 2025, 53, D609–D617. [Google Scholar] [CrossRef] [PubMed]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jankowski, J.; Chen, J.; Cho, G.; Lee, S.-G.; Liu, C.; Young, N.; Kim, J.; Hennighausen, L. Mouse Model of STAT3 Mutation Resulting in Job’s Syndrome Diverges from Human Pathology. Int. J. Mol. Sci. 2025, 26, 7675. https://doi.org/10.3390/ijms26167675
Jankowski J, Chen J, Cho G, Lee S-G, Liu C, Young N, Kim J, Hennighausen L. Mouse Model of STAT3 Mutation Resulting in Job’s Syndrome Diverges from Human Pathology. International Journal of Molecular Sciences. 2025; 26(16):7675. https://doi.org/10.3390/ijms26167675
Chicago/Turabian StyleJankowski, Jakub, Jichun Chen, Gyuhyeok Cho, Sung-Gwon Lee, Chengyu Liu, Neal Young, Jungwook Kim, and Lothar Hennighausen. 2025. "Mouse Model of STAT3 Mutation Resulting in Job’s Syndrome Diverges from Human Pathology" International Journal of Molecular Sciences 26, no. 16: 7675. https://doi.org/10.3390/ijms26167675
APA StyleJankowski, J., Chen, J., Cho, G., Lee, S.-G., Liu, C., Young, N., Kim, J., & Hennighausen, L. (2025). Mouse Model of STAT3 Mutation Resulting in Job’s Syndrome Diverges from Human Pathology. International Journal of Molecular Sciences, 26(16), 7675. https://doi.org/10.3390/ijms26167675





