Cloning, Heterologous Expression, and Antifungal Activity Evaluation of a Novel Truncated TasA Protein from Bacillus amyloliquefaciens BS-3
Abstract
1. Introduction
2. Result
2.1. Sequence and Structure Analysis
2.2. Cloning, Heterologous Expression, and Purification of TasA Gene
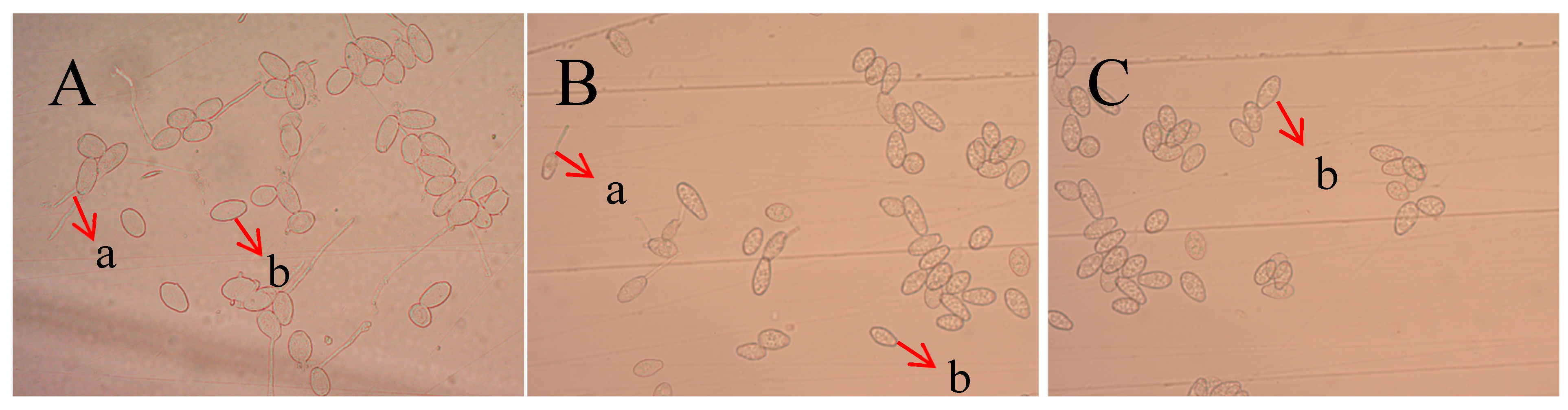
2.3. Detection of Antifungal Activity of Recombinant Protein TasA on Spores of Colletotrichum Acutatum
2.4. Inhibitory Effect of Recombinant Protein TasA on Oidium Heveae Steinmann
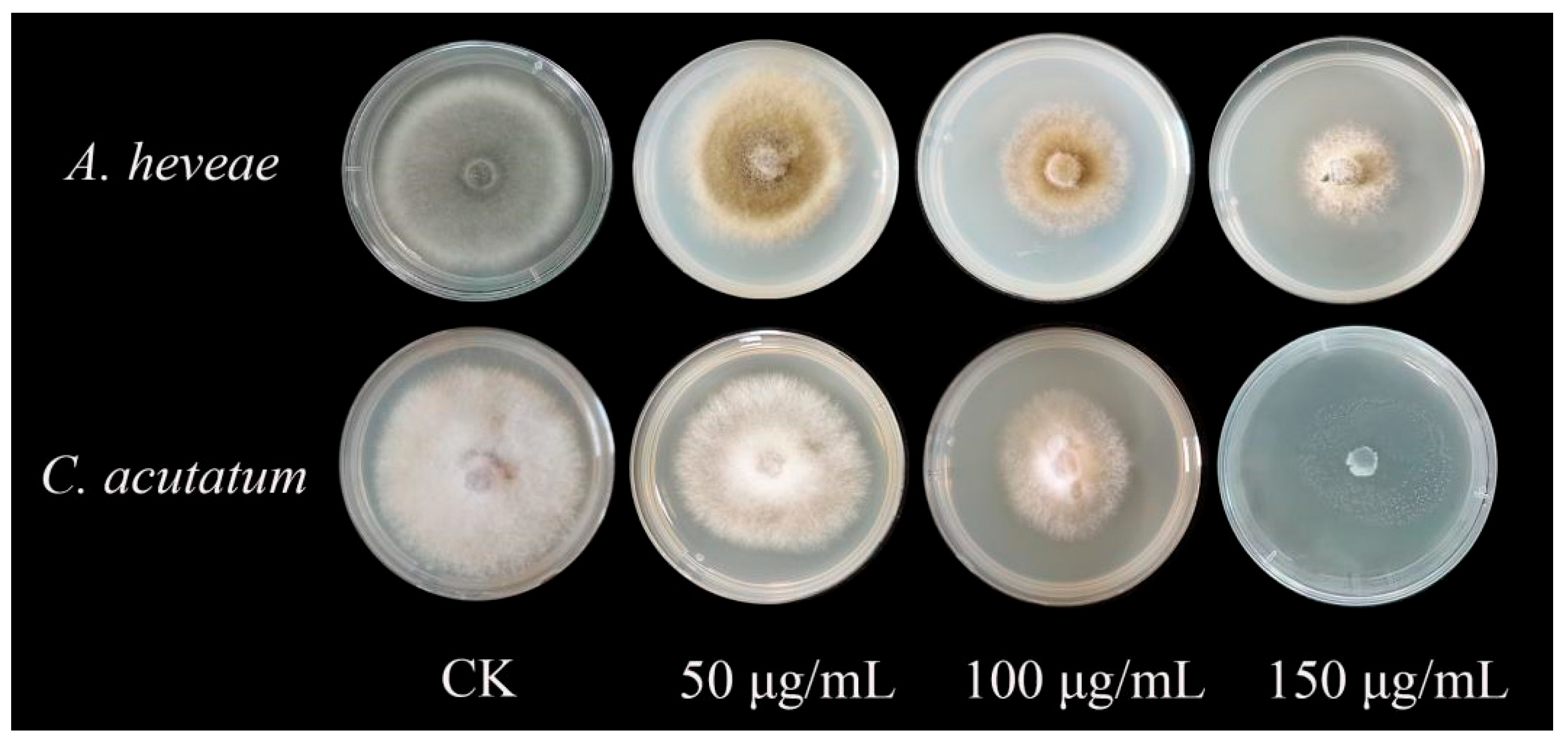
2.5. Inhibitory Effect of Recombinant Protein TasA on Plant Pathogenic Fungi
3. Discussion
4. Materials and Methods
4.1. Microbial Strainsand Chemical Reagent
4.2. Sequence Analysis
4.3. Gene Cloning
4.4. Construction of Prokaryotic Expression Vector
4.5. Expression and Purification of Protein
4.6. Inhibitory Effect of Recombinant Protein TasA on the Spores of Colletotrichum Acutatumfrom Rubber Trees
4.7. Inhibitory Effect of Recombinant Protein TasA on the Spores of Oidium Heveae Steinmann from Rubber Trees
4.8. Determination of Inhibitory Effect of Recombinant Protein TasA on Plant Pathogenic Fungi
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Chen, X.H.; Koumoutsi, A.; Scholz, R.; Eisenreich, A.; Schneider, K.; Heinemeyer, I.; Morgenstern, B.; Voss, B.; Hess, W.R.; Reva, O.; et al. Comparative analysis of the complete genome sequence of the plant growth–promoting bacterium Bacillus amyloliquefaciens FZB42. Nat. Biotechnol. 2007, 25, 1007–1014. [Google Scholar] [CrossRef]
- Wells, J.A.; Ferrari, E.; Henner, D.J.; Estell, D.A.; Chen, E.Y. Cloning, sequencing, and secretion of Bacillus amyloliquefaciens subtfllsin in Bacillus subtilis. Nucleic. Acids. Res. 1983, 11, 7911–7925. [Google Scholar] [CrossRef]
- Xu, K.Z.; Yin, L.J.; Ding, Z.W.; Wang, Y.J.; Jia, A.Q. YtnP: One novel quorum quenching enzyme from Bacillus amyloliquefaciens W11 inhibits biofilms and spoilage of white radish by Serratia marcescens. LWT 2024, 198, 116058. [Google Scholar] [CrossRef]
- Ansari, M.M.; Bisht, N.; Singh, T.; Mishra, S.K.; Anshu, A.; Singh, P.C.; Chauhan, P.S. Bacillus amyloliquefaciens modulate autophagy pathways to control Rhizoctonia solani infection in rice. Plant. Physiol. Bioch. 2025, 218, 109317. [Google Scholar] [CrossRef]
- Caldeira, A.T.; Feio, S.S.; Santos Arteiro, J.M.; Roseiro, J.C. Bacillus amyloliquefaciens CCMI 1051 in vitro activity against wood contaminant fungi. Ann. Microbiol. 2007, 57, 29–33. [Google Scholar] [CrossRef]
- Caldeira, A.T.; Feio, S.S.; Arteiro, J.M.S.; Coelho, A.V.; Roseiro, J.C. Environmental dynamics of Bacillus amyloliquefaciens CCMI 1051 antifungal activity under different nitrogen patterns. J. Appl. Microbiol. 2008, 104, 808–816. [Google Scholar] [CrossRef] [PubMed]
- Arrebola, E.; Sivakumar, D.; Korsten, L. Effect of volatile compounds produced by Bacillus strains on postharvest decay in citrus. Biol. Control 2010, 53, 122–128. [Google Scholar] [CrossRef]
- Lee, Y.K.; Senthilkumar, M.; Kim, J.H.; Swarnalakshmi, K.; Annapurna, K. Purification and partial characterization of antifungal metabolite from Paenibacilluslentimorbus WJ5. World J. Microbiol. Biotechnol. 2008, 24, 3057–3062. [Google Scholar] [CrossRef]
- Quan, C.S.; Wang, J.H.; Xun, H.T.; Fan, S.D. Isolation, identification and fermentation conditions of an antifungal Bacillus amylolyticus. Acta Microbiol. Sin. 2006, 46, 7–12. [Google Scholar] [CrossRef]
- Zhang, Y.S.; Wang, J.; Hao, J.A.; Zhang, X.Z.; Cheng, Y. Flocculating activity of Penicillium purpurogenum EL-02 and its flocculating activity. Acta Microbiol. Sin. 2010, 50, 917–922. [Google Scholar] [CrossRef]
- Abdallah, R.A.B.; Jabnoun-Khiareddine, H.; Nefzi, A.; Mokni-Tlili, S.; Daami-Remadi, M. Biocontrol of Fusarium Wilt and growth promotion of tomato plants using endophytic bacteria isolated from Solanum elaeagnifolium Stems. J. Phytopathol. 2016, 164, 811–824. [Google Scholar] [CrossRef]
- Liu, J.; Qin, D.; Huang, W.; Wang, X.Q.; Li, Y.G.; Zhang, R. Biocontrol ability and action mechanism of Bacillus amyloliquefaciens Baf1 against Fusarium incarnatum causing fruit rot in postharvest muskmelon (cv. Yugu) fruit. LWT 2023, 181, 114714. [Google Scholar] [CrossRef]
- Li, Y.L.; Cao, X.M.; Chai, Y.R.; Chen, R.F.; Zhao, Y.J.; Borriss, R.; Ding, X.L.; Wu, X.Q.; Ye, J.R.; Hao, D.J.; et al. A phosphate starvation induced small RNA promotes Bacillus biofilm formation. Npj. Biofilms. Microbi. 2024, 10, 115. [Google Scholar] [CrossRef]
- Böhning, J.; Ghrayeb, M.; Pedebos, C.; Abbas, D.K.; Khalid, S.; Chai, L.; Bharat, T.A.M. Donor-strand exchange drives assembly of the TasA scaffold in Bacillus subtilis biofilms. Nat. Commun. 2022, 13, 7082. [Google Scholar] [CrossRef] [PubMed]
- Pratt, L.A.; Kolter, R. Genetic analysis of Escherichia coli biofilm formation: Roles of flagella, motility, chemotaxis and type I pili. Mol. Microbiol. 1998, 30, 285–293. [Google Scholar] [CrossRef] [PubMed]
- Klausen, M.; Heydorn, A.; Ragas, P.; Lambertsen, L.; Aaes-Jørgensen, A.; Molin, S.; Tolker-Nielsen, T. Biofilm formation by Pseudomonas aeruginosa wild type, flagella and type IV pili mutants. Mol. Microbiol. 2003, 48, 1511–1524. [Google Scholar] [CrossRef]
- Liu, T.; Chen, Y.P.; Li, Q.J.; Gu, Z.F. Cloning and functional analyses of TasA, an antimicrobial protein gene from Bacillus pumilus DX01. J. Shanghai. Jiaotong. U. Agr. Sci. 2014, 32, 48–56. [Google Scholar] [CrossRef]
- Zhang, S.M.; Jiang, W.; Meng, L.Q.; Hu, J.H.; Li, J.; Liu, Y.S.; Gao, W. Cloning and expression of antifungal protein gene TasA from Bacillus amyloliquefaciens TF28. Biotechnology 2016, 26, 130–135. [Google Scholar] [CrossRef]
- Yang, L.R.; Wang, Z.J.; Xue, B.G.; Liu, H.Y.; Ma, J.G.; Zheng, Y.P. Cloning of antagonistic protein TasA gene in Bacillus amyloliquefaciens YN-1 and its prokaryotic expression. Genom. Appl. Biol. 2010, 29, 823–828. [Google Scholar]
- Long, X.L.; Wang, Z.K.; Chen, Y.L.; Yin, Y.P. Cloning antagonistic protein TasA gene in Bacillus CQBS03 and its prokaryotic expression. Acta Phytophylacica. Sin. 2012, 39, 438–442. [Google Scholar] [CrossRef]
- Han, Y.; Fan, J.; Zhou, Z.J.; Tan, X.Q.; Zhao, X. Cloning and efficient expression of Bacillus sp. BH072 tasA gene in Escherichia coli. Trans. Tianjin Univ. 2015, 21, 26–31. [Google Scholar] [CrossRef]
- Yin, X.T.; Fan, S.S.; Xu, L.; Xu, L.N.; Liu, Z.Y. Cloning and sequence analysis of TasA gene from mulberry endophytic bacteria Bacillussubtilis and antibancterial activity of its expressed product. Sci. Seric. 2010, 36, 319–322. [Google Scholar] [CrossRef]
- Liu, S.; Huang, J.; Zhang, C.; Wang, L.; Fan, C.; Zhong, C. Probing the growth and mechanical properties of Bacillus subtilis biofilms through genetic mutation strategies. Syn. Syst. Biotechnol. 2022, 7, 965–971. [Google Scholar] [CrossRef]
- Zhao, X.; Zhou, Z.J.; Han, Y.; Wang, Z.Z.; Fan, J.; Xiao, H.Z. Isolation and identification of antifungal peptides from Bacillus BH072, a novel bacterium isolated from honey. Microbiol. Res. 2013, 168, 598–606. [Google Scholar] [CrossRef]
- Dai, L.M.; Li, L.L.; Liu, Y.X.; Shi, Y.P.; Cai, Z.Y. Whole genome sequencing and genomics analysis of Bacillus amyloliquefaciens BS-3 with biocontrol activity. Microbiol. Chin. 2021, 48, 2073–2088. [Google Scholar] [CrossRef]
- Dai, L.M.; Li, L.L.; Cai, Z.Y.; Mu, H.J.; Liu, Y.X.; Shi, Y.P.; He, L.L.; Qiu, Y.F. An Antifungal Protein Derived from Bacillus amyloliquefaciens BS-3 and Its Applications. CN 119529040 B, 6 May 2025. [Google Scholar]
- Li, L.L.; Dai, L.M.; Cai, Z.Y.; Mu, H.J.; Liu, Y.X.; Shi, Y.P.; He, L.L.; Qiu, Y.F. Bacillus amyloliquefaciens BS-3 and Its Antibacterial Protein Derived Therefrom and Their Applications. CN 118853465 B, 7 March 2021. [Google Scholar]
- Xu, L. Molecular Identification of the Endophytic bacteria in Poplar and Cloning and Expression of TasA Gene. Master’s Thesis, Shandong Agricultural University, Taian, China, 2009. [Google Scholar]
- Webb, B.; Sali, A. Comparative Protein Structure Modeling Using MODELLER. Curr. Protoc. Bioinform. 2016, 54, 5.6.1–5.6.37. [Google Scholar] [CrossRef]
- Cámara-Almirón, J.; Domínguez-García, L.; El Mammeri, N.; Lends, A.; Habenstein, B.; de Vicente, A.; Loquet, A.; Romero, D. Molecular characterization of the N-terminal half of TasA during amyloid-like assembly and its contribution to Bacillus subtilis biofilm formation. Npj. Biofilms. Microbi. 2023, 9, 68. [Google Scholar] [CrossRef]
- Zhao, K.; Li, H.Y.; Song, W.; Wu, J.; Gao, C.; Guo, L.; Chen, X.L. Combinatorial mutagenesis of Bacillus amyloliquefaciens for efficient production of protease. Syst. Microbiol. Biomanuf. 2023, 3, 457–468. [Google Scholar] [CrossRef]
- Zhang, P.; Hao, H.T.; Wang, L.H.; Liu, Z.H.; Ma, L. Endophytes Bacillus amyloliquefaciens AW3 (CGMCC1.16683) improves the growth of Populus davidiana × Populusbolleana (PdPap) and induces its resistance to wilt disease by Fusarium oxysporum Fox68 (CFCC86068). Eur. J. Plant. Pathol. 2022, 162, 1–17. [Google Scholar] [CrossRef]
- Luo, L.; Zhao, C.Z.; Wang, E.T.; Raza, A.; Yin, C.Y. Bacillus amyloliquefaciensas an excellent agent for biofertilizer and biocontrol in agriculture: An overview for its mechanisms. Microbiol. Res. 2022, 259, 127016. [Google Scholar] [CrossRef] [PubMed]
- Liao, F.P.; Xu, X.; Wang, H.X.; Huang, L.R.; Li, K.T. Antifungal lipopeptides from the marine Bacillus amyloliquefaciens HY2–1: A potential biocontrol agent exhibiting in vitro and in vivo antagonistic activities against Penicillium digitatum. Int. J. Biol. Macromol. 2025, 287, 138583. [Google Scholar] [CrossRef]
- Nan, J.; Qiu, J.H.; Tian, D.G.; Shi, H.B.; Liu, Z.Q.; Wen, H.; Xie, S.W.; Chen, H.Z.; Wu, M.; Kou, Y.J. Mixture of Bacillus Amyloliquefaciens and Bacillus Pumilus Modulates Community Structures of Rice Rhizosphere Soil to Suppress Rice Seedling Blight. Rice Sci. 2024, 32, 118–130. [Google Scholar] [CrossRef]
- Erskine, E.; Morris, R.J.; Schor, M.; Earl, C.; Gillespie, R.M.C.; Bromley, K.M.; Sukhodub, T.; Clark, L.; Fyfe, P.K.; Serpell, L.C.; et al. Formation of functional, non-amyloidogenic fibres by recombinant Bacillus subtilis TasA. Mol. Microbiol. 2018, 110, 897–913. [Google Scholar] [CrossRef]
- Steinberg, N.; Keren-Paz, A.; Hou, Q.H.; Doron, S.; Yanuka-Golub, K.; Olender, T.; Hadar, R.; Rosenberg, G.; Jain, R.; Cámara-Almirón, J.; et al. The extracellular matrix protein TasA is a developmental cue that maintains a motile subpopulation within Bacillus subtilis biofilms. Sci. Signal. 2020, 13, 1–15. [Google Scholar] [CrossRef]
- Cámara-Almirón, J.; Navarro, Y.; Díaz-Martínez, L.; Magno-Pérez-Bryan, M.C.; Molina-Santiago, C.; Pearson, J.R.; de Vicente, A.; Pérez-García, A.; Romero, D. Dual functionality of the amyloid protein TasA in Bacillus physiology and fitness on the phylloplane. Nat. Commun. 2020, 11, 1859. [Google Scholar] [CrossRef]
- Zhu, M.L.; Wang, Y.H.; Dai, Y.; Wu, X.Q.; Ye, J.R. Effects of different culture conditions on the biofilm formation of Bacillus pumilus HR10. Curr. Microbiol. 2020, 77, 1405–1411. [Google Scholar] [CrossRef]
- Fu, H.H.; Chen, F.F.; Liu, W.H.; Kong, W.L.; Wang, C.E.; Fang, X.Q.; Ye, J.R. Adding nutrients to the biocontrol strain JK-SH007 promotes biofilm formation and improves resistance to stress. AMB Express 2020, 10, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Liu, Y.; Wu, G.; Zhang, N.; Shen, Q.; Zhang, R. Beneficial Rhizobacterium Bacillus amyloliquefaciens SQR9 induces plant salt tolerance through Spermidine Production. Mol. Plant-Microbe Interact. 2017, 30, 423–432. [Google Scholar] [CrossRef] [PubMed]
- Gulyás, D.; Kocsis, B.; Szabó, D. Plasmid copy number and qnr gene expression in selection of Fluoroquinolone-resistant Escherichia coli. Acta Microbiol. Immunol. Hung. 2019, 66, 169–178. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.R.; Wang, Y.M.; Wang, Q.; Song, H.; Huang, Z.; Huang, L.; Zhong, M.; Peng, S.Y. Biocontrol effects of antifungal protein TasA on Tobacco black shank. Biol. Disaster Sci. 2023, 46, 425–431. [Google Scholar] [CrossRef]
- Zhang, Q. Study on the Mechanism of Bacillus amyloliquefaciens Resistant to Fusarium graminearumand the Functi on of Antimicrobial Gene TasA. Master’s Thesis, Nanjing Agricultural University, Nanjing, China, 2020. [Google Scholar]
- Diehl, A.; Roske, Y.; Ball, L.; Chowdhury, A.; Hiller, M.; Molière, N.; Regina, K.; Daniel, S.; Catherine, W.; Brigitte, L.S.; et al. Structural changes of TasA in biofilm formation of Bacillus subtilis. Proc. Natl. Acad. Sci. USA 2018, 115, 3237–3242. [Google Scholar] [CrossRef] [PubMed]
- Petersen, T.N.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 4.0: Discriminating signal peptides from transmembrane regions. Nat. Methods 2011, 8, 785–786. [Google Scholar] [CrossRef]
- Sapay, N.; Guermeur, Y.; Deléage, G. Prediction of amphipathic in-plane membrane anchors in monotopic proteins using a SVM classifier. BMC Bioinform. 2006, 7, 255. [Google Scholar] [CrossRef]
- Homma, F.; Huang, J.; van der Hoorn, R.A.L. AlphaFold-Multimer predicts cross-kingdom interactions at the plant-pathogen interface. Nat. Commun. 2023, 14, 6040. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Hall, T.; Biosciences, I.; Carlsbad, C.J. BioEdit An important software for molecular biology. GERF Bull. Biosci. 2011, 2, 60–61. [Google Scholar]
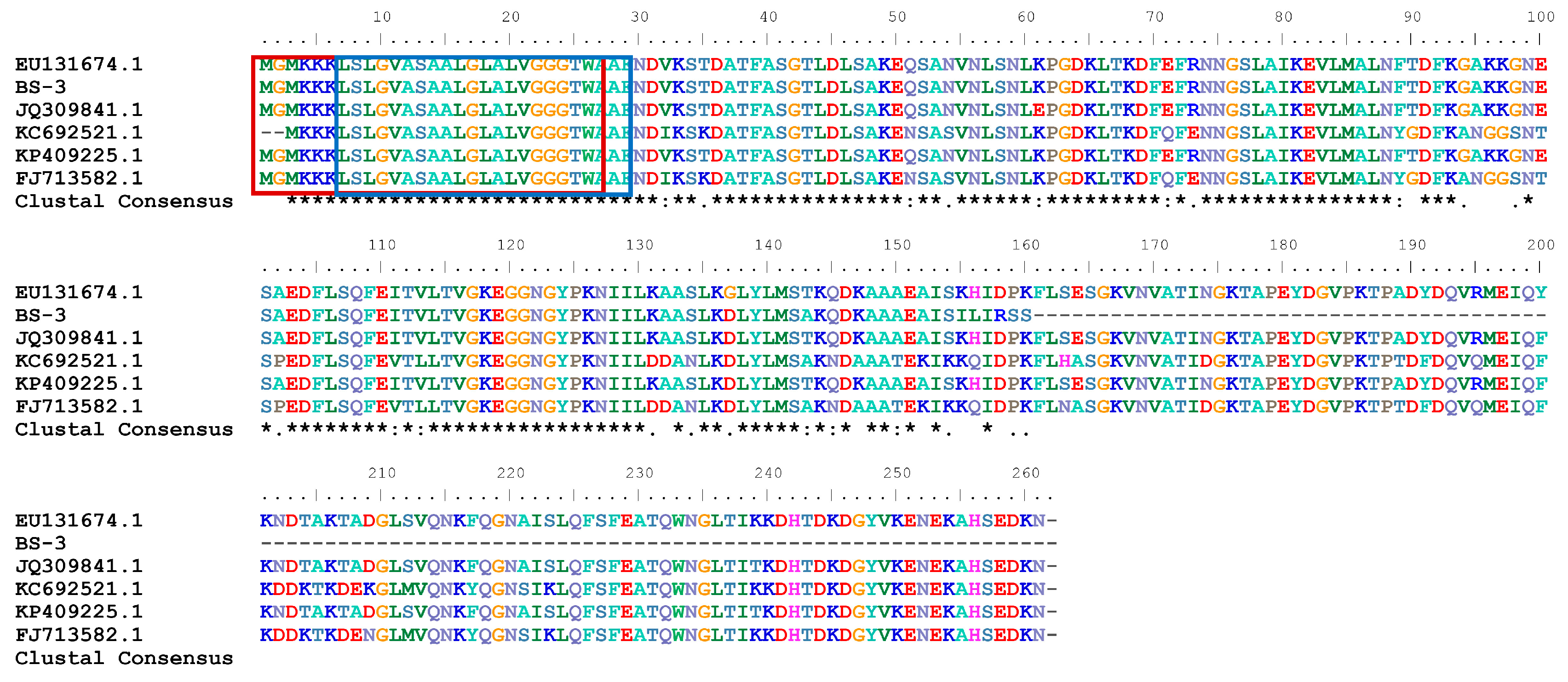
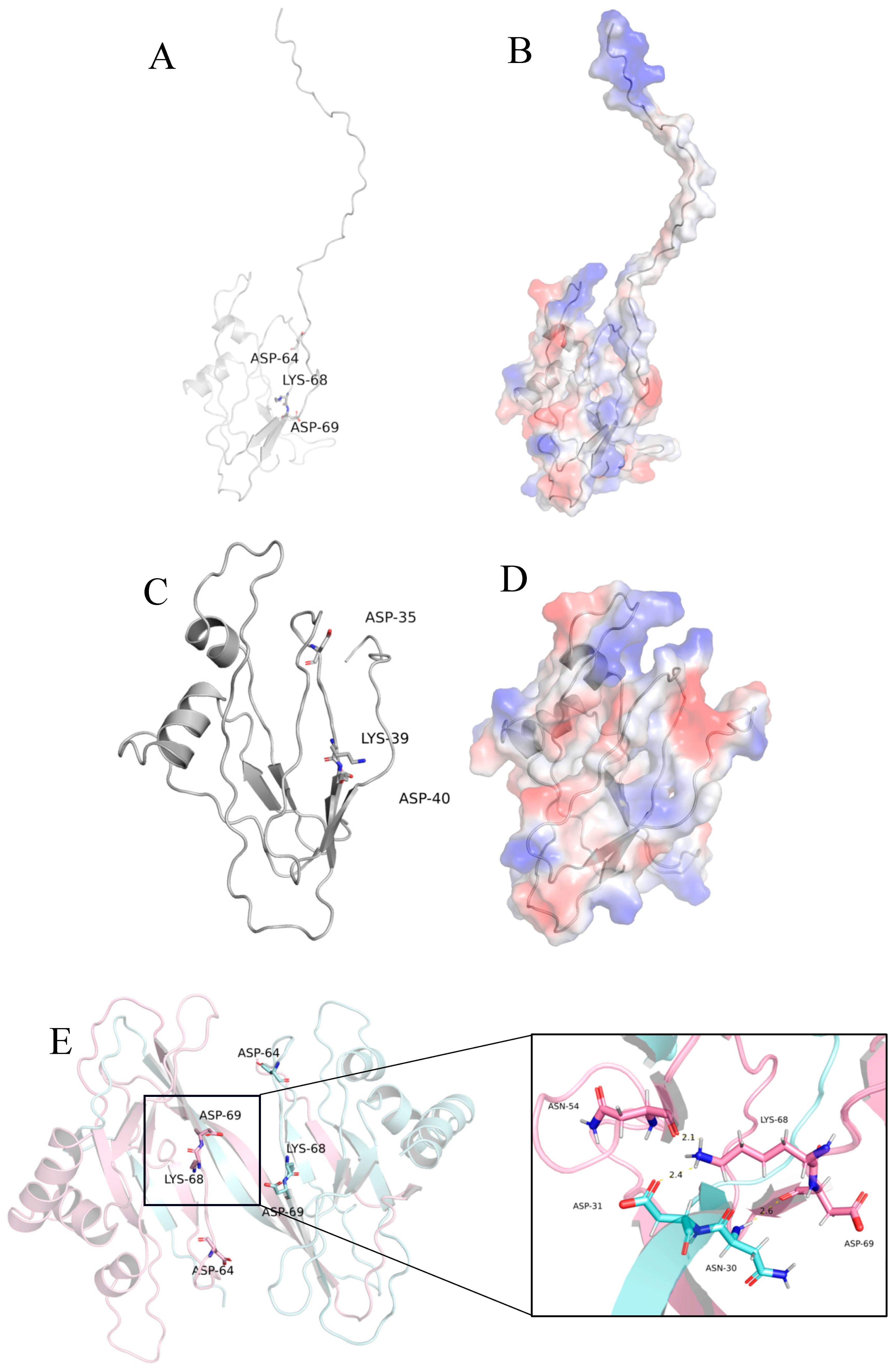

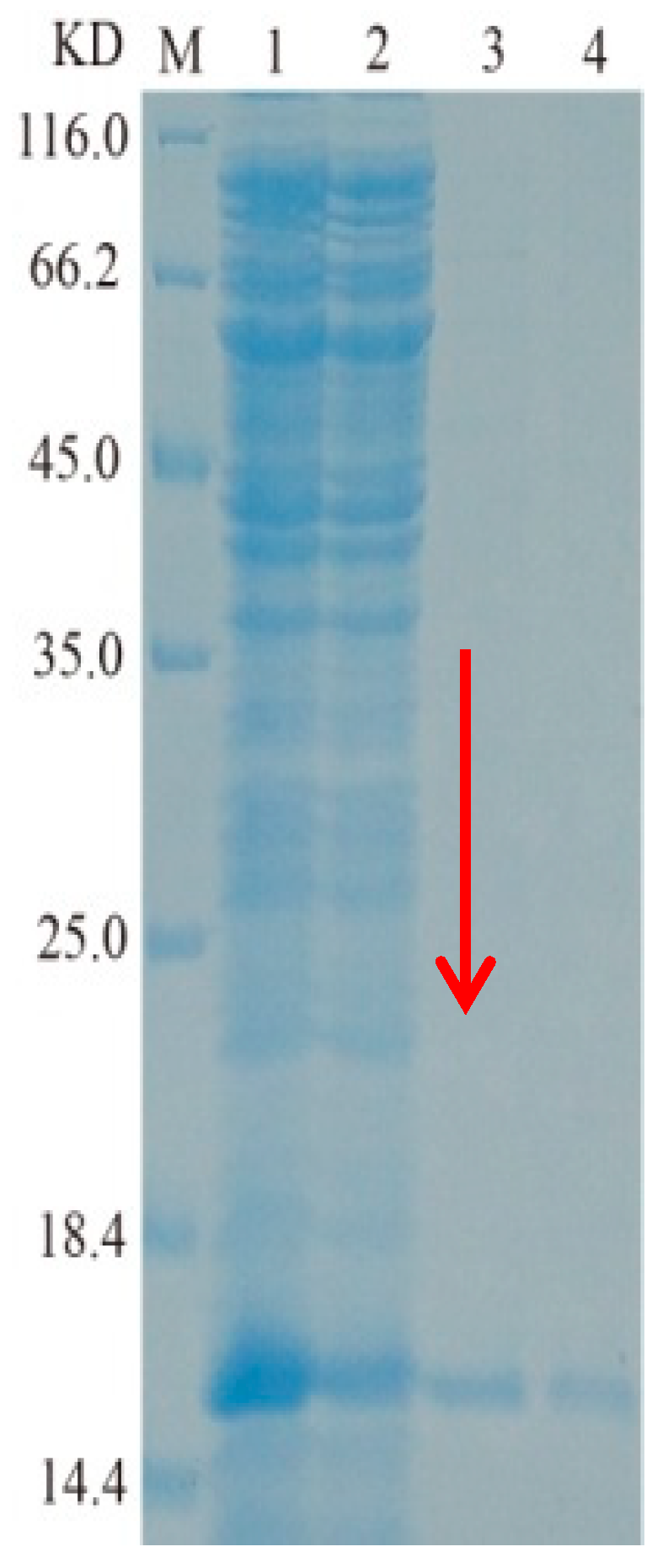
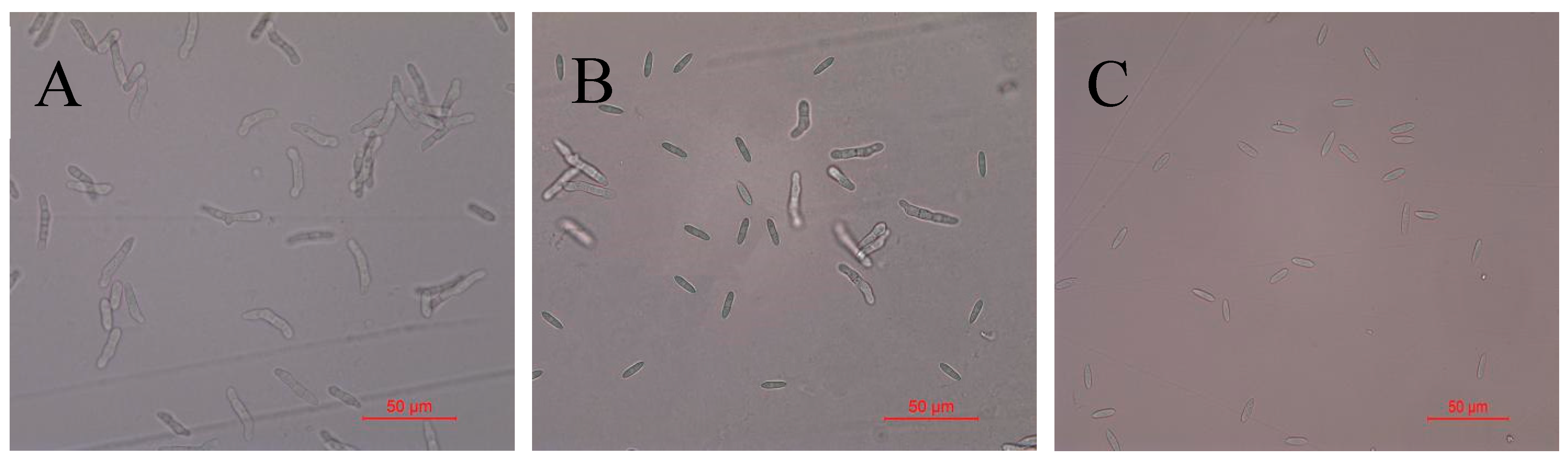
| Number | GenBank | Query Coverage (%) | Similarity (%) | |
|---|---|---|---|---|
| B. amyloliquefaciens | 205 | CP054415.1 | 100 | 99.18 |
| H | CP041693.1 | 100 | 99.18 | |
| HK1 | CP018902.1 | 100 | 99.18 | |
| SRCM101267 | CP021505.1 | 100 | 99.18 | |
| SRCM123386 | CP128501.1 | 100 | 99.18 | |
| SRCM124317 | CP116011.1 | 100 | 99.18 | |
| B. subtilis | ATCC 13952 | CP009748.1 | 100 | 99.18 |
| Sequence | Positions in Master Proteins | Theo. MH + Da | Charge: Sequest HT | m/z Da: Sequest HT |
|---|---|---|---|---|
| VHHHHHHMNDVK | 5–16 | 1527.71346 | 3 | 509.91019 |
| EQSANVNLSNLKPGDKLTK | 33–51 | 2056.10331 | 3 | 686.04144 |
| AASLKDLYLMSAK | 115–127 | 1426.76103 | 3 | 476.25912 |
| EVLMALNFTDFKGAK | 65–79 | 1683.87745 | 2 | 842.44513 |
| VHHHHHHMNDVKSTDATFASGTLDLSAK | 5–32 | 3093.47077 | 4 | 774.12451 |
| EGGNGYPKNIILK | 102–114 | 1402.76889 | 2 | 701.88715 |
| NNGSLAIKEVLMALNFTDFK | 57–76 | 2225.16347 | 3 | 742.38464 |
| QDKAAAEAISILIR | 128–141 | 1498.85877 | 2 | 749.93365 |
| NIILKAASLK | 110–119 | 1070.69321 | 2 | 535.85052 |
| GNESAEDFLSQFEITVLTVGK | 81–101 | 2284.13433 | 2 | 1142.57556 |
| QDKAAAEAISILIRSS | 128–143 | 1672.92282 | 3 | 558.31409 |
| EQSANVNLSNLKPGDKLTKDFEFR | 33–56 | 2750.41078 | 4 | 688.35828 |
| STDATFASGTLDLSAKEQSANVNLSNLKPGDKLTK | 17–51 | 3621.86062 | 4 | 906.22137 |
| STDATFASGTLDLSAKEQSANVNLSNLKPGDK | 17–48 | 3279.63392 | 4 | 820.66321 |
| MNHKVHHHHHHMNDVKSTDATFASGTLDLSAK | 1–32 | 3645.71863 | 5 | 729.94092 |
| DFEFRNNGSLAIK | 52–64 | 1510.76487 | 2 | 755.88629 |
| NIILKAASLKDLYLMSAK | 110–127 | 1992.1562 | 3 | 664.72455 |
| DLYLMSAKQDK | 120–130 | 1327.65623 | 2 | 664.33289 |
| EQSANVNLSNLKPGDK | 33–48 | 1713.8766 | 2 | 857.44183 |
| VHHHHHHMNDVK | 5–16 | 1543.70837 | 4 | 386.68253 |
| MNHKVHHHHHHMNDVK | 1–16 | 2095.95622 | 3 | 699.32373 |
| DLYLMSAK | 120–127 | 940.48083 | 1 | 940.48022 |
| EGGNGYPK | 102–109 | 821.37881 | 2 | 411.19736 |
| EVLMALNFTDFK | 65–76 | 1443.71883 | 2 | 722.36908 |
| STDATFASGTLDLSAK | 17–32 | 1584.77516 | 2 | 792.8924 |
| AASLKDLYLMSAKQDK | 115–130 | 1781.9466 | 4 | 446.24292 |
| NNGSLAIK | 57–64 | 816.45739 | 2 | 408.73315 |
| LTKDFEFR | 49–56 | 1055.55202 | 3 | 352.52206 |
| AAAEAISILIR | 131–141 | 1127.67828 | 2 | 564.34387 |
| DLYLMSAKQDK | 120–130 | 1311.66131 | 2 | 656.33484 |
| KGNESAEDFLSQFEITVLTVGK | 80–101 | 2412.2293 | 3 | 804.75092 |
| Concentrate/Time | 4.5 h | 6 h | 12 h |
|---|---|---|---|
| CK | 59.5% ± 0.36% | 35% ± 0.15% | 0% ± 0.00% |
| 30 μg/mL | 93.91% ± 1.35% ** | 91.95% ± 0.01% ** | 87.4% ± 0.42% ** |
| 60 μg/mL | 100% ± 0.00% ** | 100% ± 0.00% ** | 100% ± 0.00% ** |
| Concentrate/Time | 6 h |
|---|---|
| CK | 38.75% ± 0.35% |
| 30 μg/mL | 88.38% ± 0.14% ** |
| 60 μg/mL | 100% ± 0.00% ** |
| Pathogen | Concentrate (μg/mL) | Diameter (mm) | Inhibition Rate (%) |
|---|---|---|---|
| A. heveae | 150 | 31.29 ± 0.63 | 64.77 ± 1.34 ** |
| 100 | 19.14 ± 0.14 | 39.62 ± 0.87 * | |
| 50 | 12.62 ± 0.48 | 26.11 ± 1.26 * | |
| C. acutatum | 150 | 48.63 ± 0.15 | 98.6 ± 1.09 ** |
| 100 | 23.07 ± 0.51 | 52.47 ± 1.19 * | |
| 50 | 18.17 ± 0.24 | 36.83 ± 1.87 * |
| Primer | Restriction Enzyme | Sequences a |
|---|---|---|
| TasAS | F: 5′-ATGGGTATGAAAAAGAA-3′ | |
| R: 5′-AGAACTTCGGATCAATATGCT′-3′ | ||
| pET28a-TasA | EcoRΙ | F:5′-CAAATGGGTCGCGGATCCGAATTCGCATTTAATGATGTGAAGTCCAC-3′ |
| NotI | R:5′-GTGGTGGTGCTCGAGTGCGGCCGCAGAACTTCGGATCAATATGCTGAT-3′ | |
| pCZN1-TasA | NdeI | F:5′-ATGAATCACAAAGTGCATCATCATCATCATCATATGAACGATGTGAAAAGCA-3′ |
| XbaI | R:5′-CTTTTAAGCAGAGATTACCTATCTAGATTAGCTGCTACGAATCA-3′ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dai, L.-M.; He, L.-L.; Li, L.-L.; Liu, Y.-X.; Shi, Y.-P.; Su, H.-P.; Cai, Z.-Y. Cloning, Heterologous Expression, and Antifungal Activity Evaluation of a Novel Truncated TasA Protein from Bacillus amyloliquefaciens BS-3. Int. J. Mol. Sci. 2025, 26, 7529. https://doi.org/10.3390/ijms26157529
Dai L-M, He L-L, Li L-L, Liu Y-X, Shi Y-P, Su H-P, Cai Z-Y. Cloning, Heterologous Expression, and Antifungal Activity Evaluation of a Novel Truncated TasA Protein from Bacillus amyloliquefaciens BS-3. International Journal of Molecular Sciences. 2025; 26(15):7529. https://doi.org/10.3390/ijms26157529
Chicago/Turabian StyleDai, Li-Ming, Li-Li He, Lan-Lan Li, Yi-Xian Liu, Yu-Ping Shi, Hai-Peng Su, and Zhi-Ying Cai. 2025. "Cloning, Heterologous Expression, and Antifungal Activity Evaluation of a Novel Truncated TasA Protein from Bacillus amyloliquefaciens BS-3" International Journal of Molecular Sciences 26, no. 15: 7529. https://doi.org/10.3390/ijms26157529
APA StyleDai, L.-M., He, L.-L., Li, L.-L., Liu, Y.-X., Shi, Y.-P., Su, H.-P., & Cai, Z.-Y. (2025). Cloning, Heterologous Expression, and Antifungal Activity Evaluation of a Novel Truncated TasA Protein from Bacillus amyloliquefaciens BS-3. International Journal of Molecular Sciences, 26(15), 7529. https://doi.org/10.3390/ijms26157529






