Loss of SPRED3 Causes Primary Hypothyroidism and Alters Thyroidal Expression of Autophagy Regulators LC3, p62, and ATG5 in Mice
Abstract
1. Introduction
2. Results
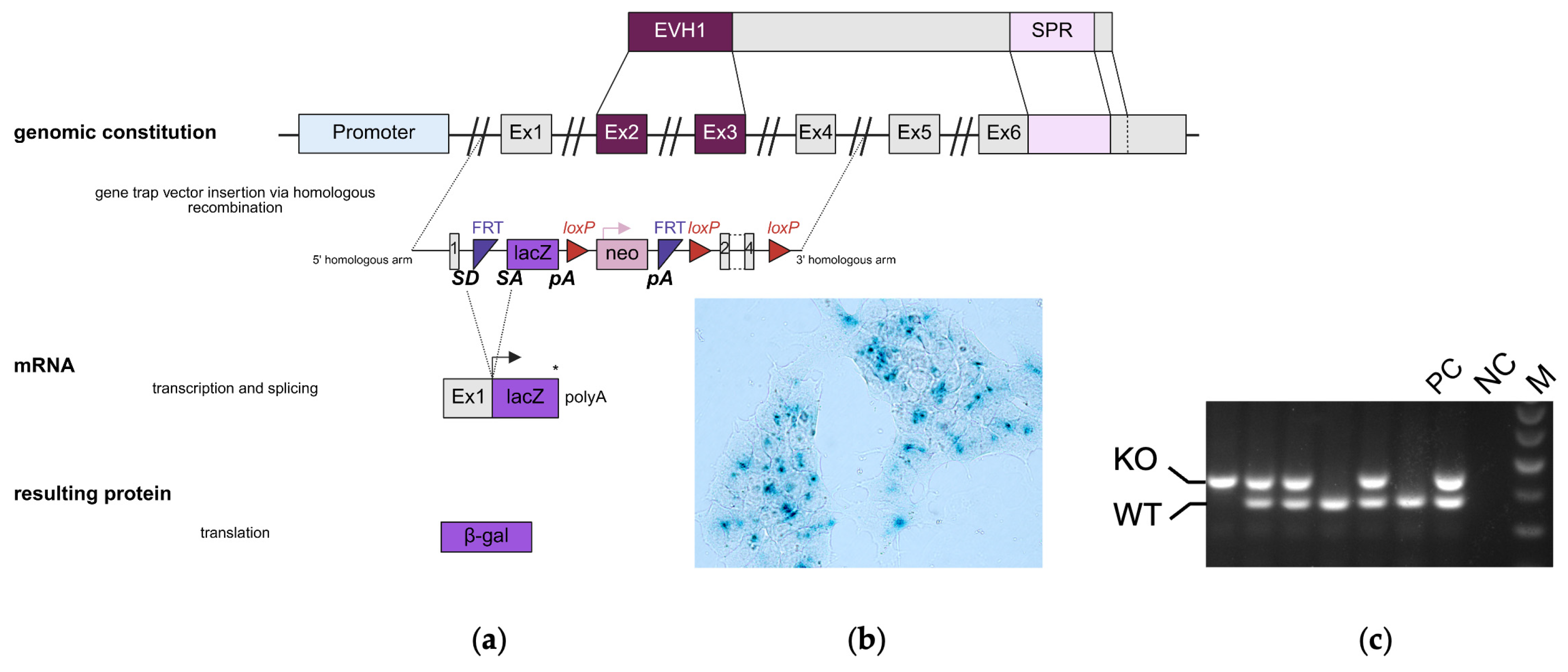
2.1. Generation and Validation of SPRED3 Knockout Mice
- A reporter trap element including:
- ○
- A splice acceptor (SA) sequence from the mouse engrailed-2 gene.
- ○
- A lacZ reporter gene for β-galactosidase expression.
- ○
- A polyadenylation (Poly (A)) signal derived from Simian Virus 40, which truncates the endogenous transcription.
- A neomycin resistance gene (neo) driven by a phosphoglycerate kinase (PGK) promoter, flanked by FRT sites for conditional removal.
- Three loxP sites enabling the potential for conditional gene deletion [17].
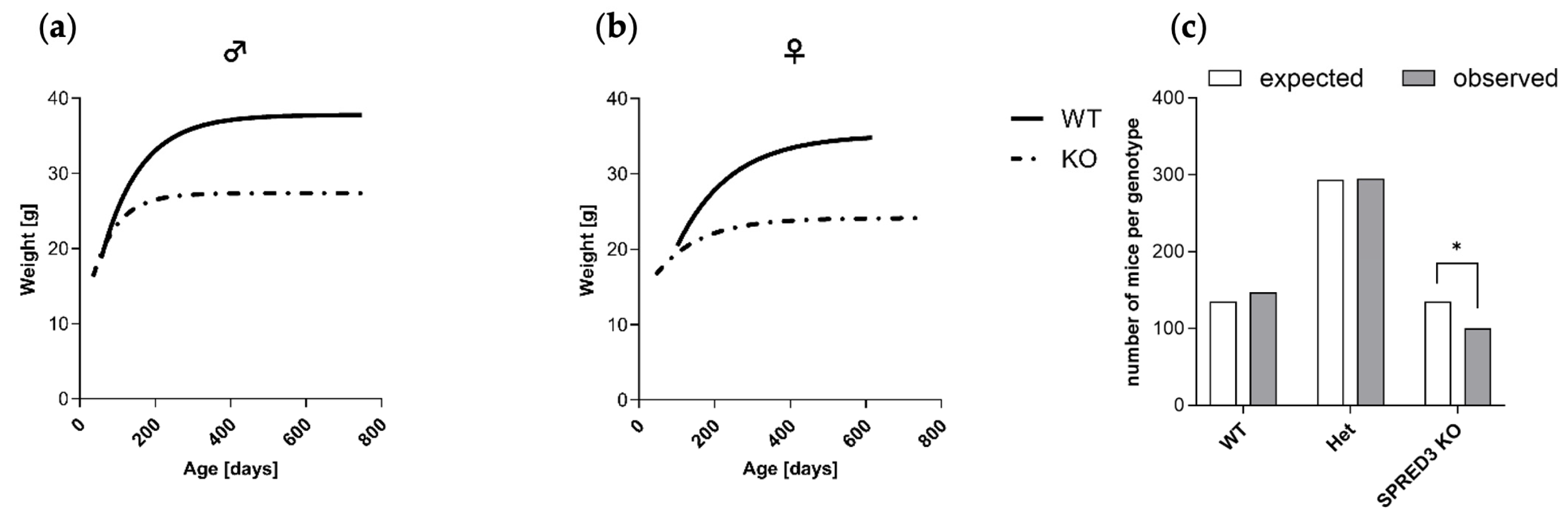
2.2. SPRED3 Deficiency Causes Growth Retardation and Deviations from Mendelian Ratios
2.3. SPRED3 Deficiency Does Not Alter Cardiac Performance
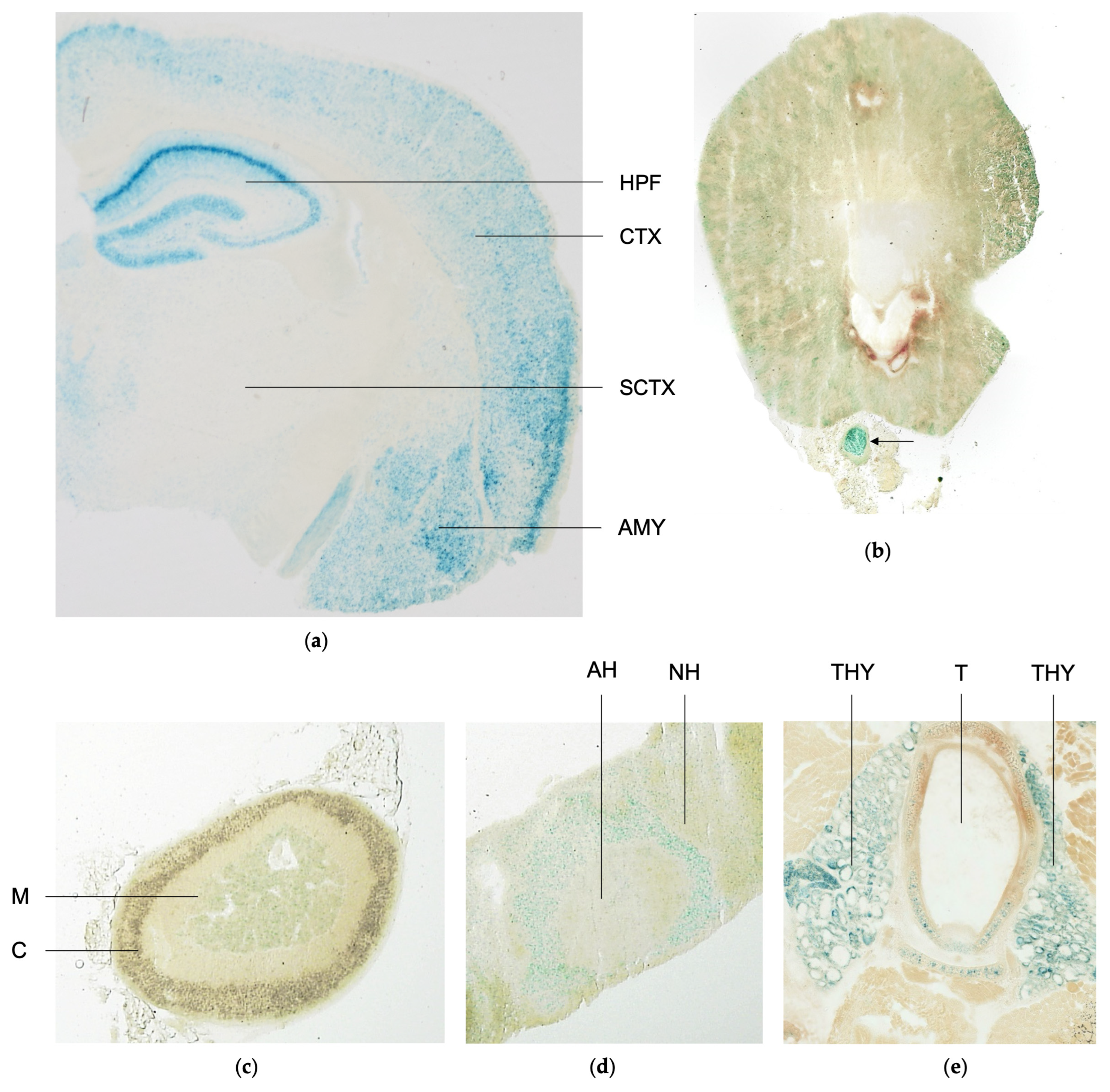
2.4. Scrutiny of Tissue-Specific Spred3 Promoter Activity
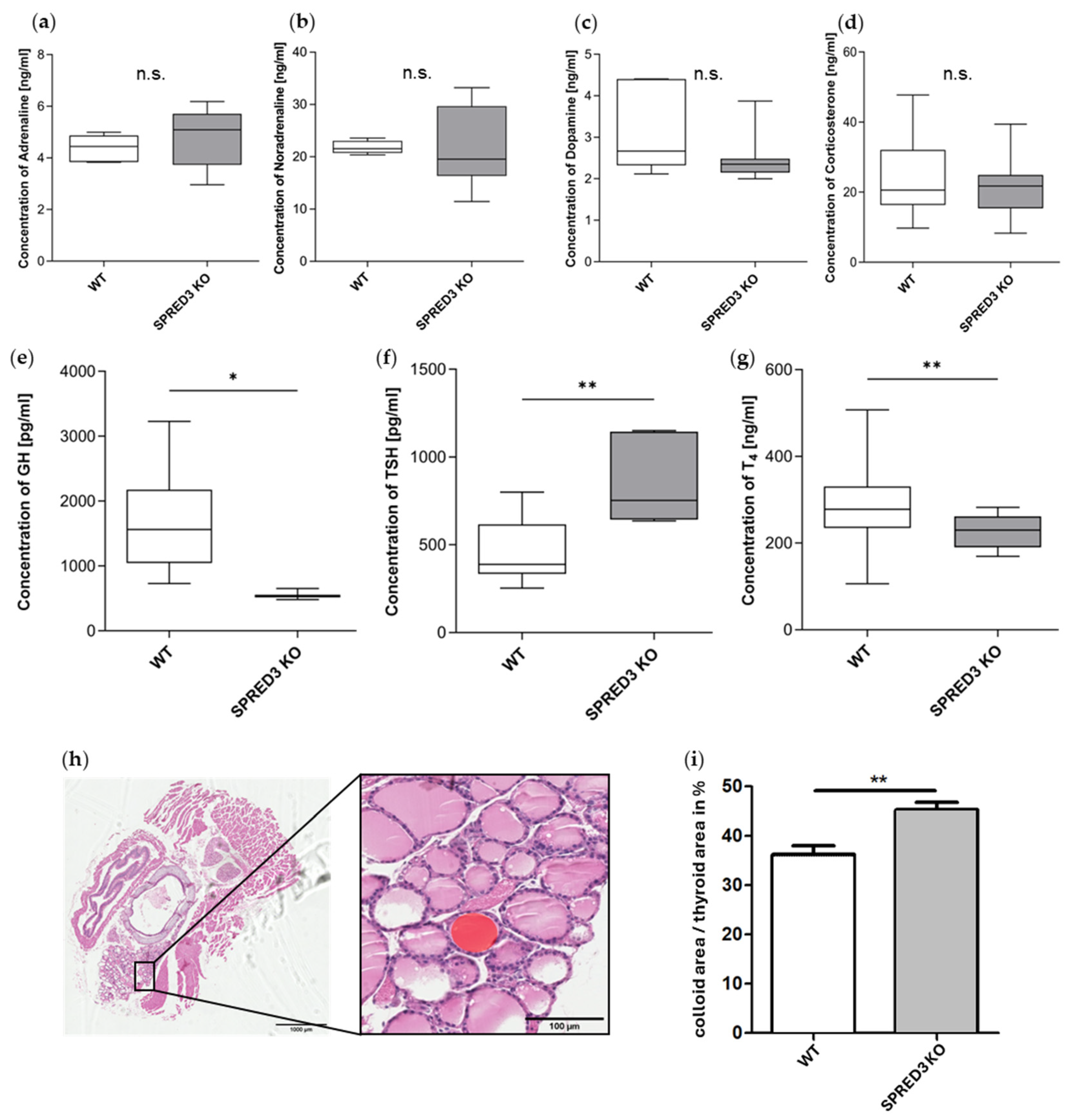
2.5. Hormonal Profiling Reveals Primary Hypothyroidism in SPRED3 KO Mice
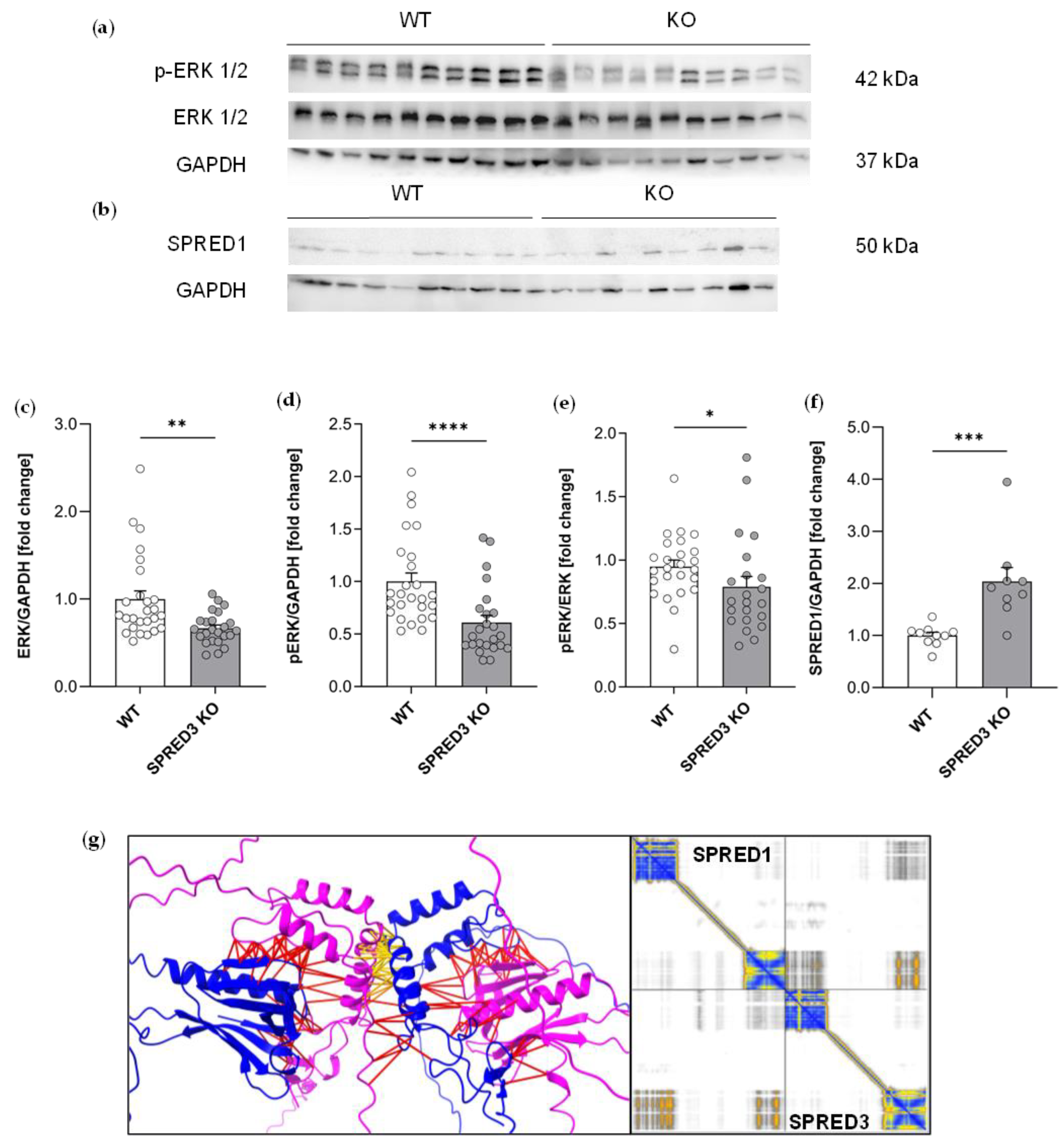
2.6. ERK Signaling in the Thyroid Gland Is Reduced in SPRED3 KO Mice
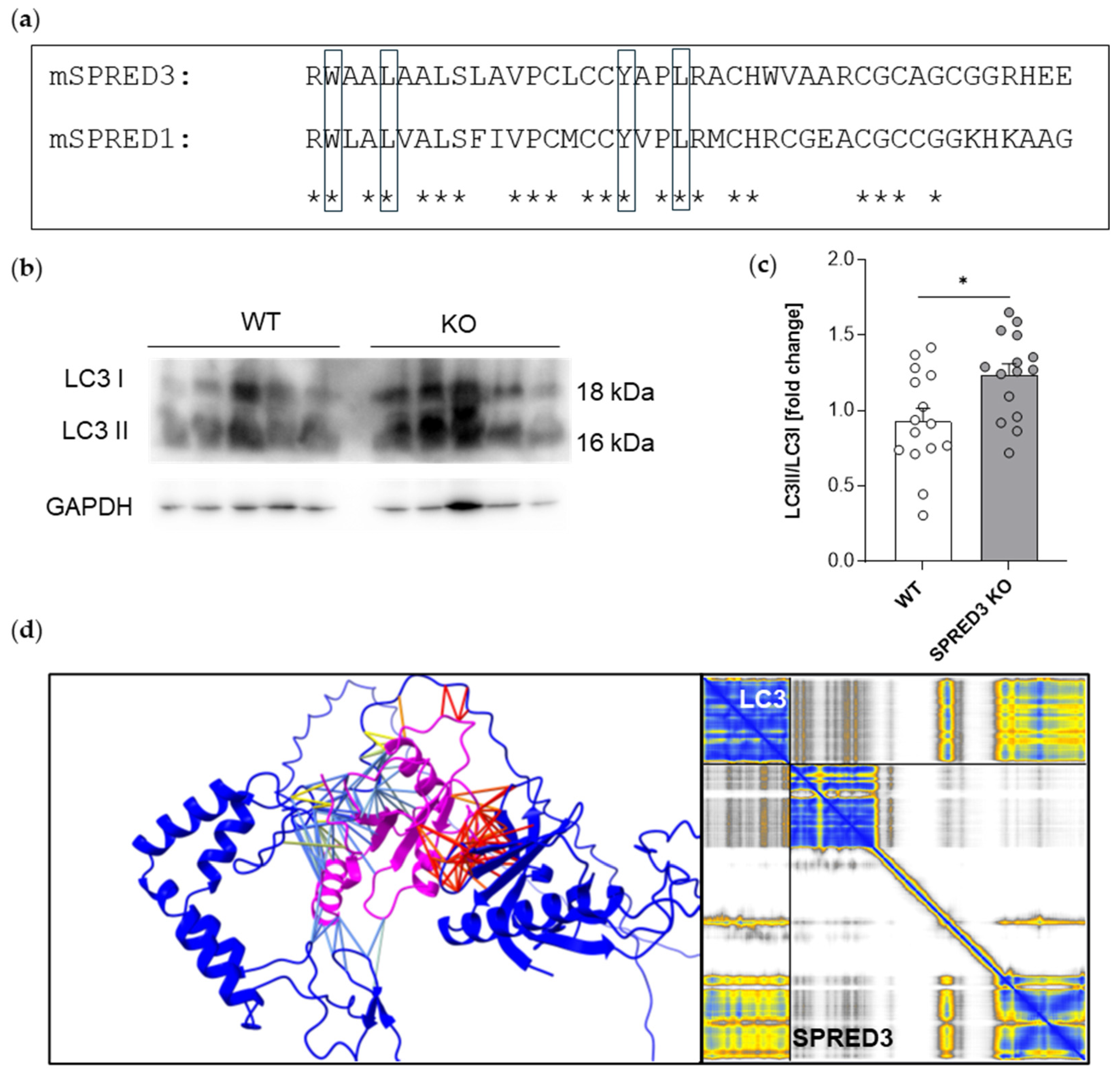
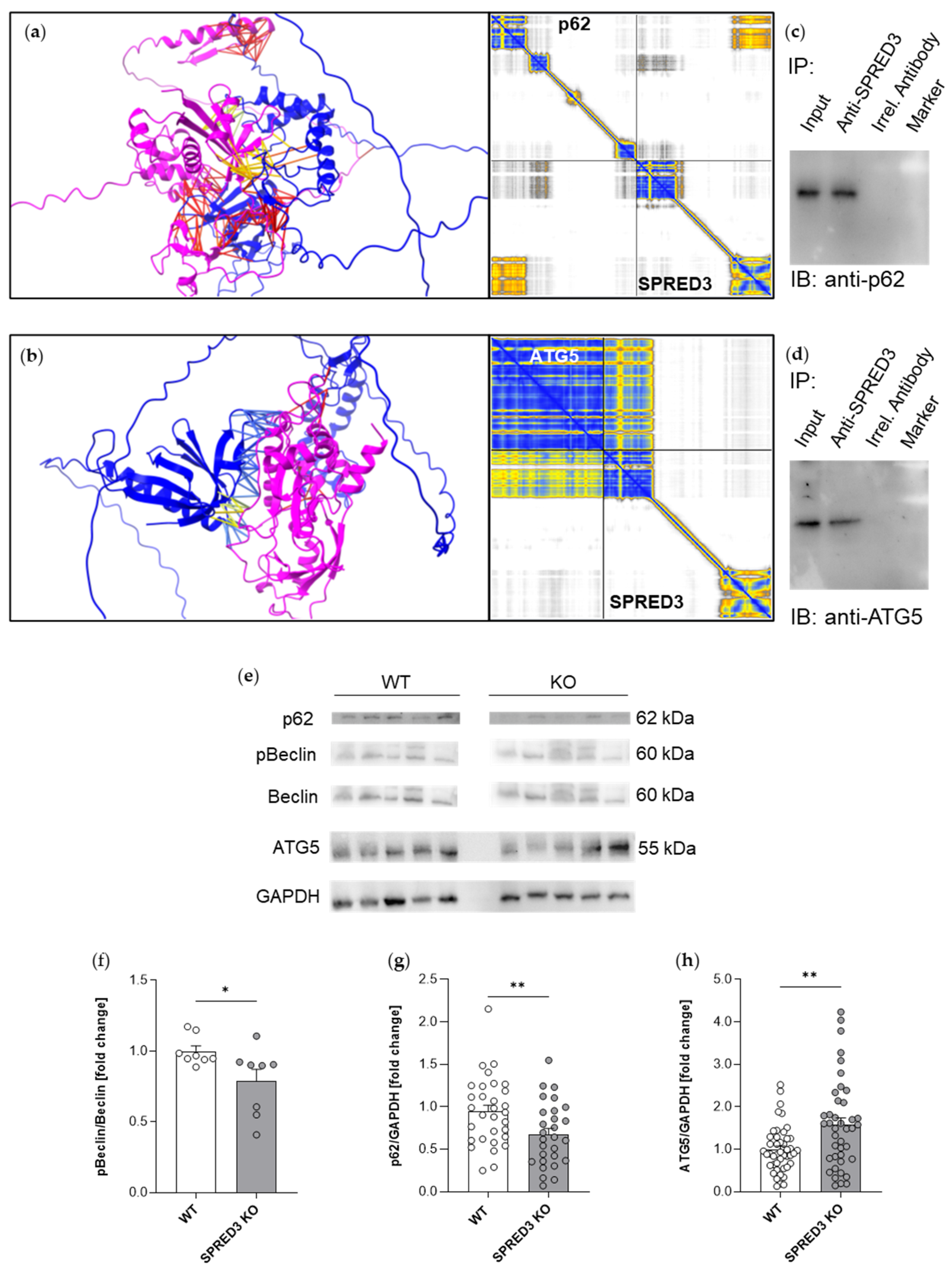
2.7. SPRED3 Deficiency Alters Autophagy-Related Protein Expression in the Thyroid Gland
3. Discussion
4. Materials and Methods
4.1. SPRED3 Mutant Mice
4.2. X-Gal Staining
4.3. Genotyping of Mice
4.4. RNA Extraction and RT-qPCR Analyses
4.5. Western Blotting and Densitometric Quantification of Detected Bands
4.6. Examination of Serum Hormone Levels
4.7. Invasive Hemodynamics
4.8. Co-Immunoprecipitation Assays
4.9. ChimeraX
4.10. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wakioka, T.; Sasaki, A.; Kato, R.; Shouda, T.; Matsumoto, A.; Miyoshi, K.; Tsuneoka, M.; Komiya, S.; Baron, R.; Yoshimura, A. Spred is a Sprouty-related suppressor of Ras signalling. Nature 2001, 412, 647–651. [Google Scholar] [CrossRef]
- Plotnikov, A.; Zehorai, E.; Procaccia, S.; Seger, R. The MAPK cascades: Signaling components, nuclear roles and mechanisms of nuclear translocation. Biochim. Biophys. Acta 2011, 1813, 1619–1633. [Google Scholar] [CrossRef] [PubMed]
- Yoon, S.; Seger, R. The extracellular signal-regulated kinase: Multiple substrates regulate diverse cellular functions. Growth Factors 2006, 24, 21–44. [Google Scholar] [CrossRef] [PubMed]
- Gong, J.; Yan, Z.; Liu, Q. Progress in experimental research on SPRED protein family. J. Int. Med. Res. 2020, 48, 300060520929170. [Google Scholar] [CrossRef]
- Motta, M.; Fasano, G.; Gredy, S.; Brinkmann, J.; Bonnard, A.A.; Simsek-Kiper, P.O.; Gulec, E.Y.; Essaddam, L.; Utine, G.E.; Guarnetti Prandi, I.; et al. SPRED2 loss-of-function causes a recessive Noonan syndrome-like phenotype. Am. J. Hum. Genet. 2021, 108, 2112–2129. [Google Scholar] [CrossRef]
- Lorenzo, C.; McCormick, F. SPRED proteins and their roles in signal transduction, development, and malignancy. Genes Dev. 2020, 34, 1410–1421. [Google Scholar] [CrossRef]
- Kato, R.; Nonami, A.; Taketomi, T.; Wakioka, T.; Kuroiwa, A.; Matsuda, Y.; Yoshimura, A. Molecular cloning of mammalian Spred-3 which suppresses tyrosine kinase-mediated Erk activation. Biochem. Biophys. Res. Commun. 2003, 302, 767–772. [Google Scholar] [CrossRef] [PubMed]
- Decock, A.; Ongenaert, M.; Cannoodt, R.; Verniers, K.; De Wilde, B.; Laureys, G.; Van Roy, N.; Berbegall, A.P.; Bienertova-Vasku, J.; Bown, N.; et al. Methyl-CpG-binding domain sequencing reveals a prognostic methylation signature in neuroblastoma. Oncotarget 2016, 7, 1960–1972. [Google Scholar] [CrossRef]
- Mizoguchi, M.; Nutt, C.L.; Louis, D.N. Mutation analysis of CBL-C and SPRED3 on 19q in human glioblastoma. Neurogenetics 2004, 5, 81–82. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Wang, C.; Li, M.; Cai, S.; Liu, X. SPRED3 regulates the NF-kappaB signaling pathway in thyroid cancer and promotes the proliferation. Sci. Rep. 2024, 14, 20506. [Google Scholar] [CrossRef]
- Zhang, X.; Meng, X.; Wang, P.; Luan, C.; Wang, H. Bioinformatics analysis for the identification of Sprouty-related EVH1 domain-containing protein 3 expression and its clinical significance in thyroid carcinoma. Sci. Rep. 2024, 14, 4549. [Google Scholar] [CrossRef]
- Rosina, E.; Pezzani, L.; Apuril, E.; Pezzoli, L.; Marchetti, D.; Bellini, M.; Lucca, C.; Meossi, C.; Massimello, M.; Mariani, M.; et al. Comparison of first-tier whole-exome sequencing with a multi-step traditional approach for diagnosing paediatric outpatients: An Italian prospective study. Mol. Genet. Genom. Med. 2024, 12, e2316. [Google Scholar] [CrossRef]
- Carvalho, D.P.; Dupuy, C. Thyroid hormone biosynthesis and release. Mol. Cell. Endocrinol. 2017, 458, 6–15. [Google Scholar] [CrossRef] [PubMed]
- Glick, D.; Barth, S.; Macleod, K.F. Autophagy: Cellular and molecular mechanisms. J. Pathol. 2010, 221, 3–12. [Google Scholar] [CrossRef]
- Robertson, E.; Bradley, A.; Kuehn, M.; Evans, M. Germ-line transmission of genes introduced into cultured pluripotential cells by retroviral vector. Nature 1986, 323, 445–448. [Google Scholar] [CrossRef]
- Thomas, K.R.; Capecchi, M.R. Site-directed mutagenesis by gene targeting in mouse embryo-derived stem cells. Cell 1987, 51, 503–512. [Google Scholar] [CrossRef]
- Skarnes, W.C.; Rosen, B.; West, A.P.; Koutsourakis, M.; Bushell, W.; Iyer, V.; Mujica, A.O.; Thomas, M.; Harrow, J.; Cox, T.; et al. A conditional knockout resource for the genome-wide study of mouse gene function. Nature 2011, 474, 337–342. [Google Scholar] [CrossRef] [PubMed]
- Bundschu, K.; Knobeloch, K.P.; Ullrich, M.; Schinke, T.; Amling, M.; Engelhardt, C.M.; Renne, T.; Walter, U.; Schuh, K. Gene disruption of Spred-2 causes dwarfism. J. Biol. Chem. 2005, 280, 28572–28580. [Google Scholar] [CrossRef]
- Ullrich, M.; Assmus, B.; Augustin, A.M.; Habich, H.; Abesser, M.; Martin Machado, J.; Werner, F.; Erkens, R.; Arias-Loza, A.P.; Umbenhauer, S.; et al. SPRED2 deficiency elicits cardiac arrhythmias and premature death via impaired autophagy. J. Mol. Cell. Cardiol. 2019, 129, 13–26. [Google Scholar] [CrossRef] [PubMed]
- Burn, S.F. Detection of beta-galactosidase activity: X-gal staining. Methods Mol. Biol. 2012, 886, 241–250. [Google Scholar] [CrossRef]
- Eichenbaum, H. The role of the hippocampus in navigation is memory. J. Neurophysiol. 2017, 117, 1785–1796. [Google Scholar] [CrossRef]
- Passingham, R.E.; Stephan, K.E.; Kotter, R. The anatomical basis of functional localization in the cortex. Nat. Rev. Neurosci. 2002, 3, 606–616. [Google Scholar] [CrossRef]
- Phillips, C.G.; Zeki, S.; Barlow, H.B. Localization of function in the cerebral cortex. Past, present and future. Brain 1984, 107, 327–361. [Google Scholar] [CrossRef]
- Bombardi, C. Neuronal localization of the 5-HT2 receptor family in the amygdaloid complex. Front. Pharmacol. 2014, 5, 68. [Google Scholar] [CrossRef]
- Wallace, M.A. Anatomy and physiology of the kidney. AORN J. 1998, 68, 800, 803–816, 819–820, quiz 821-4. [Google Scholar] [CrossRef] [PubMed]
- Brun, J.H. Physiology of the adrenal gland. Br. J. Anaesth. 1956, 28, 459–469. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Choi, M.H. Embryonic Development and Adult Regeneration of the Adrenal Gland. Endocrinol. Metab. 2020, 35, 765–773. [Google Scholar] [CrossRef] [PubMed]
- Engelhardt, C.M.; Bundschu, K.; Messerschmitt, M.; Renne, T.; Walter, U.; Reinhard, M.; Schuh, K. Expression and subcellular localization of Spred proteins in mouse and human tissues. Histochem. Cell Biol. 2004, 122, 527–538. [Google Scholar] [CrossRef]
- Ullrich, M.; Bundschu, K.; Benz, P.M.; Abesser, M.; Freudinger, R.; Fischer, T.; Ullrich, J.; Renne, T.; Walter, U.; Schuh, K. Identification of SPRED2 (sprouty-related protein with EVH1 domain 2) as a negative regulator of the hypothalamic-pituitary-adrenal axis. J. Biol. Chem. 2011, 286, 9477–9488. [Google Scholar] [CrossRef]
- Cuttler, L. The regulation of growth hormone secretion. Endocrinol. Metab. Clin. N. Am. 1996, 25, 541–571. [Google Scholar] [CrossRef] [PubMed]
- Zaballos, M.A.; Santisteban, P. Key signaling pathways in thyroid cancer. J. Endocrinol. 2017, 235, R43–R61. [Google Scholar] [CrossRef] [PubMed]
- Jiang, K.; Liu, M.; Lin, G.; Mao, B.; Cheng, W.; Liu, H.; Gal, J.; Zhu, H.; Yuan, Z.; Deng, W.; et al. Tumor suppressor Spred2 interaction with LC3 promotes autophagosome maturation and induces autophagy-dependent cell death. Oncotarget 2016, 7, 25652–25667. [Google Scholar] [CrossRef]
- Mizushima, N.; Komatsu, M. Autophagy: Renovation of cells and tissues. Cell 2011, 147, 728–741. [Google Scholar] [CrossRef] [PubMed]
- Murrow, L.; Debnath, J. Autophagy as a stress-response and quality-control mechanism: Implications for cell injury and human disease. Annu. Rev. Pathol. 2013, 8, 105–137. [Google Scholar] [CrossRef]
- Cohen, M.S.; Hadjivassiliou, H.; Taunton, J. A clickable inhibitor reveals context-dependent autoactivation of p90 RSK. Nat. Chem. Biol. 2007, 3, 156–160. [Google Scholar] [CrossRef]
- Sarg, N.H.; Zaher, D.M.; Abu Jayab, N.N.; Mostafa, S.H.; Ismail, H.H.; Omar, H.A. The interplay of p38 MAPK signaling and mitochondrial metabolism, a dynamic target in cancer and pathological contexts. Biochem. Pharmacol. 2024, 225, 116307. [Google Scholar] [CrossRef]
- Keyse, S.M. Dual-specificity MAP kinase phosphatases (MKPs) and cancer. Cancer Metastasis Rev. 2008, 27, 253–261. [Google Scholar] [CrossRef] [PubMed]
- Klionsky, D.J.; Abdel-Aziz, A.K.; Abdelfatah, S.; Abdellatif, M.; Abdoli, A.; Abel, S.; Abeliovich, H.; Abildgaard, M.H.; Abudu, Y.P.; Acevedo-Arozena, A.; et al. Guidelines for the use and interpretation of assays for monitoring autophagy (4th edition)(1). Autophagy 2021, 17, 1–382. [Google Scholar] [CrossRef] [PubMed]
- Maday, S.; Wallace, K.E.; Holzbaur, E.L. Autophagosomes initiate distally and mature during transport toward the cell soma in primary neurons. J. Cell Biol. 2012, 196, 407–417. [Google Scholar] [CrossRef]
- Pankiv, S.; Clausen, T.H.; Lamark, T.; Brech, A.; Bruun, J.A.; Outzen, H.; Overvatn, A.; Bjorkoy, G.; Johansen, T. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J. Biol. Chem. 2007, 282, 24131–24145. [Google Scholar] [CrossRef] [PubMed]
- Margineanu, A.; Chan, J.J.; Kelly, D.J.; Warren, S.C.; Flatters, D.; Kumar, S.; Katan, M.; Dunsby, C.W.; French, P.M. Screening for protein-protein interactions using Forster resonance energy transfer (FRET) and fluorescence lifetime imaging microscopy (FLIM). Sci. Rep. 2016, 6, 28186. [Google Scholar] [CrossRef] [PubMed]
- Ullrich, M.; Schuh, K. Gene trap: Knockout on the fast lane. Methods Mol. Biol. 2009, 561, 145–159. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Meng, E.C.; Couch, G.S.; Croll, T.I.; Morris, J.H.; Ferrin, T.E. UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci. 2021, 30, 70–82. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapen, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]

| Hemodynamic Parameters (Isoflurane Anesthesia, ±SEM) | WT (n = 13) | SPRED3-KO (n = 12) |
|---|---|---|
| HR (min−1) | 488 ± 12.5 | 497 ± 17.8 |
| SV (µL) | 15.3 ± 0.6 | 15.5 ± 0.7 |
| ESV (µL) | 17.4 ± 1.2 | 16.5 ± 1.5 |
| EDV (µL) | 30.8 ± 1.6 | 30.5 ± 1.3 |
| EF (%) | 49.6 ± 2.1 | 51.9 ± 2.9 |
| CO (µL min−1) | 7426.6 ± 212.9 | 7807.6 ± 542.2 |
| ESP (mmHg) | 95.6 ± 2.7 | 94.8 ± 3.5 |
| EDP (mmHg) | 8.5 ± 1.3 | 6.4 ± 0.7 |
| SW (mmHg × µL) | 1208.0 ± 63.6 | 1255.3 ± 70.8 |
| V@dP/dt max (µL) | 31.3 ± 1.4 | 30.4 ± 1.2 |
| V@dP/dt min (µL) | 16.9 ± 1.2 | 16.0 ± 1.3 |
| Ea (mmHg µL−1) | 6.3 ± 0.3 | 6.2 ± 0.4 |
| Systolic indices | ||
| dP/dt max (mmHg s−1) | 7657.1 ± 263.5 | 8465.8 ± 682.4 |
| dV/dt max (µL s−1) | 642.9 ± 25.3 | 693.2 ± 37.8 |
| Diastolic indices | ||
| -dP/dt min (-mmHg s−1) | 8385.2 ± 426.8 | 9284.7 ± 637.0 |
| Tau (W) (ms) | 6.7 ± 0.4 | 5.9 ± 0.4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dogan, C.; Haas, L.; Holzapfel, R.; Schmitt, F.; Hepbasli, D.; Ullrich, M.; Bösl, M.R.; Abeßer, M.; Schuh, K.; Gredy, S. Loss of SPRED3 Causes Primary Hypothyroidism and Alters Thyroidal Expression of Autophagy Regulators LC3, p62, and ATG5 in Mice. Int. J. Mol. Sci. 2025, 26, 7660. https://doi.org/10.3390/ijms26157660
Dogan C, Haas L, Holzapfel R, Schmitt F, Hepbasli D, Ullrich M, Bösl MR, Abeßer M, Schuh K, Gredy S. Loss of SPRED3 Causes Primary Hypothyroidism and Alters Thyroidal Expression of Autophagy Regulators LC3, p62, and ATG5 in Mice. International Journal of Molecular Sciences. 2025; 26(15):7660. https://doi.org/10.3390/ijms26157660
Chicago/Turabian StyleDogan, Celine, Luisa Haas, Rebecca Holzapfel, Franziska Schmitt, Denis Hepbasli, Melanie Ullrich, Michael R. Bösl, Marco Abeßer, Kai Schuh, and Sina Gredy. 2025. "Loss of SPRED3 Causes Primary Hypothyroidism and Alters Thyroidal Expression of Autophagy Regulators LC3, p62, and ATG5 in Mice" International Journal of Molecular Sciences 26, no. 15: 7660. https://doi.org/10.3390/ijms26157660
APA StyleDogan, C., Haas, L., Holzapfel, R., Schmitt, F., Hepbasli, D., Ullrich, M., Bösl, M. R., Abeßer, M., Schuh, K., & Gredy, S. (2025). Loss of SPRED3 Causes Primary Hypothyroidism and Alters Thyroidal Expression of Autophagy Regulators LC3, p62, and ATG5 in Mice. International Journal of Molecular Sciences, 26(15), 7660. https://doi.org/10.3390/ijms26157660






