DNA Methylation in Bladder Cancer: Diagnostic and Therapeutic Perspectives—A Narrative Review
Abstract
1. Introduction
2. Approach to Literature Selection
3. Molecular Landscape of DNA Methylation and Demethylation
3.1. DNA Methylation Patterns in Bladder Cancer
3.1.1. Active Demethylation Pathways
3.1.2. Passive Demethylation Pathways
4. DNA Methylation in Bladder Cancer
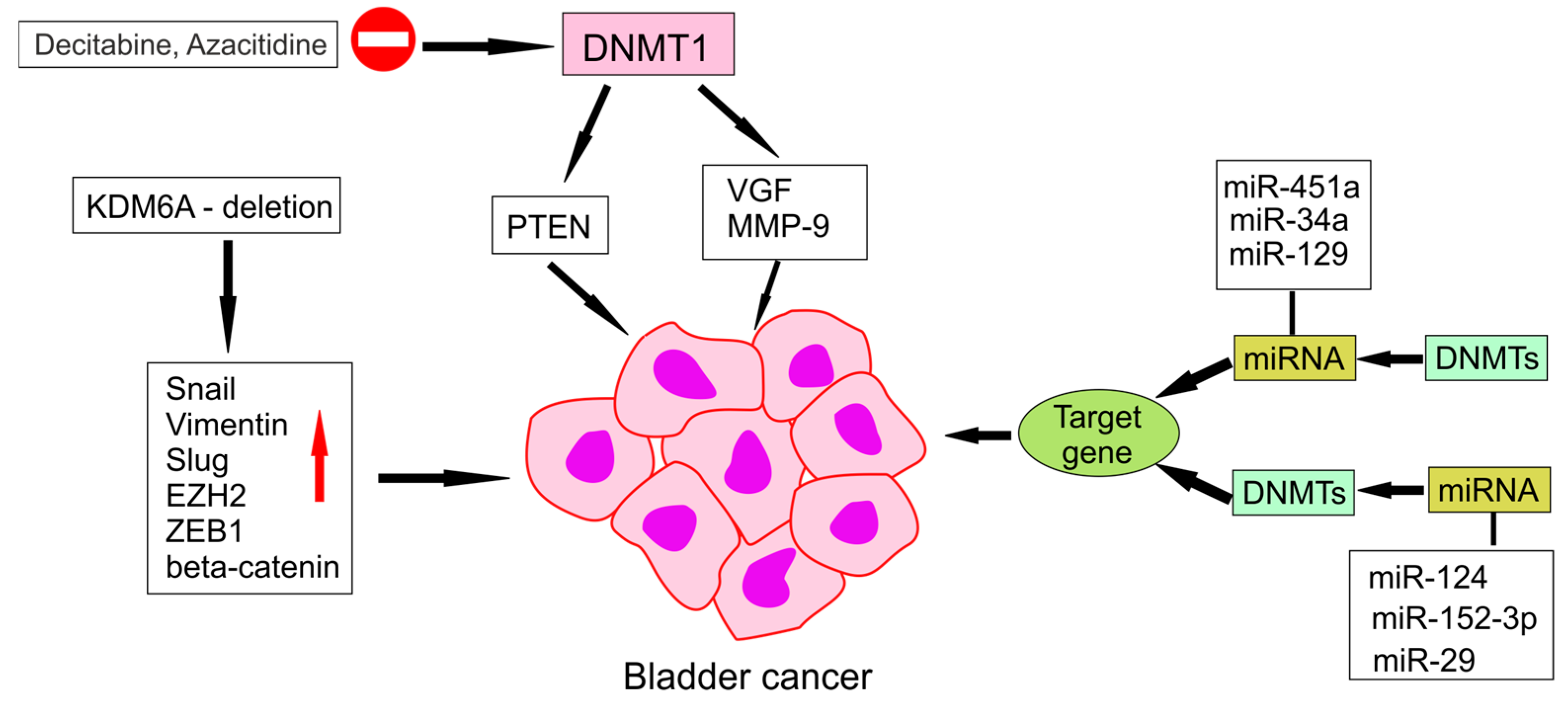
4.1. Hypermethylation of Tumours Suppressor Genes
4.2. Hypomethylation of Tumours Suppressor Genes
4.3. Impact of DNA Demethylation on Tumour Development and Progression
4.4. Clinical Implications of DNA Methylation Changes
5. Therapeutic Applications and Clinical Studies
5.1. Potential for Reversing Aberrant Methylation Patterns
5.2. Demethylating Agents and Their Mechanisms of Action
5.2.1. FDA-Approved Demethylating Agents
5.2.2. Experimental Demethylating Compounds
5.2.3. Challenges and Limitations in Therapy Development
5.3. Prognostic Value of DNA Demethylation in Bladder Cancer
5.4. Clinical Trials and Studies Focusing on DNA Methylation Markers
5.4.1. Clinical Evidence and Translational Insights into DNA Demethylation in Bladder Cancer
5.4.2. Ongoing Clinical Trials
6. Limitations and Future Directions
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| NAT2 | N-acetyltransferase 2 |
| GSTM1 | Glutathione S-Transferase Mu 1 |
| TURBT | transurethral resection of bladder tumours |
| SOX11 | SRY-Box Transcription Factor 11 |
| HSPB9 | Heat Shock Protein Beta-9 |
| CpG | Cytosine-phosphate-Guanine dinucleotide |
| AI | Artificial Intelligence |
| TET | Ten-Eleven Translocation |
| 5mC | 5-methylcytosine |
| 5hmC | 5-hydroxymethylcytosine |
| 5fC | 5-formylcytosine |
| 5caC | 5-carboxylcytosine |
| TDG | thymine DNA glycosylase |
| BER | base excision repair |
| HDACi | Histone deacetylase inhibitors |
| TSA | trichostatin A |
| 3′-UTR | 3′-untranslated region |
| NLUTD | neurogenic lower urinary tract dysfunction |
| PCR | Polymerase Chain Reaction |
| KIM-1 | Kidney Injury Molecule-1 |
| FDA | Food and Drug Administration |
| DNMTs | DNA methyltransferases |
| 5-aza | 5-azacytidine |
| MeDEGs | methylation-driven differentially expressed |
| miRNAs | silenced microRNAs |
References
- Thompson, D.; Lawrentschuk, N.; Bolton, D. New Approaches to Targeting Epigenetic Regulation in Bladder Cancer. Cancers 2023, 15, 1856. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Chung, W.; Bondaruk, J.; Jelinek, J.; Lotan, Y.; Liang, S.; Czerniak, B.; Issa, J.-P.J. Detection of bladder cancer using novel DNA methylation biomarkers in urine sediments. Cancer Epidemiol. Biomark. Prev. 2011, 20, 1483–1491. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Jaszek, N.; Bogdanowicz, A.; Siwiec, J.; Starownik, R.; Kwaśniewski, W.; Mlak, R. Epigenetic Biomarkers as a New Diagnostic Tool in Bladder Cancer-From Early Detection to Prognosis. J. Clin. Med. 2024, 13, 7159. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Besaratinia, A.; Cockburn, M.; Tommasi, S. Alterations of DNA methylome in human bladder cancer. Epigenetics 2013, 8, 1013–1022. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Ando, M.; Saito, Y.; Xu, G.; Bui, N.Q.; Medetgul-Ernar, K.; Pu, M.; Fisch, K.; Ren, S.; Sakai, A.; Fukusumi, T.; et al. Chromatin Dysregulation and DNA Methylation at Transcription Start Sites Associated with Transcriptional Repression in Cancers. Nat. Commun. 2019, 10, 2188. [Google Scholar] [CrossRef]
- Nguyen, C.T.; Gonzales, F.A.; Jones, P.A. Altered Chromatin Structure Associated with Methylation-Induced Gene Silencing in Cancer Cells: Correlation of Accessibility, Methylation, MeCP2 Binding and Acetylation. Nucleic Acids Res. 2001, 29, 4598–4606. [Google Scholar] [CrossRef]
- Zhang, J.; Xu, H.; He, Y.; Zheng, X.; Lin, T.; Yang, L.; Tan, P.; Wei, Q. Inhibition of KDM4A restricts SQLE transcription and induces oxidative stress imbalance to suppress bladder cancer. Redox Biol. 2024, 77, 103407. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Singh, P.; Raychaudhuri, D.; Chakraborty, B.; Meher, S.; Tannir, A.J.; Majumdar, A.; Hawkins, J.; Xiong, Y.; Lorenzi, P.; Sharma, P.; et al. Epigenetic modulation shapes tumor immunity and therapeutic response in bladder cancer. bioRxiv 2024. [Google Scholar] [CrossRef]
- Pereira, W.O.; De Carvalho, D.D.; Zenteno, M.E.; Ribeiro, B.F.; Jacysyn, J.F.; Sardinha, L.R.; Zanichelli, M.A.; Hamerschlak, N.; Jones, G.E.; Pagnano, K.B.; et al. BCR-ABL1-induced downregulation of WASP in chronic myeloid leukemia involves epigenetic modification and contributes to malignancy. Cell Death Dis. 2017, 8, e3114. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Su, S.F.; de Castro Abreu, A.L.; Chihara, Y.; Tsai, Y.; Andreu-Vieyra, C.; Daneshmand, S.; Skinner, E.C.; Jones, P.A.; Siegmund, K.D.; Liang, G. A panel of three markers hyper- and hypomethylated in urine sediments accurately predicts bladder cancer recurrence. Clin. Cancer Res. 2014, 20, 1978–1989. [Google Scholar] [CrossRef] [PubMed]
- Bosschieter, J.; Lutz, C.; Segerink, L.I.; Vis, A.N.; Zwarthoff, E.C.; van Moorselaar, R.J.A.; van Rhijn, B.W.; Heymans, M.W.; Jansma, E.P.; Steenbergen, R.D.; et al. The diagnostic accuracy of methylation markers in urine for the detection of bladder cancer: A systematic review. Epigenomics 2018, 10, 673–687. [Google Scholar] [CrossRef] [PubMed]
- Loo Yau, H.; Bell, E.; Ettayebi, I.; de Almeida, F.C.; Boukhaled, G.M.; Shen, S.Y.; Allard, D.; Morancho, B.; Marhon, S.A.; Ishak, C.A.; et al. DNA hypomethylating agents increase activation and cytolytic activity of CD8+ T cells. Mol. Cell 2021, 81, 1469–1483.e8. [Google Scholar] [CrossRef] [PubMed]
- Piatti, P.; Chew, Y.C.; Suwoto, M.; Yamada, T.; Jara, B.; Jia, X.Y.; Guo, W.; Ghodoussipour, S.; Daneshmand, S.; Ahmadi, H.; et al. Clinical evaluation of Bladder CARE, a new epigenetic test for bladder cancer detection in urine samples. Clin. Epigenet. 2021, 13, 84. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Koukourikis, P.; Papaioannou, M.; Pervana, S.; Apostolidis, A. Exploring the DNA Methylation Profile of Genes Associated with Bladder Cancer in Bladder Tissue of Patients with Neurogenic Lower Urinary Tract Dysfunction. Int. J. Mol. Sci. 2024, 25, 5660. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Jiang, Y.H.; Liu, Y.S.; Wei, Y.C.; Jhang, J.F.; Kuo, H.C.; Huang, H.H.; Chan, M.W.Y.; Lin, G.L.; Cheng, W.C.; Lin, S.C.; et al. Hypermethylation Loci of ZNF671, IRF8, and OTX1 as Potential Urine-Based Predictive Biomarkers for Bladder Cancer. Diagnostics 2024, 14, 468. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Shen, C.-H.; Li, P.-Y.; Wang, S.-C.; Wu, S.-R.; Hsieh, C.-Y.; Dai, Y.-C.; Liu, Y.-W. Epigenetic regulation of human WIF1 and DNA methylation situation of WIF1 and GSTM5 in urothelial carcinoma. Heliyon 2023, 9, e16004. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Zhang, C.; Xu, X.; Wang, T.; Yang, M.; Sun, J.; He, Q.; Zhao, H.; Xie, W.; Yuan, J.; Wang, J. Clinical performance and utility of a noninvasive urine-based methylation biomarker: TWIST1/Vimentin to detect urothelial carcinoma of the bladder. Sci. Rep. 2024, 14, 7941. [Google Scholar] [CrossRef]
- Ruan, W.; Chen, X.; Huang, M.; Wang, H.; Chen, J.; Liang, Z.; Zhang, J.; Yu, Y.; Chen, S.; Xu, S.; et al. A urine-based DNA methylation assay to facilitate early detection and risk stratification of bladder cancer. Clin. Epigenet. 2021, 13, 91. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Beijert, I.J.; van den Burgt, Y.; Hentschel, A.E.; Bosschieter, J.; Kauer, P.C.; Lissenberg-Witte, B.I.; van Moorselaar, R.J.A.; Nieuwenhuijzen, J.A.; Steenbergen, R.D.M. Bladder cancer detection by urinary methylation markers GHSR/MAL: A validation study. World J. Urol. 2024, 42, 578. [Google Scholar] [CrossRef]
- Ilijazi, D.; Pulverer, W.; Ertl, I.E.; Lemberger, U.; Kimura, S.; Abufaraj, M.; D’Andrea, D.; Pradere, B.; Bruchbacher, A.; Graf, A.; et al. Discovery of Molecular DNA Methylation-Based Biomarkers through Genome-Wide Analysis of Response Patterns to BCG for Bladder Cancer. Cells 2020, 9, 1839. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Silva-Ferreira, M.; Carvalho, J.A.; Salta, S.; Henriques, T.S.; Rodrigues, P.P.; Monteiro-Reis, S.; Henriquea, R.; Jerónimo, C. Diagnostic test accuracy of urinary DNA methylation-based biomarkers for the detection of primary and recurrent bladder cancer: A systematic review and meta-analysis. Eur. Urol. Focus 2024, 10, 922–934. [Google Scholar] [CrossRef]
- Zhang, S.; Zhang, J.; Zhang, Q.; Liang, Y.; Du, Y.; Wang, G. Identification of Prognostic Biomarkers for Bladder Cancer Based on DNA Methylation Profile. Front. Cell Dev. Biol. 2022, 9, 817086. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Zhou, Q.; Chen, Q.; Chen, X.; Hao, L. Bioinformatics analysis to screen DNA methylation-driven genes for prognosis of patients with bladder cancer. Transl. Androl. Urol. 2021, 10, 3604–3619. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Kisseljova, N.P.; Kisseljov, F.L. DNA demethylation and carcinogenesis. Biochemistry (Moscow) 2005, 70, 743–752. [Google Scholar] [CrossRef]
- Martinez, V.G.; Rubio, C.; Martínez-Fernández, M.; Segovia, C.; López-Calderón, F.; Garín, M.; Villacampa, F.; Paramio, J.M. Epigenetics of bladder cancer: Where biomarkers and therapeutic targets meet. Front. Genet. 2019, 10, 1125. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Koukourikis, P.; Papaioannou, M.; Georgopoulos, P.; Tsotridou, E.; Tzortzis, V.; Chaldoupis, G.; Chatzidimitriou, D.; Giannakis, D.; Apostolidis, A. A study of DNA methylation of bladder cancer biomarkers in the urine of patients with neurogenic lower urinary tract dysfunction. Biology 2023, 12, 1126. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Kim, C.; Lee, H.; Park, S.; Jung, H.; Choi, Y.; Kim, Y.; Seo, J.; Han, D. Unveiling bladder cancer prognostic insights by integrating patient-matched sample and CpG methylation analysis. Medicina 2024, 60, 1175. [Google Scholar] [CrossRef] [PubMed]
- Nunes, S.P.; Henrique, R.; Jerónimo, C.; Paramio, J.M. DNA Methylation as a Therapeutic Target for Bladder Cancer. Cells 2020, 9, 1850. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Dan, H.; Zhang, S.; Zhou, Y.; Guan, Q. DNA Methyltransferase Inhibitors: Catalysts for Antitumour Immune Responses. OncoTargets Ther. 2019, 12, 10903–10916. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Xu, B.-S.; Jiang, Y.-Y.; Liao, C.; Yan, M.-B. Heparanase gene hypomethylation as a potential biomarker for precision screening of bladder cancer. Dis. Diagn. 2022, 11, 100–104. [Google Scholar] [CrossRef]
- Chen, X.; Zhang, J.; Ruan, W.; Huang, M.; Wang, C.; Wang, H.; Jiang, Z.; Wang, S.; Liu, Z.; Liu, C.; et al. Urine DNA methylation assay enables early detection and recurrence monitoring for bladder cancer. J. Clin. Investig. 2020, 130, 6278–6289. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Qiu, H.; Makarov, V.; Bolzenius, J.K.; Halstead, A.; Parker, Y.; Wang, A.; Iyer, G.V.; Wise, H.; Kim, D.; Thayaparan, V.; et al. KDM6A Loss Triggers an Epigenetic Switch That Disrupts Urothelial Differentiation and Drives Cell Proliferation in Bladder Cancer. Cancer Res. 2023, 83, 814–829. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Ramakrishnan, S.; Hu, Q.; Krishnan, N.; Wang, D.; Smit, E.; Granger, V.; Rak, M.; Attwood, K.; Johnson, C.; Morrison, C.; et al. Decitabine, a DNA-demethylating agent, promotes differentiation via NOTCH1 signaling and alters immune-related pathways in muscle-invasive bladder cancer. Cell Death Dis. 2017, 8, 3217. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Jung, I.; An, J.; Ko, M. Epigenetic Regulators of DNA Cytosine Modification: Promising Targets for Cancer Therapy. Biomedicines 2023, 11, 654. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Puia, D.; Ivănuță, M.; Pricop, C. Kidney Injury Molecule-1 as a Biomarker for Renal Cancer: Current Insights and Future Perspectives—A Narrative Review. Int. J. Mol. Sci. 2025, 26, 3431. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Kandimalla, R.; van Tilborg, A.A.; Kompier, L.C.; Stumpel, D.J.; Stam, R.W.; Bangma, C.H.; Zwarthoff, E.C. Genome-wide analysis of CpG island methylation in bladder cancer identified TBX2, TBX3, GATA2, and ZIC4 as pTa-specific prognostic markers. Eur. Urol. 2012, 61, 1245–1256. [Google Scholar] [CrossRef]
- Lin, H.H.; Ke, H.L.; Huang, S.P.; Wu, W.J.; Chen, Y.K.; Chang, L.L. Increase sensitivity in detecting superficial, low grade bladder cancer by combination analysis of hypermethylation of E-cadherin, p16, p14, RASSF1A genes in urine. Urol. Oncol. 2010, 28, 597–602. [Google Scholar] [CrossRef] [PubMed]
- Maruyama, R.; Toyooka, S.; Toyooka, K.O.; Harada, K.; Virmani, A.K.; Zöchbauer-Müller, S.; Farinas, A.J.; Vakar-Lopez, F.; Minna, J.D.; Sagalowsky, A.; et al. Aberrant promoter methylation profile of bladder cancer and its relationship to clinicopathological features. Cancer Res. 2001, 61, 8659–8663. [Google Scholar] [PubMed]
- Ivănuță, M.; Puia, D.; Cimpoeșu, D.C.; Ivănuță, A.M.; Bîcă, O.D.; Pricop, C. Longitudinal Evaluation of Renal Function in Patients with Acquired Solitary Kidney-Urological Perspectives Post-Nephrectomy. J. Clin. Med. 2024, 13, 7470. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Mancini, M.; Righetto, M.; Zumerle, S.; Montopoli, M.; Zattoni, F. The Bladder EpiCheck Test as a Non-Invasive Tool Based on the Identification of DNA Methylation in Bladder Cancer Cells in the Urine: A Review of Published Evidence. Int. J. Mol. Sci. 2020, 21, 6542. [Google Scholar] [CrossRef]
- Deng, L.; Chao, H.; Deng, H.; Yu, Z.; Zhao, R.; Huang, L.; Gong, Y.; Zhu, Y.; Wang, Q.; Li, F.; et al. A novel and sensitive DNA methylation marker for the urine-based liquid biopsies to detect bladder cancer. BMC Cancer 2022, 22, 510. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Laranjeira, A.B.A.; Hollingshead, M.G.; Nguyen, D.; Kinders, R.J.; Doroshow, J.H.; Yang, S.X. DNA damage, demethylation and anticancer activity of DNA methyltransferase (DNMT) inhibitors. Sci. Rep. 2023, 13, 5964. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Cheishvili, D.; Boureau, L.; Szyf, M. DNA demethylation and invasive cancer: Implications for therapeutics. Br. J. Pharmacol. 2015, 172, 2705–2715. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Kim, S.; Kim, Y.; Kong, J.; Kim, E.; Choi, J.H.; Yuk, H.D.; Lee, H.; Kim, H.R.; Lee, K.H.; Kang, M.; et al. Epigenetic regulation of mammalian Hedgehog signaling to the stroma determines the molecular subtype of bladder cancer. eLife 2019, 8, e43024. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Kim, E.; Choi, S.; Kang, B.; Kong, J.; Kim, Y.; Yoon, W.H.; Lee, H.R.; Kim, S.; Kim, H.M.; Lee, H.; et al. Creation of bladder assembloids mimicking tissue regeneration and cancer. Nature 2020, 588, 664–669. [Google Scholar] [CrossRef] [PubMed]
- Bošković, M.; Roje, B.; Chung, F.F.; Gelemanović, A.; Cahais, V.; Cuenin, C.; Khoueiry, R.; Vilović, K.; Herceg, Z.; Terzić, J. DNA Methylome Changes of Muscle- and Neuronal-Related Processes Precede Bladder Cancer Invasiveness. Cancers 2022, 14, 487. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Lee, K.H.; Song, C.G. Epigenetic regulation in bladder cancer: Development of new prognostic targets and therapeutic implications. Transl. Cancer Res. 2017, 6 (Suppl. 4), S677–S688. [Google Scholar] [CrossRef]
- Siddiqui, A.S.; Alshehri, F.A.; Yaqinuddin, A. Aberrant DNA methylation in bladder cancer among Saudi Arabia population. J. Health Allied Sci. NU 2021, 11, 164–169. [Google Scholar] [CrossRef]
- Xu, W.; Gaborieau, V.; Niman, S.M.; Mukeria, A.; Liu, X.; Maremanda, K.P.; Takakura, A.; Zaridze, D.; Freedman, M.L.; Xie, W.; et al. Plasma Kidney Injury Molecule-1 for Preoperative Prediction of Renal Cell Carcinoma Versus Benign Renal Masses, and Association with Clinical Outcomes. J. Clin. Oncol. 2024, 42, 2691–2701. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Shivakumar, M.; Lee, Y.; Bang, L.; Garg, T.; Sohn, K.A.; Kim, D. Identification of epigenetic interactions between miRNA and DNA methylation associated with gene expression as potential prognostic markers in bladder cancer. BMC Med. Genom. 2017, 10 (Suppl. 1), 30. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Fantony, J.J.; Abern, M.R.; Gopalakrishna, A.; Owusu, R.; Tay, J.K.; Lance, R.S.; Inman, B.A. Multi-institutional external validation of urinary TWIST1 and NID2 methylation as a diagnostic test for bladder cancer. Urol. Oncol. 2015, 33, 387.e1–387.e6. [Google Scholar] [CrossRef] [PubMed]

| Author | Gene | Methylation Status | Clinical Relevance | Detection Method |
|---|---|---|---|---|
| Su [10] | SOX11 | Hypermethylated | Early detection marker; frequently methylated in non-invasive bladder tumours, aiding in differentiation from benign conditions | Bisulfite pyrosequencing |
| HSPB9 | Hypermethylated | Potential urinary biomarker for early-stage bladder cancer; supports non-invasive diagnosis | Bisulfite pyrosequencing | |
| Jung [34] | CDKN2A (p16) | Hypermethylated | Commonly hypermethylated in high-grade tumours; may predict response to DNMT inhibitors | qMSP, literature review |
| Chung [2] | NPTX2 | Hypermethylated | Silencing contributes to reduced synaptic activity; potential diagnostic and prognostic marker | qMSP |
| PENK | Hypermethylated | Methylation correlates with tumour aggressiveness and recurrence risk | qMSP | |
| NKX6-2 | Hypermethylated | Frequently silenced in bladder cancer; loss of function linked to decreased apoptotic signalling | qMSP | |
| MYO3A | Hypermethylated | Emerging prognostic biomarker: methylation status associated with tumour subtype differentiation | qMSP | |
| CA10 | Hypermethylated | May influence tumour cell motility; associated with adverse histopathological features | qMSP | |
| Ruan [18] | WIF1 | Reactivation after demethylation | Potential diagnostic value; differential methylation observed between tumour and adjacent normal tissue | In vitro cell line assay |
| Kandimalla [36] | MEIS1 | Hypermethylated | Highly methylated in MIBC, poor prognosis | Tissue |
| SYNPO2 | Hypermethylated | Associated with BCG resistance | Tissue | |
| Lin [37] | APC | Hypermethylated | Diagnostic marker; high detection rate with FGFR3 | Urine/Tissue |
| RASSF1A | Hypermethylated | Diagnostic/prognostic; high detection with FGFR3 | Urine/Tissue | |
| SFRP2 | Hypermethylated | High detection accuracy when combined with FGFR3 mutation | Urine/Tissue | |
| Maruyama [38] | CDH1 | Hypermethylated | Associated with poor survival; independent predictor | Urine/Tissue |
| FHIT | Hypermethylated | Associated with poor survival | Urine/Tissue |
| Demethylation Type | Key Enzymes | Mechanism Description | Example Genes Affected | Clinical Relevance | Key Enzymes |
|---|---|---|---|---|---|
| Active | TET1/2/3, TDG | 5mC → 5hmC/5fC/5caC → BER-mediated replacement | WIF1, CDKN2A | Tumour suppressor reactivation | TET1/2/3, TDG |
| Passive | DNMT1 inhibition | Failure of maintenance during replication | Prognostic markers (e.g., SOX11) | Associated with tumour progression | DNMT1 inhibition |
| Author | Intervention/Agent | Mechanism | Phase | Outcomes |
|---|---|---|---|---|
| Laranjeira et al. [42] | Decitabine, Azacitidine | DNMT inhibition | Preclinical | Effective demethylation and tumour inhibition |
| Ramakrishnan et al. [33] | Low-dose Decitabine | NOTCH1 upregulation | Preclinical | Reversal of gene silencing |
| Thompson et al. [1] | DNMT1 silencing | Epigenetic therapy | Review | Growth suppression via DNMT1 inhibition |
| Qiu et al. [32] | — | KDM6A pathway | Translational | TF network disruption in KDM6A-mutant tumours |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Puia, D.; Ivănuță, M.; Pricop, C. DNA Methylation in Bladder Cancer: Diagnostic and Therapeutic Perspectives—A Narrative Review. Int. J. Mol. Sci. 2025, 26, 7507. https://doi.org/10.3390/ijms26157507
Puia D, Ivănuță M, Pricop C. DNA Methylation in Bladder Cancer: Diagnostic and Therapeutic Perspectives—A Narrative Review. International Journal of Molecular Sciences. 2025; 26(15):7507. https://doi.org/10.3390/ijms26157507
Chicago/Turabian StylePuia, Dragoş, Marius Ivănuță, and Cătălin Pricop. 2025. "DNA Methylation in Bladder Cancer: Diagnostic and Therapeutic Perspectives—A Narrative Review" International Journal of Molecular Sciences 26, no. 15: 7507. https://doi.org/10.3390/ijms26157507
APA StylePuia, D., Ivănuță, M., & Pricop, C. (2025). DNA Methylation in Bladder Cancer: Diagnostic and Therapeutic Perspectives—A Narrative Review. International Journal of Molecular Sciences, 26(15), 7507. https://doi.org/10.3390/ijms26157507