High-Resolution Mass Spectrometry Method for Targeted Screening and Monitoring of Fabry, Gaucher and ASMD Using Dried Blood Spots and Capitainers: Impact of Sample Matrix on Measurement Results
Abstract
1. Introduction
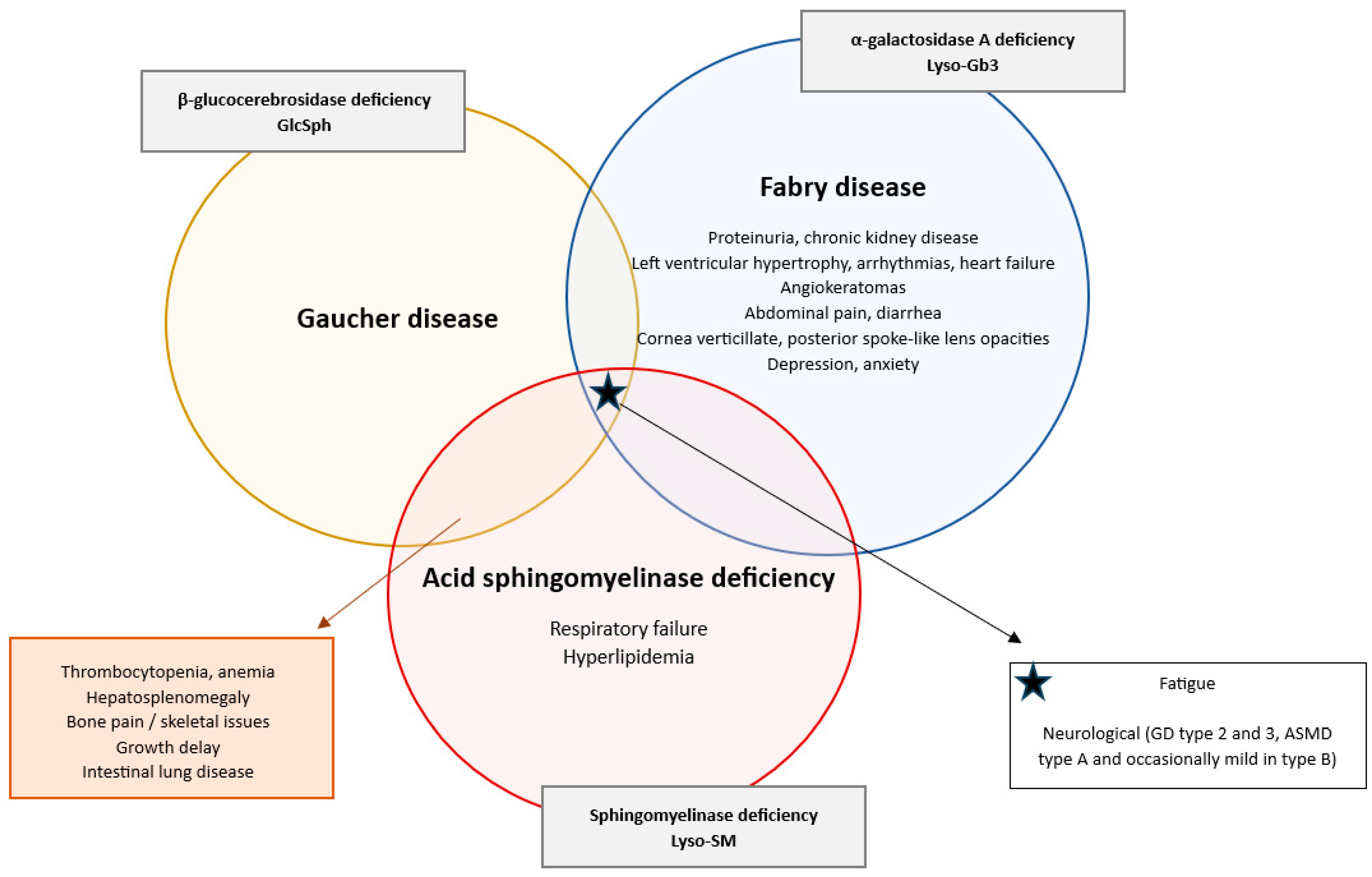
2. Results
2.1. The LC-MS/MS Method
2.2. Analytical Validation
2.3. Case Finding Results
3. Discussion
4. Materials and Methods
4.1. Chemicals and Reagents
4.2. Standards, Internal Standards and Quality Controls
4.3. Sample Preparation
4.4. Liquid Chromatography with Tandem Mass Spectrometry
4.5. Method Validation
4.5.1. Linearity
4.5.2. Precision
4.5.3. Accuracy
4.5.4. Carry-Over
4.5.5. Lower Limit of Quantification
4.5.6. Case Finding
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
List of Abbreviations
| LC-MS/MS | Liquid chromatography with tandem mass spectrometry |
| CLSI | Clinical and Laboratory Standards Institute |
| LSD | Lysosomal storage diseases |
| GD | Gaucher disease |
| FD | Fabry disease |
| ASMD | Acid sphingomyelin deficiency |
| Gb1 | Glycosphingolipid glucosylceramide |
| GlcSph | Lyso-glucosylsphingosine |
| Gb3 | Globotriaosylceramide |
| Lyso-Gb3 | Globotriaosylsphingosine |
| Lyso-SM | Lyso-sphingomyelin |
| ERT | Enzyme replacement therapy |
| SRT | Substrate reduction therapy |
| DNA | Desoxyribonucleic acid |
| CCL18 | C-C motif chemokine ligand 18 |
| DBS | Dried blood spot |
| QC | Quality control |
| DMSO | Dimethylsulfoxide |
| MeOH | Methanol |
| RBCs | Red blood cells |
| K3-EDTA | K3 potassium salt of ethylene diamine tetra acetic acid |
| WBCs | White blood cells |
| ESI | Electrospray ionization |
| MRM | Multiple reaction monitoring |
| IS | Internal standard |
| LOQ | Limit of quantification |
| CV% | Coefficient of variance |
| SD | Standard deviation |
| S/N | Signal-to-noise ratio |
List of Human Genes
| GBA1 | Glucosylceramidase beta 1, HGNC ID 4177 |
| Alias symbols | GBA, GCB, GLUC, alias names: glucocerebrosidase |
| GLA | Galactosidase alpha, HGNC ID 4296 |
| Alias symbols | GALA |
| SMPD1 | Sphingomyelin phosphodiesterase 1, HGNC:11120, OMIM 607616 |
| Alias symbols | ASM alias names: acid sphingomyelinase, Niemann–Pick type A/B |
References
- Cozma, C.; Cullufi, P.; Kramp, G.; Hovakimyan, M.; Velmishi, V.; Gjikopulli, A.; Tomori, S.; Fischer, S.; Oppermann, S.; Grittner, U.; et al. Treatment Efficiency in Gaucher Patients Can Reliably Be Monitored by Quantification of Lyso-Gb1 Concentrations in Dried Blood Spots. Int. J. Mol. Sci. 2020, 21, 4577. [Google Scholar] [CrossRef]
- Dinur, T.; Bauer, P.; Beetz, C.; Kramp, G.; Cozma, C.; Iurașcu, M.-I.; Becker-Cohen, M.; Istaiti, M.; Rolfs, A.; Zimran, A.; et al. Gaucher Disease Diagnosis Using Lyso-Gb1 on Dry Blood Spot Samples: Time to Change the Paradigm? Int. J. Mol. Sci. 2022, 23, 1627. [Google Scholar] [CrossRef]
- Revel-Vilk, S.; Fuller, M.; Zimran, A. Value of Glucosylsphingosine (Lyso-Gb1) as a Biomarker in Gaucher Disease: A Systematic Literature Review. Int. J. Mol. Sci. 2020, 21, 7159. [Google Scholar] [CrossRef]
- Talbot, A.; Nicholls, K.; Fletcher, J.M.; Fuller, M. A simple method for quantification of plasma globotriaosylsphingosine: Utility for Fabry disease. Mol. Genet. Metab. 2017, 122, 121–125. [Google Scholar] [CrossRef]
- Nowak, A.; Mechtler, T.P.; Desnick, R.J.; Kasper, D.C. Plasma LysoGb3: A useful biomarker for the diagnosis and treatment of Fabry disease heterozygotes. Mol. Genet. Metab. 2017, 120, 57–61. [Google Scholar] [CrossRef]
- Breilyn, M.S.; Zhang, W.; Yu, C.; Wasserstein, M.P. Plasma lyso-sphingomyelin levels are positively associated with clinical severity in acid sphingomyelinase deficiency. Mol. Genet. Metab. Rep. 2021, 28, 100780. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Breiden, B.; Sandhoff, K. Acid Sphingomyelinase, a Lysosomal and Secretory Phospholipase C, Is Key for Cellular Phospholipid Catabolism. Int. J. Mol. Sci. 2021, 22, 9001. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Wenger, D.A.; Coppola, S.; Liu, S.L. Lysosomal storage disorders: Diagnostic dilemmas and prospects for therapy. Genet. Med. 2002, 4, 412–419. [Google Scholar] [CrossRef] [PubMed]
- Beutler, E.; Grabowski, G.A. Gaucher Disease. In The Metabolic and Molecular Bases of Inherited Disease, 8th ed.; Scriver, C., Beaudet, A.L., Sly, W.S., Valle, D., Eds.; McGraw-Hill: New York, NY, USA, 2001; Volume 3, pp. 3635–3668. [Google Scholar] [CrossRef]
- Stirnemann, J.; Belmatoug, N.; Camou, F.; Serratrice, C.; Froissart, R.; Caillaud, C.; Levade, T.; Astudillo, L.; Serratrice, J.; Brassier, A.; et al. A review of gaucher disease pathophysiology, clinical presentation and treatments. Int. J. Mol. Sci. 2017, 18, 441. [Google Scholar] [CrossRef] [PubMed]
- Desnick, R.J.; Ioannou, Y.A.; Eng, C.M. α-Falactosidase Deficiency: Fabry Disease. In The Metabolic and Molecular Bases of Inherited Disease, 8th ed.; Scriver, C., Beaudet, A.L., Sly, W.S., Valle, D., Eds.; McGraw-Hill: New York, NY, USA, 2001; Volume 3, pp. 3733–3774. [Google Scholar] [CrossRef]
- Bernardes, T.; Foresto, R.; Kirsztajn, G. Fabry disease: Genetics, pathology, and treatment. Rev. Assoc. Med. Bras. 2020, 66, 10–16. [Google Scholar] [CrossRef] [PubMed]
- Gragnaniello, V.; Burlina, A.P.; Polo, G.; Giuliani, A.; Salviati, L.; Duro, G.; Cazzorla, C.; Rubert, L.; Maines, E.; Germain, D.P.; et al. Newborn screening for fabry disease in northeastern Italy: Results of five years of experience. Biomolecules 2021, 11, 951. [Google Scholar] [CrossRef]
- Gragnaniello, V.; Burlina, A.P.; Polo, G.; Giuliani, A.; Salviati, L.; Duro, G.; Cazzorla, C.; Rubert, L.; Maines, E.; Germain, D.P.; et al. Fabry disease: A review of current management strategies. QJM 2010, 103, 641–659. [Google Scholar] [CrossRef]
- Delarosa-Rodríguez, R.; Santotoribio, J.D.; Paula, H.A.; González-Meneses, A.; García-Morillo, S.; Jiménez-Arriscado, P.; Guerrero, J.M.; Macher, H.C. Accuracy diagnosis improvement of fabry disease from dried blood spots: Enzyme activity, lyso-Gb3 accumulation and GLA gene sequencing. Clin. Genet. 2021, 99, 761–771. [Google Scholar] [CrossRef] [PubMed]
- De Brabander, I.; Yperzeele, L.; Ceuterick-De Groote, C.; Brouns, R.; Baker, R.; Belachew, S.; Delbecq, J.; De Keulenaer, G.; Dethy, S.; Eyskens, F.; et al. Phenotypical characterization of α-galactosidase A gene mutations identified in a large fabry disease screening program in stroke in the young. Clin. Neurol. Neurosurg. 2013, 115, 1088–1093. [Google Scholar] [CrossRef] [PubMed]
- Schuchman, E.H.; Desnick, R.J. Types A and B Niemann-Pick disease. Mol. Genet. Metab. 2017, 120, 27–33. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Sandra, D.K.K.; Bodamer, O.A.; Wijburg, F.A. Epidemiology and diagnosis of lysosomal storage disorders; challenges of screening. Best Pract. Res. Clin. Endocrinol. Metab. 2015, 29, 145–157. [Google Scholar] [CrossRef]
- McGovern, M.M.; Avetisyan, R.; Sanson, B.J.; Lidove, O. Disease manifestations and burden of illness in patients with acid sphingomyelinase deficiency (ASMD). Orphanet J. Rare Dis. 2017, 12, 41. [Google Scholar] [CrossRef]
- Giacomarra, M.; Colomba, P.; Francofonte, D.; Zora, M.; Caocci, G.; Diomede, D.; Giuffrida, G.; Fiori, L.; Montanari, C.; Sapuppo, A.; et al. Gaucher disease or acid sphingomyelinase deficiency? The importance of differential diagnosis. J. Clin. Med. 2024, 13, 1487. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Cappellini, M.D.; Motta, I.; Barbato, A.; Giuffrida, G.; Manna, R.; Carubbi, F.; Giona, F. Similarities and differences between Gaucher disease and acid sphingomyelinase deficiency: An algorithm to support the diagnosis. Eur. J. Intern. Med. 2023, 108, 81–84. [Google Scholar] [CrossRef] [PubMed]
- Hollak, C.E.; de Sonnaville, E.S.; Cassiman, D.; Linthorst, G.E.; Groener, J.E.; Morava, E.; Wevers, R.A.; Mannens, M.; Aerts, J.M.; Meersseman, W.; et al. Acid sphingomyelinase (Asm) deficiency patients in The Netherlands and Belgium: Disease spectrum and natural course in attenuated patients. Mol. Genet. Metab. 2012, 107, 526–533. [Google Scholar] [CrossRef] [PubMed]
- Chuang, W.L.; Pacheco, J.; Cooper, S.; McGovern, M.M.; Cox, G.F.; Keutzer, J.; Zhang, X.K. Lyso-sphingomyelin is elevated in dried blood spots of Niemann-Pick B patients. Mol. Genet. Metab. 2014, 111, 209–211. [Google Scholar] [CrossRef]
- Deodato, F.; Boenzi, S.; Taurisano, R.; Semeraro, M.; Sacchetti, E.; Carrozzo, R.; Dionisi-Vici, C. The impact of biomarkers analysis in the diagnosis of Niemann-Pick C disease and acid sphingomyelinase deficiency. Clin. Chim. Acta. 2018, 486, 387–394. [Google Scholar] [CrossRef] [PubMed]
- Thomas; Mehta, A.; Hughes, D. Diagnosing Gaucher disease: An on-going need for increased awareness amongst haematologists. Blood Cells Mol. Dis. 2013, 50, 212–217. [Google Scholar] [CrossRef] [PubMed]
- Hurvitz, N.; Dinur, T.; Becker-Cohen, M.; Cozma, C.; Hovakimyan, M.; Oppermann, S.; Demuth, L.; Rolfs, A.; Abramov, A.; Zimran, A.; et al. Glucosylsphingosine (lyso-gb1) as a biomarker for monitoring treated and untreated children with gaucher disease. Int. J. Mol. Sci. 2019, 20, 3033. [Google Scholar] [CrossRef]
- Dekker, N.; van Dussen, L.; Hollak, C.E.; Overkleeft, H.; Scheij, S.; Ghauharali, K.; van Breemen, M.J.; Ferraz, M.J.; Groener, J.E.; Maas, M.; et al. Elevated plasma glucosylsphingosine in gaucher disease: Relation to phenotype, storage cell markers, and therapeutic response. Blood 2011, 118, 118–127. [Google Scholar] [CrossRef]
- Kleytman, N.; Ruan, J.; Ruan, A.; Zhang, B.; Murugesan, V.; Lin, H.; Guo, L.; Klinger, K.; Mistry, P.K. Incremental biomarker and clinical outcomes after switch from enzyme therapy to eliglustat substrate reduction therapy in gaucher disease. Mol. Genet. Metab. Rep. 2021, 29, 100798. [Google Scholar] [CrossRef]
- Rolfs, A.; Giese, A.K.; Grittner, U.; Mascher, D.; Elstein, D.; Zimran, A.; Böttcher, T.; Lukas, J.; Hübner, R.; Gölnitz, U.; et al. Glucosylsphingosine is a highly sensitive and specific biomarker for primary diagnostic and follow-up monitoring in gaucher disease in a non-jewish, caucasian cohort of gaucher disease patients. PLoS ONE 2013, 8, e79732. [Google Scholar] [CrossRef]
- Smid, B.E.; van der Tol, L.; Biegstraaten, M.; Linthorst, G.E.; Hollak, C.E.; Poorthuis, B.J. Plasma globotriaosylsphingosine in relation to phenotypes of fabry disease. J. Med. Genet. 2015, 52, 262–268. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, Y.; Chen, B.; Pan, X.; Wang, Z.; Ren, H.; Xu, Y.; Ni, L.; Yu, X.; Yang, L.; Chen, N. Clinical significance of plasma globotriaosylsphingosine levels in chinese patients with fabry disease. Exp. Ther. Med. 2018, 15, 3733–3742. [Google Scholar] [CrossRef]
- Malvagia, S.; Ferri, L.; Della Bona, M.; Borsini, W.; Cirami, C.L.; Dervishi, E.; Feriozzi, S.; Gasperini, S.; Motta, S.; Mignani, R.; et al. Multicenter evaluation of use of dried blood spot compared to conventional plasma in measurements of globotriaosylsphingosine (lysoGb3) concentration in 104 fabry patients. Clin. Chem. Lab. Med. 2021, 59, 1516–1526. [Google Scholar] [CrossRef]
- Burlina, A.B.; Polo, G.; Rubert, L.; Gueraldi, D.; Cazzorla, C.; Duro, G.; Salviati, L.; Burlina, A.P. Implementation of second-tier tests in newborn screening for lysosomal disorders in north eastern Italy. Int. J. Neonatal Screen 2019, 5, 24. [Google Scholar] [CrossRef]
- Hannon, W.H.; Ronald, W.; Fernhoff, P.; Halonen, T.; Lavochkin, M. Blood Collection on Filter Paper for Newborn Screening Programs; Approved Standard-Sixth Edition. CLSI Document NBS01-A6 from the Clinical & Laboratory Standards Institute 33, 2013, pp. 1–52. NBS01-A6: Blood Collection on Filter Paper for Newborn Screening Programs; Approved Standard—Sixth Edition (clsi.org). Available online: https://www.researchgate.net/publication/288949889_Blood_Collection_on_Filter_Paper_for_Newborn_Screening_Programs_Approved_Standard-Fifth_Edition (accessed on 14 November 2023).
- Capitainer®B–Quantitative Dried Blood Spot Sampling. Available online: https://capitainer.com/capitainerb-in-multi-panel-drug-testing/ (accessed on 27 May 2025).
- Van Baelen, A.; Roosens, L.; Devos, S.; Verhulst, S.; Eyskens, F. A new multiplex analysis of glucosylsphingosine and globotriaosylsphingosine in dried blood spots by tandem mass spectrometry. Mol. Genet. Metab. Rep. 2023, 37, 100993. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Van Baelen, A.; Verhulst, S.; Eyskens, F. Unexplained splenomegaly as a diagnostic marker for a rare but severe disease with an innovative and highly effective new treatment option: A case report. Mol. Genet. Metab. Rep. 2024, 41, 101144. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Van Kuilenburg, A.B.P.; Pop, A.; Poorthuis, B.J.H.M.; Goorden, S.M.I. Sphingolipids. In Laboratory Guide to the Methods in Biochemical Genetics; Blau, N., Vaz, F.M., Eds.; Springer: Cham, Switzeland, 2024; pp. 211–220. [Google Scholar] [CrossRef]
- Stiles, A.R.; Zhang, H.; Dai, J.; McCaw, P.; Beasley, J.; Rehder, C.; Koeberl, D.D.; McDonald, M.; Bali, D.S.; Young, S.P. A comprehensive testing algorithm for the diagnosis of Fabry disease in males and females. Mol. Genet. Metab. 2020, 130, 209–214. [Google Scholar] [CrossRef] [PubMed]
- Baydakova, G.; Ilyushkina, A.; Gaffke, L.; Pierzynowska, K.; Bychkov, I.; Ługowska, A.; Wegrzyn, G.; Tylki-Szymanska, A.; Zakharova, E. Elevated LysoGb3 in the neuronopathic forms of mucopolysaccharidoses. Diagnostics 2020, 10, 155. [Google Scholar] [CrossRef]
- Geberhiwot, T.; Wasserstein, M.; Wanninayake, S.; Bolton, S.C.; Dardis, A.; Lehman, A.; Lidove, O.; Dawson, C.; Giugliani, R.; Imrie, J.; et al. Consensus clinical management guidelines for acid sphingomyelinase deficiency (Niemann-Pick disease types A, B and A/B). Orphanet. J. Rare Dis. 2023, 18, 85. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Wasserstein, M.P.; Lachmann, R.; Hollak, C.; Barbato, A.; Gallagher, R.C.; Giugliani, R.; Guelbert, N.B.; Hennermann, J.B.; Ikezoe, T.; Lidove, O.; et al. Continued improvement in disease manifestations of acid sphingomyelinase deficiency for adults with up to 2 years of olipudase alfa treatment: Open-label extension of the ASCEND trial. Orphanet. J. Rare Dis. 2023, 18, 378. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Thangavelu, M.U.; Wouters, B.; Kindt, A.; Reiss, I.K.M.; Hankemeier, T. Blood microsampling technologies: Innovations and applications in 2022. Anal. Sci. Adv. 2023, 4, 154–180. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Boemer, F.; Hovhannesyan, K.; Piazzon, F.; Minner, F.; Mni, M.; Jacquemin, V.; Mashhadizadeh, D.; Benmhammed, N.; Bours, V.; Jacquinet, A.; et al. Population-based, first-tier genomic newborn screening in the maternity ward. Nat. Med. 2025, 31, 1339–1350. [Google Scholar] [CrossRef]
- Gragnaniello, V.; Cazzorla, C.; Gueraldi, D.; Puma, A.; Loro, C.; Porcù, E.; Stornaiuolo, M.; Miglioranza, P.; Salviati, L.; Burlina, A.P.; et al. Light and Shadows in Newborn Screening for Lysosomal Storage Disorders: Eight Years of Experience in Northeast Italy. Int. J. Neonatal. Screen. 2023, 10, 3. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
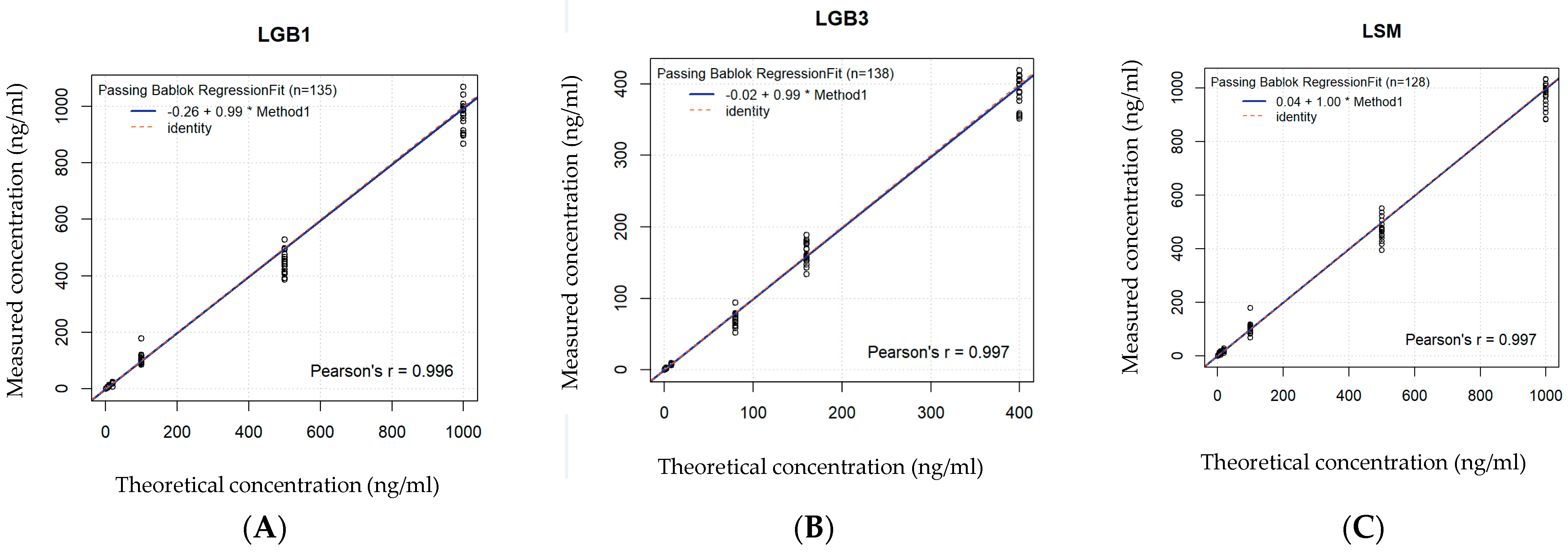

| DBS | R2 | Spearman’s p | Significant with | Slope | Regression Equation | Conclusion |
|---|---|---|---|---|---|---|
| GlcSph | 0.977 | 0.988 | <0.001 | 0.99 | 0.99x − 0.26 | Good significant correlation |
| Lyso-Gb3 | 0.973 | 0.986 | <0.001 | 0.99 | 0.99x − 0.02 | Good significant correlation |
| Lyso-SM 509 | 0.963 | 0.981 | <0.001 | 1 | 1x + 0.04 | Good significant correlation |
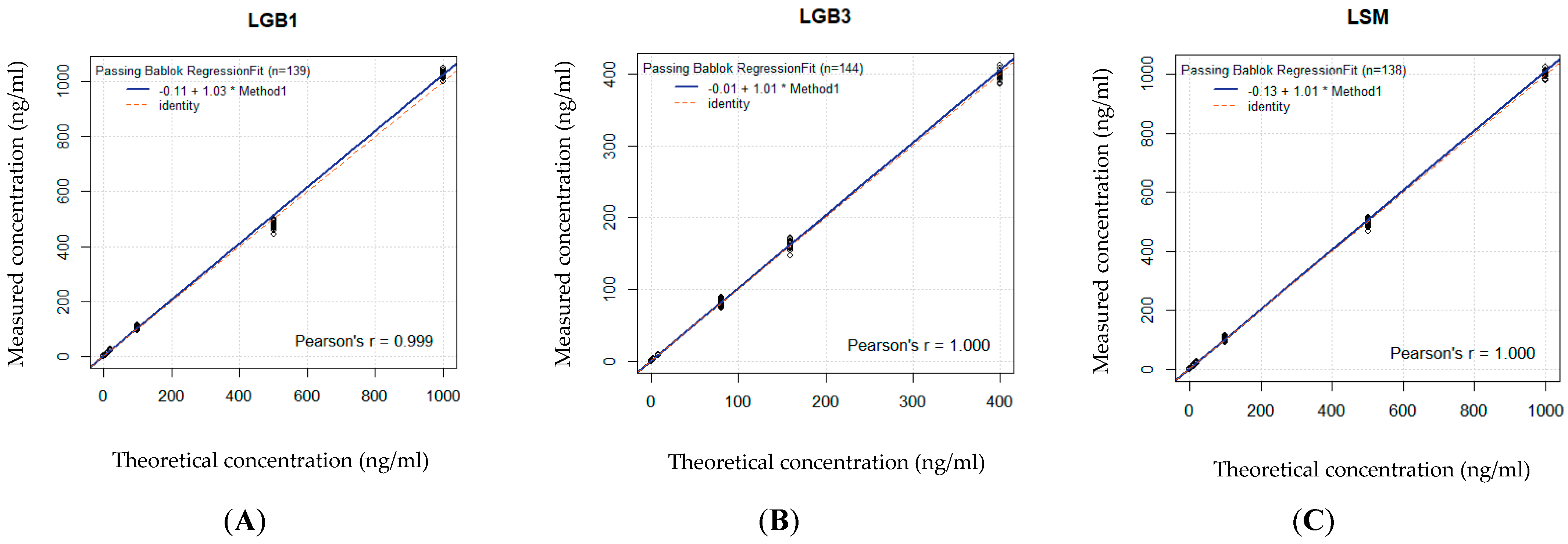
| CAP | R2 | Spearman’s p | Significant with | Slope | Regression equation | Conclusion |
| GlcSph | 0.979 | 0.989 | <0.001 | 1.03 | 1.03x − 0.11 | Good significant correlation |
| Lyso-Gb3 | 0.978 | 0.989 | <0.001 | 1.01 | 1.01x − 0.01 | Good significant correlation |
| Lyso-SM 509 | 0.979 | 0.989 | <0.001 | 1.01 | 1.01x − 0.13 | Good significant correlation |
| GlcSph | QC Level | Precision, CV% Intra-Assay DBS | Precision, CV% Inter-Assay DBS | Precision, CV% Intra-Assay CAP | Precision, CV% Inter-Assay CAP |
|---|---|---|---|---|---|
| QC 1 | 12.4 | 10 | 10 | 4.9 | |
| QC 2 | 9.7 | 7.9 | 5.3 | 6.2 | |
| QC 3 | 9.6 | 6.6 | 4.6 | 9.3 | |
| QC 4 | 5.5 | 5 | 5.1 | 7 | |
| QC 5 | 6.6 | 3.9 | 4.3 | 10.5 | |
| Lyso-Gb3 | QC level | Precision, CV% Intra-Assay DBS | Precision, CV% Inter-Assay DBS | Precision, CV% Intra-Assay CAP | Precision, CV% Inter-Assay CAP |
| QC 1 | 12.7 | 9.3 | 22 | 0 | |
| QC 2 | 8 | 13.6 | 8.3 | 4.5 | |
| QC 3 | 7.1 | 5.5 | 7.3 | 2.9 | |
| QC 4 | 4.7 | 8 | 5.4 | 7.2 | |
| QC 5 | 6 | 0 | 4.9 | 9.3 | |
| Lyso-SM | QC level | Precision, CV% Intra-Assay DBS | Precision, CV% Inter-Assay DBS | Precision, CV% Intra-Assay CAP | Precision, CV% Inter-Assay CAP |
| QC 1 | 30.8 | 52.8 | 20.6 | 12.7 | |
| QC 2 | 19.7 | 33.7 | 9.5 | 4.8 | |
| QC 3 | 10.4 | 10.5 | 5.8 | 6.2 | |
| QC 4 | 6 | 5.7 | 5.2 | 8.6 | |
| QC 5 | 7.8 | 6.3 | 4.8 | 9.8 |
| GlcSph | QC Level | Accuracy, RE% Inter-Assay DBS | Accuracy, RE% Inter-Assay CAP |
|---|---|---|---|
| QC 1 | 0.5 | 3 | |
| QC 2 | 13.2 | 6.9 | |
| QC 3 | −2 | 9.9 | |
| QC 4 | 5.8 | 5.6 | |
| QC 5 | −4.5 | 5.4 | |
| Lyso-Gb3 | QC Level | Accuracy, RE% Inter-Assay DBS | Accuracy, RE% Inter-Assay CAP |
| QC 1 | 25 | 5.8 | |
| QC 2 | 24.7 | 4.7 | |
| QC 3 | 1.8 | 8.2 | |
| QC 4 | 14.7 | 13.7 | |
| QC 5 | 0.2 | 11.8 | |
| Lyso-SM | QC Level | Accuracy, RE% Inter-Assay DBS | Accuracy, RE% Inter-Assay CAP |
| QC 1 | −3.5 | 0.5 | |
| QC 2 | 39.3 | 0.2 | |
| QC 3 | −5.2 | 2.8 | |
| QC 4 | 3.3 | 6.4 | |
| QC 5 | −11 | 3.9 |
| Cut-Off | Low-Low Mean | High-Low Mean | Carry-Over Absolute | Conclusion | |
|---|---|---|---|---|---|
| GlcSph | 1.34 | 8.68 | 9.39 | 0.71 | Below cut-off |
| Lyso-Gb3 | 2.65 | 2.74 | 3.033 | 0.29 | Below cut-off |
| Lyso-SM | 7.60 | 8.44 | 10.36 | 1.92 | Below cut-off |
| DBS | Cut-Off | Limit for LOQ | Obtained LOQ |
|---|---|---|---|
| GlcSph | 320 | 3200 | 14,400 |
| Lyso-Gb3 | 76 | 760 | 1560 |
| Lyso-SM | 490 | 4900 | 86,200 |
| CAP | Cut-Off | Limit for LOQ | Obtained LOQ |
| GlcSph | 600 | 6000 | 32,800 |
| Lyso-Gb3 | 125 | 1250 | 30,750 |
| Lyso-SM | 330 | 3300 | 2,428,500 |
| GlcSph (ng/mL) | Lyso-Gb3 (ng/mL) | Lyso-SM (ng/mL) | Diagnosis | |
|---|---|---|---|---|
| Patient sample 1 | 0.76 | 0.04 | 266.94 | ASMD |
| Patient sample 2 | 0.93 | 73.16 | 37.1 | Fabry |
| Patient sample 3 | 524.5 | 1.52 | 34.24 | Gaucher |
| Patient sample 4 | 211 | 0.72 | 26.64 | Gaucher |
| Patient sample 5 | 1.03 | 0.52 | 300.39 | ASMD |
| Patient sample 6 | 3.94 | 0.11 | 205 | ASMD |
| Patient sample 7 | 1.67 | 79.1 | 20.17 | Fabry |
| Patient sample 8 | 286.65 | 1.53 | 0 | Gaucher |
| Patient sample 9 | 1.12 | 1.04 | 400.37 | ASMD |
| Patient sample 10 | 397.79 | 2.25 | 0 | Gaucher |
| Patient sample 11 | 0.18 | 30.5 | 0.49 | Fabry |
| Patient sample 12 | 0.4 | 22.97 | 6.11 | Fabry |
| Patient sample 13 | 0.63 | 1.44 | 617.45 | ASMD |
| Patient sample 14 | 1.23 | 10.9 | 18.96 | Fabry |
| Patient sample 15 | 68.94 | 0.94 | 19.19 | Gaucher |
| Patient sample 16 | 2.67 | 2.34 | 1367.31 | ASMD |
| Patient sample 17 | 2 | 1.29 | 114.90 | ASMD |
| Patient sample 18 | 0.4 | 22.97 | 6.11 | Fabry |
| Patient sample 19 | 440 | 2.69 | 52.76 | Gaucher |
| Patient sample 20 | 2.06 | 1.49 | 110.87 | ASMD |
| Patient sample 21 | 166.89 | 1.55 | 44.00 | Gaucher |
| Patient sample 22 | 1.09 | 7.55 | 3.86 | Fabry |
| Patient sample 23 | 90.75 | 0.52 | 27.66 | Gaucher |
| Patient sample 24 | 0.49 | 0.55 | 479.63 | ASMD |
| Patient sample 25 | 78 | 0.25 | 54.39 | Gaucher |
| Patient sample 26 | 1.12 | 1.04 | 400.37 | ASMD |
| Patient sample 27 | 0.63 | 0.19 | 35.34 | Healthy |
| Patient sample 28 | 3.30 | 0.42 | 24.78 | Healthy |
| Patient sample 29 | 2.32 | 0.36 | 26.16 | Healthy |
| Patient sample 30 | 1.88 | 0.16 | 33.11 | Healthy |
| Patient sample 31 | 2,65 | 0.57 | 40.65 | Healthy |
| Patient sample 32 | 1.12 | 0.08 | 30.27 | Healthy |
| Patient sample 33 | 1.015 | 0.17 | 38.78 | Healthy |
| Patient sample 34 | 2.022 | 0.20 | 16.13 | Healthy |
| Patient sample 35 | 0.54 | 0.08 | 7.078 | Healthy |
| Patient sample 36 | 3.45 | 0.42 | 14.87 | Healthy |
| Patient sample 37 | 2.27 | 0.19 | 30.46 | Healthy |
| Patient sample 38 | 1.66 | 0.41 | 40.93 | Healthy |
| Patient sample 39 | 3.09 | 0.19 | 33.36 | Healthy |
| Patient sample 40 | 0.86 | 0.51 | 45.90 | Healthy |
| Patient sample 41 | 1.83 | 0.28 | 43.16 | Healthy |
| Standard (STD) | STD1 | STD2 | STD3 | STD4 | STD5 | STD6 | STD7 |
|---|---|---|---|---|---|---|---|
| GlcSph (ng/mL) | 1 | 5 | 10 | 20 | 100 | 500 | 1000 |
| Lyso-Gb3 (ng/mL) | 0.2 | 0.5 | 2 | 8 | 80 | 160 | 400 |
| Lyso-SM (ng/mL) | 1 | 5 | 10 | 20 | 100 | 500 | 1000 |
| Quality Concentration | QC1 | QC2 | QC3 | QC4 | QC5 |
|---|---|---|---|---|---|
| GlcSph (ng/mL) | 4 | 10 | 40 | 200 | 800 |
| Lyso-Gb3 (ng/mL) | 1.2 | 3 | 10 | 50 | 200 |
| Lyso-SM (ng/mL) | 4 | 10 | 40 | 200 | 800 |
| Compound Parameters | |||||||
|---|---|---|---|---|---|---|---|
| MRM | Test | Dwell Time (ms) | DP (Volts) | CE (Volts) | CXP (Volts) | ||
| 1 | GlcSph-IS | 30.0 | 176.0 | 31.0 | 20.0 | ||
| 2 | GlcSph | 30.0 | 176.0 | 31.0 | 20.0 | ||
| 3 | Lyso-Gb3-IS | 60.0 | 171.0 | 45.0 | 20.0 | ||
| 4 | Lyso-Gb3 | 60.0 | 171.0 | 45.0 | 20.0 | ||
| 5 | Lyso-SM d7 | 30.0 | 176.0 | 31.0 | 20.0 | ||
| 6 | Lyso-SM | 30.0 | 176.0 | 31.0 | 20.0 | ||
| LC-MS/MS source settings | |||||||
| Total Flow | Pressure Limits | Needle Stroke | Sampling Speed | Cooler Temp | Oven Temp | ESI Needle | Rinsing Volume |
| 0.50 mL/min | 14,000 psi | 50 mm | 2.0 µL/sec | 15 °C | 40 °C | 50 mm | 500 µL |
| EP | CUR | CAD | IS* | TEM | GS1 | GS2 | |
| 10 V | 35 psi | medium | 5500 V | 600 °C | 60 psi | 50 psi | |
| Working Solution | Extraction Method |
|---|---|
| 50% MeOH | 30 min sonication at 30 °C |
| +50% DMSO | +20 min shaker at 37 °C |
| +30 µL IS on 9.07ml | +10 min centrifuging at 4750 rpm |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Van Baelen, A.; Verhulst, S.; Eyskens, F. High-Resolution Mass Spectrometry Method for Targeted Screening and Monitoring of Fabry, Gaucher and ASMD Using Dried Blood Spots and Capitainers: Impact of Sample Matrix on Measurement Results. Int. J. Mol. Sci. 2025, 26, 7641. https://doi.org/10.3390/ijms26157641
Van Baelen A, Verhulst S, Eyskens F. High-Resolution Mass Spectrometry Method for Targeted Screening and Monitoring of Fabry, Gaucher and ASMD Using Dried Blood Spots and Capitainers: Impact of Sample Matrix on Measurement Results. International Journal of Molecular Sciences. 2025; 26(15):7641. https://doi.org/10.3390/ijms26157641
Chicago/Turabian StyleVan Baelen, Amber, Stijn Verhulst, and François Eyskens. 2025. "High-Resolution Mass Spectrometry Method for Targeted Screening and Monitoring of Fabry, Gaucher and ASMD Using Dried Blood Spots and Capitainers: Impact of Sample Matrix on Measurement Results" International Journal of Molecular Sciences 26, no. 15: 7641. https://doi.org/10.3390/ijms26157641
APA StyleVan Baelen, A., Verhulst, S., & Eyskens, F. (2025). High-Resolution Mass Spectrometry Method for Targeted Screening and Monitoring of Fabry, Gaucher and ASMD Using Dried Blood Spots and Capitainers: Impact of Sample Matrix on Measurement Results. International Journal of Molecular Sciences, 26(15), 7641. https://doi.org/10.3390/ijms26157641






