Efficient Genome Editing Using the T2A-Coupled Co-Expression of Two ZFN Monomers
Abstract
1. Introduction
2. Results
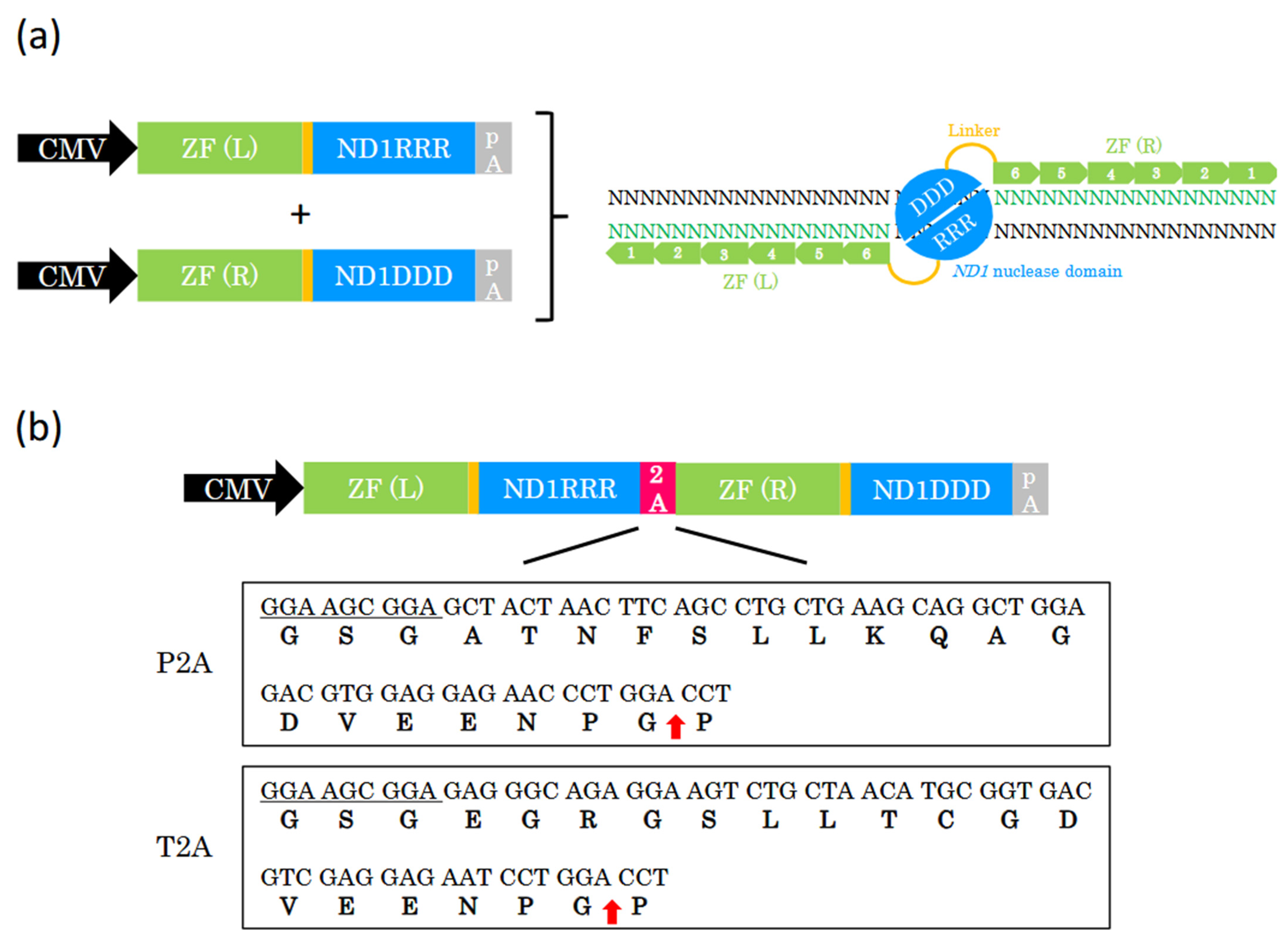
2.1. Design of ZF-ND1 Expression
2.2. T2A Peptide Enables Efficient Genome Editing with a Single Plasmid
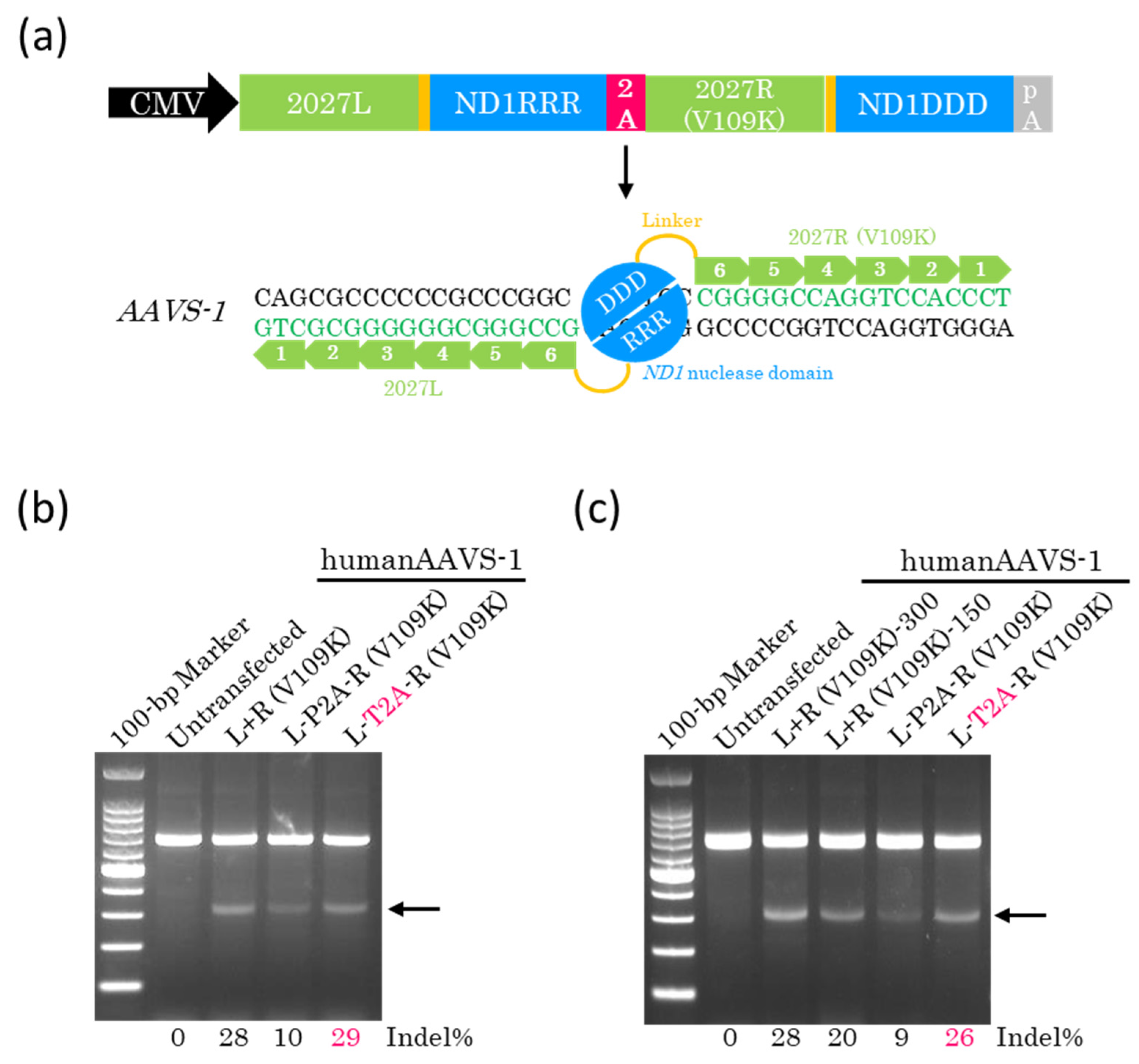
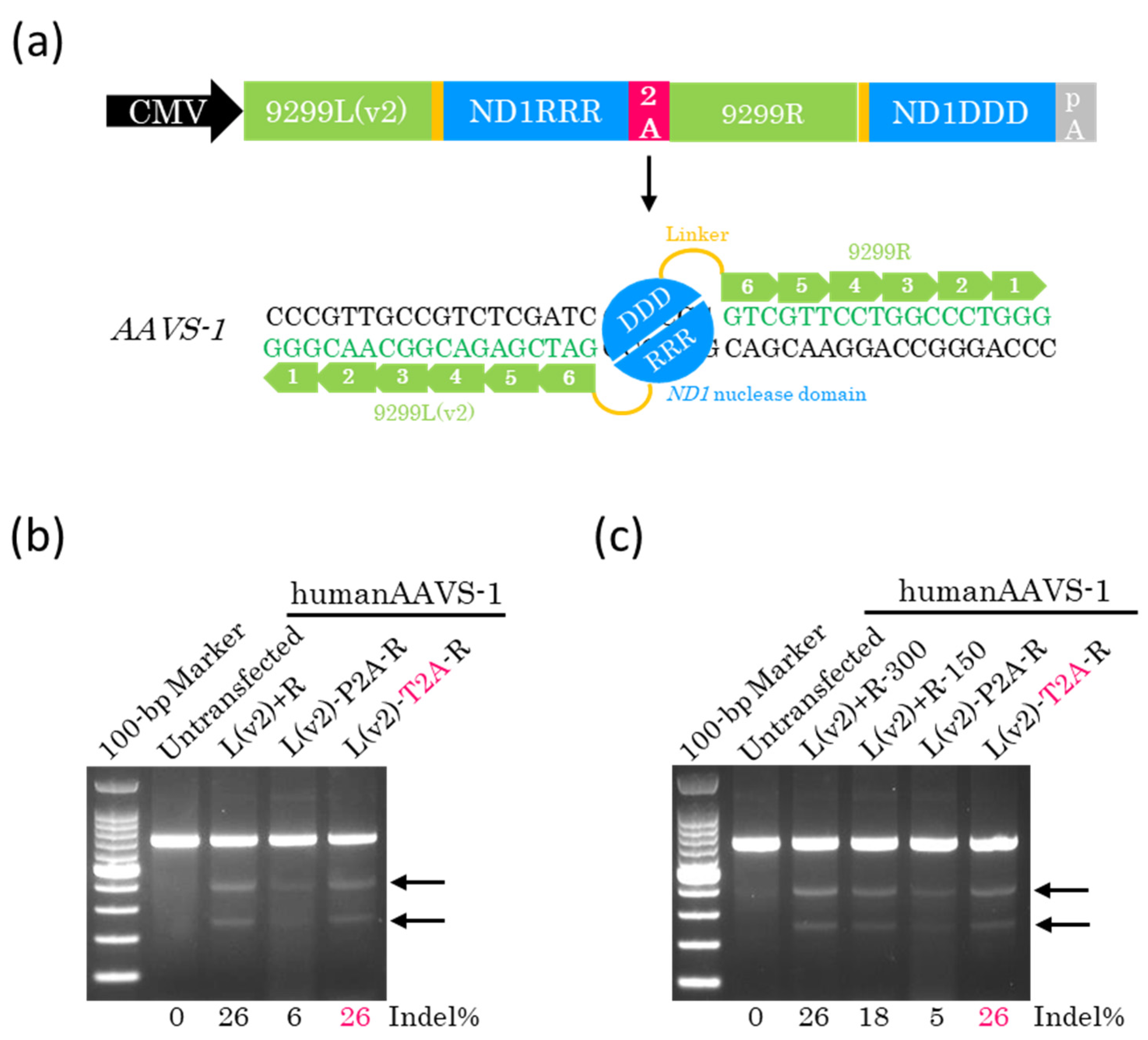
2.3. Efficient Genome Editing with a Single Plasmid Encoding Other ZF-ND1s
2.4. Lower Doses of ZF-ND1 Plasmids and Another Cell Line
3. Discussion
4. Materials and Methods
4.1. ZF Design
4.2. Vector Construction
4.3. Cell Culture, DNA Transfection, and DNA Electroporation
4.4. Transient Cold Shock
4.5. T7EI Assay
4.6. Off-Target Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Sander, J.D.; Joung, J.K. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat. Biotechnol. 2014, 32, 347–355. [Google Scholar] [CrossRef] [PubMed]
- Charpentier, E.; Jinek, M.; Chylinski, K.; Doudna Cate, J.H.; Lim, W.A.; Qi, L.S. Methods and Compositions for RNA-Directed Target DNA Modification and for RNA-Directed Modulation of Transcription. International Patent WO2013176772, 15 March 2013. [Google Scholar]
- Zhang, F. CRISPR-Cas Systems and Methods for Altering Expression of Gene Products. International Patent WO2014093661, 12 December 2013. [Google Scholar]
- Scott, C.T. The zinc finger nuclease monopoly. Nat. Biotechnol. 2005, 23, 915–918. [Google Scholar] [CrossRef] [PubMed]
- Cong, L.; Ran, F.A.; Cox, D.; Lin, S.; Barretto, R.; Habib, N.; Hsu, P.D.; Wu, X.; Jiang, W.; Marraffini, L.A.; et al. Multiplex Genome Engineering Using CRISPR/Cas Systems. Science 2013, 339, 819–823. [Google Scholar] [CrossRef] [PubMed]
- Colella, P.; Ronzitti, G.; Mingozzi, F. Emerging Issues in AAV-Mediated In Vivo Gene Therapy. Mol. Ther. Methods Clin. Dev. 2018, 8, 87–104. [Google Scholar] [CrossRef] [PubMed]
- Katayama, S.; Sato, K.; Nakazawa, T. In vivo and in vitro knockout system labelled using fluorescent protein via microhomology-mediated end joining. Life Sci. Alliance 2020, 3, e201900528. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, T.; Sakuma, T.; Saito, M. Novel nuclease domain and uses thereof. International Patent WO2020045281, 23 August 2019. [Google Scholar]
- Katayama, S.; Watanabe, M.; Kato, Y.; Nomura, W.; Yamamoto, T. Engineering of Zinc Finger Nucleases Through Structural Modeling Improves Genome Editing Efficiency in Cells. Adv. Sci. 2024, 11, e2310255. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Chen, O.; Wall, J.B.J.; Zheng, M.; Zhou, Y.; Wang, L.; Vaseghi, H.R.; Qian, L.; Liu, J. Systematic comparison of 2A peptides for cloning multi-genes in a polycistronic vector. Sci. Rep. 2017, 7, 2193. [Google Scholar] [CrossRef] [PubMed]
- Szymczak, A.L.; Workman, C.J.; Wang, Y.; Vignali, K.M.; Dilioglou, S.; Vanin, E.F.; Vignali, D.A. Correction of multi-gene deficiency in vivo using a single ‘self-cleaving’ 2A peptide-based retroviral vector. Nat. Biotechnol. 2004, 22, 589–594. [Google Scholar] [CrossRef] [PubMed]
- Osborn, M.J.; Panoskaltsis-Mortari, A.; McElmurry, R.T.; Bell, S.K.; Vignali, D.A.; Ryan, M.D.; Wilber, A.C.; McIvor, R.S.; Tolar, J.; Blazar, B.R. A picornaviral 2A-like sequence-based tricistronic vector allowing for high-level therapeutic gene expression coupled to a dual-reporter system. Mol. Ther. 2005, 12, 569–574. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Lee, S.R.; Li, L.H.; Park, H.J.; Park, J.H.; Lee, K.Y.; Kim, M.K.; Shin, B.A.; Choi, S.Y. High cleavage efficiency of a 2A peptide derived from porcine teschovirus-1 in human cell lines, zebrafish and mice. PLoS ONE 2011, 6, e18556. [Google Scholar] [CrossRef] [PubMed]
- Chng, J.; Wang, T.; Nian, R.; Lau, A.; Hoi, K.M.; Ho, S.C.; Gagnon, P.; Bi, X.; Yang, Y. Cleavage efficient 2A peptides for high level monoclonal antibody expression in CHO cells. mAbs 2015, 7, 403–412. [Google Scholar] [CrossRef] [PubMed]
- Doyon, Y.; Vo, T.D.; Mendel, M.C.; Greenberg, S.G.; Wang, J.; Xia, D.F.; Miller, J.C.; Urnov, F.D.; Gregory, P.D.; Holmes, M.C. Enhancing zinc-finger-nuclease activity with improved obligate heterodimeric architectures. Nat. Methods 2011, 8, 74–79. [Google Scholar] [CrossRef] [PubMed]
- Osborn, M.J.; DeFeo, A.P.; Blazar, B.R.; Tolar, J. Synthetic zinc finger nuclease design and rapid assembly. Hum. Gene Ther. 2011, 22, 1155–1165. [Google Scholar] [CrossRef] [PubMed]
- Pattanayak, V.; Ramirez, C.L.; Joung, J.K.; Liu, D.R. Revealing off-target cleavage specificities of zinc-finger nucleases by in vitro selection. Nat. Methods 2011, 8, 765–770. [Google Scholar] [CrossRef] [PubMed]
- Nash, P.A.; Turner, K.M.; Powell, C.A.; Van Haute, L.; Silva-Pinheiro, P.; Bubeck, F.; Wiedtke, E.; Marques, E.; Ryan, D.G.; Grimm, D.; et al. Clinically translatable mitochondrial gene therapy in muscle using tandem mtZFN architecture. EMBO Mol. Med. 2025, 17, 1222–1237. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Katayama, S.; Yamamoto, T. Efficient Genome Editing Using the T2A-Coupled Co-Expression of Two ZFN Monomers. Int. J. Mol. Sci. 2025, 26, 7602. https://doi.org/10.3390/ijms26157602
Katayama S, Yamamoto T. Efficient Genome Editing Using the T2A-Coupled Co-Expression of Two ZFN Monomers. International Journal of Molecular Sciences. 2025; 26(15):7602. https://doi.org/10.3390/ijms26157602
Chicago/Turabian StyleKatayama, Shota, and Takashi Yamamoto. 2025. "Efficient Genome Editing Using the T2A-Coupled Co-Expression of Two ZFN Monomers" International Journal of Molecular Sciences 26, no. 15: 7602. https://doi.org/10.3390/ijms26157602
APA StyleKatayama, S., & Yamamoto, T. (2025). Efficient Genome Editing Using the T2A-Coupled Co-Expression of Two ZFN Monomers. International Journal of Molecular Sciences, 26(15), 7602. https://doi.org/10.3390/ijms26157602




