Complete Mitochondrial (mtDNA) Genome Analysis of Economically Significant Fish Cirrhinus cirrhosus in Bangladesh
Abstract
1. Introduction
2. Results
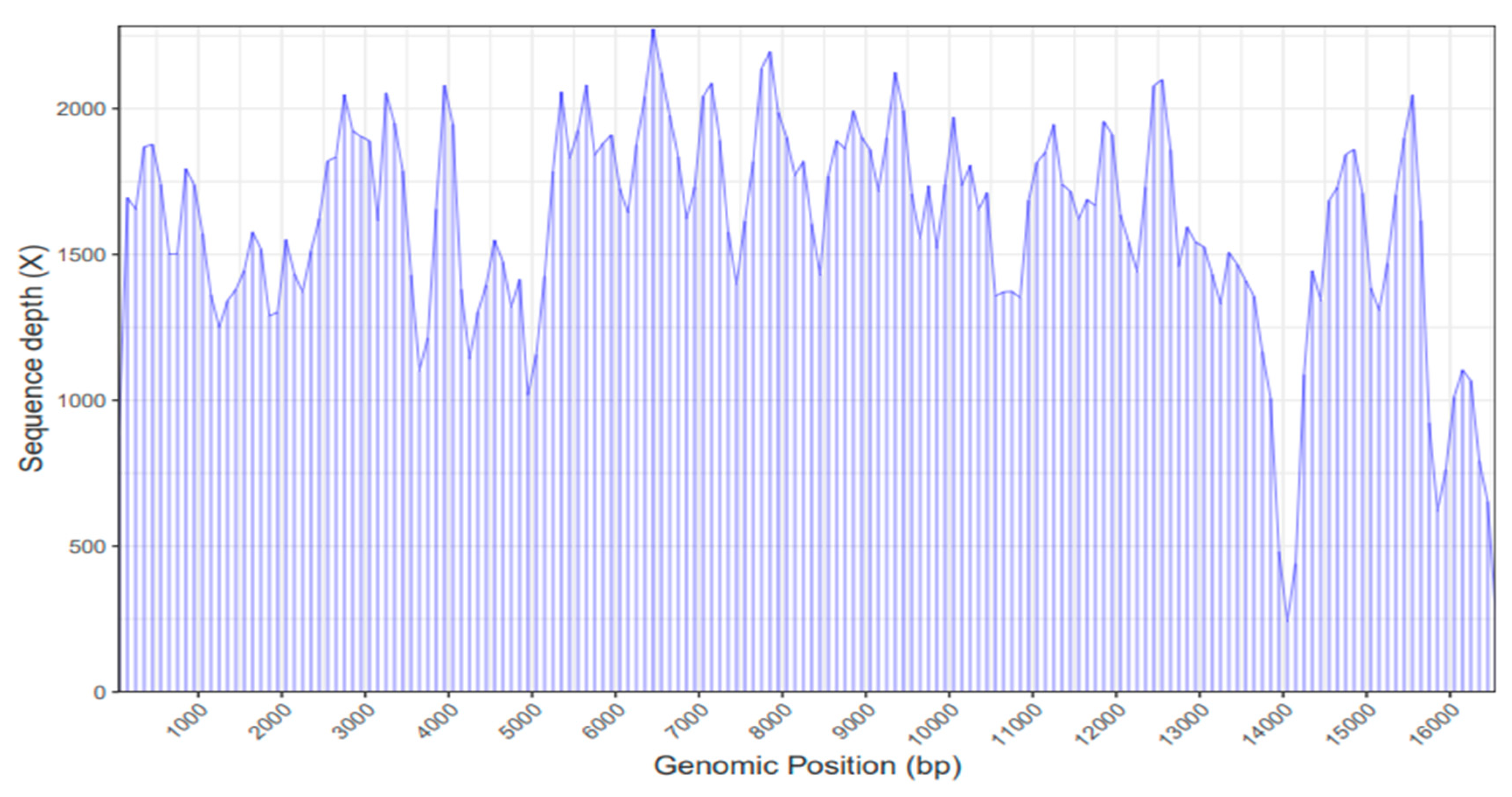
2.1. Mitochondrial Genome Assembly
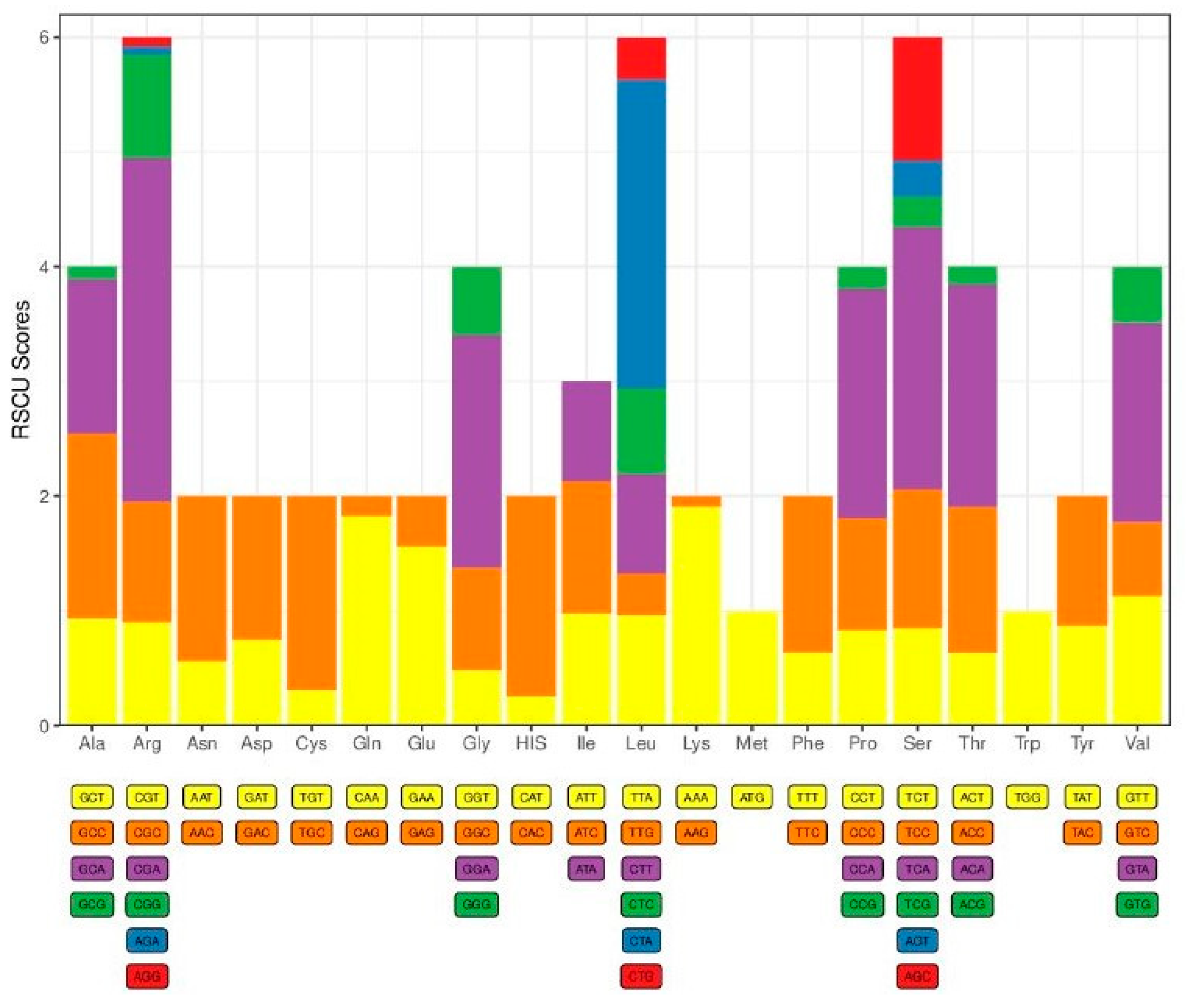
2.2. Protein-Coding Genes
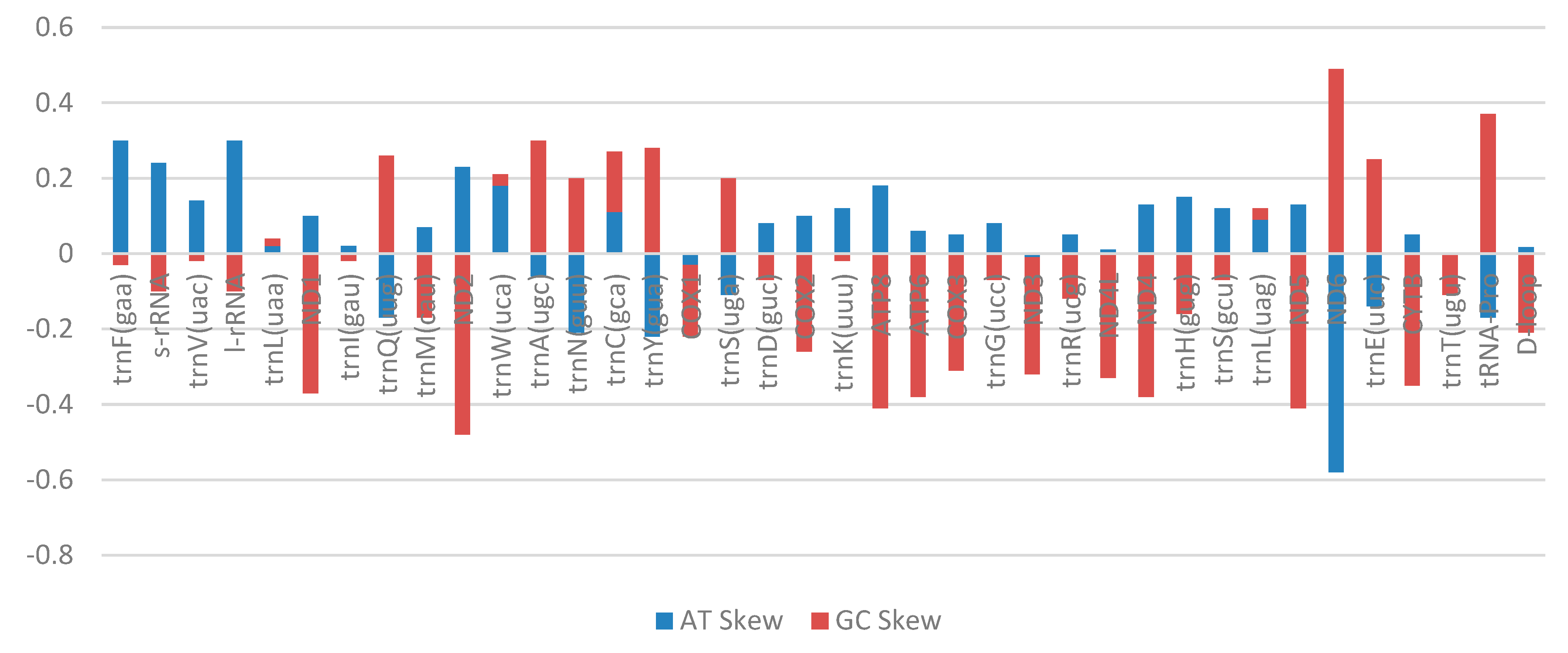
2.3. Transfer RNA Genes, Ribosomal RNA Genes, and Control Region
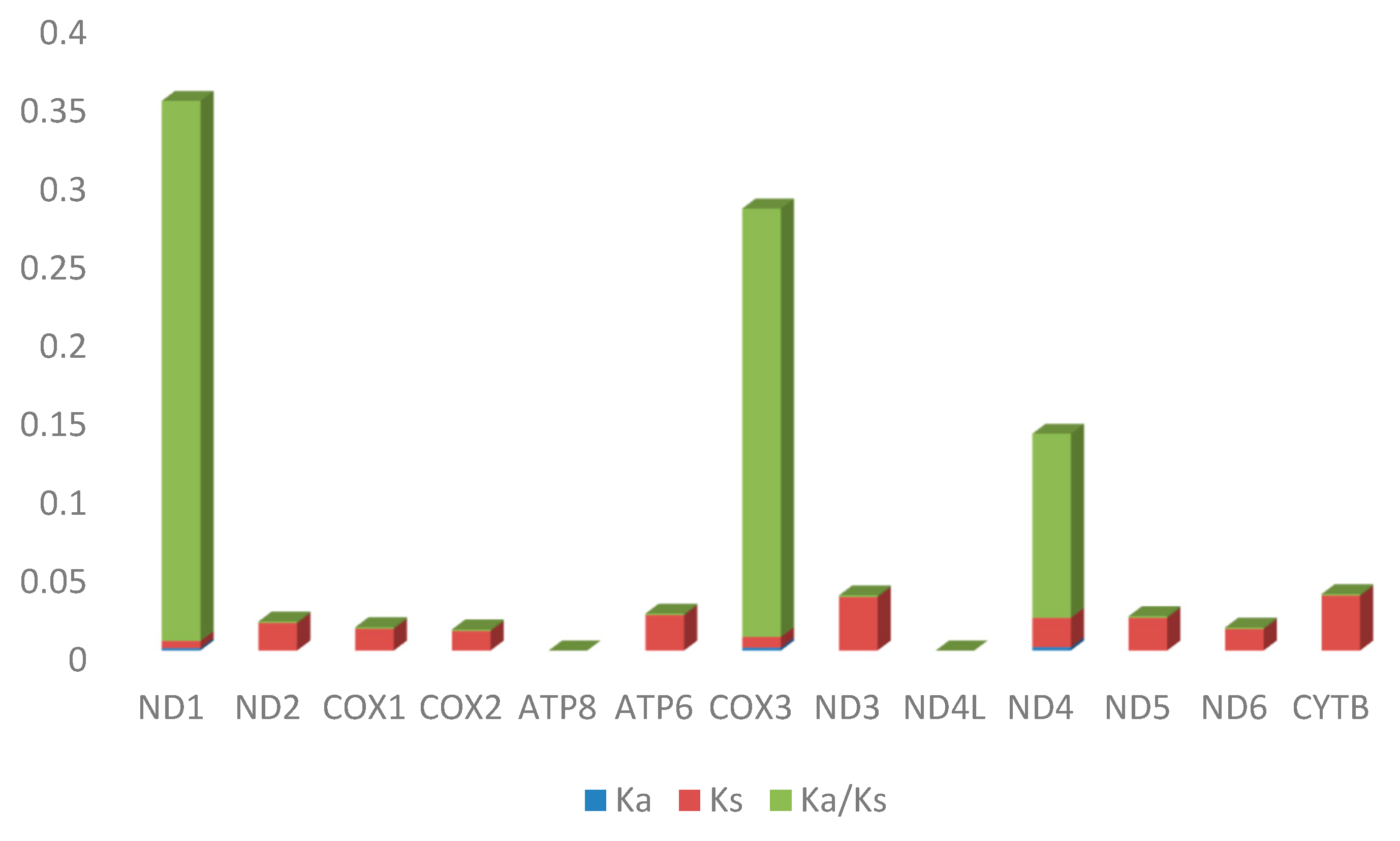
2.4. Selective Pressure Analysis
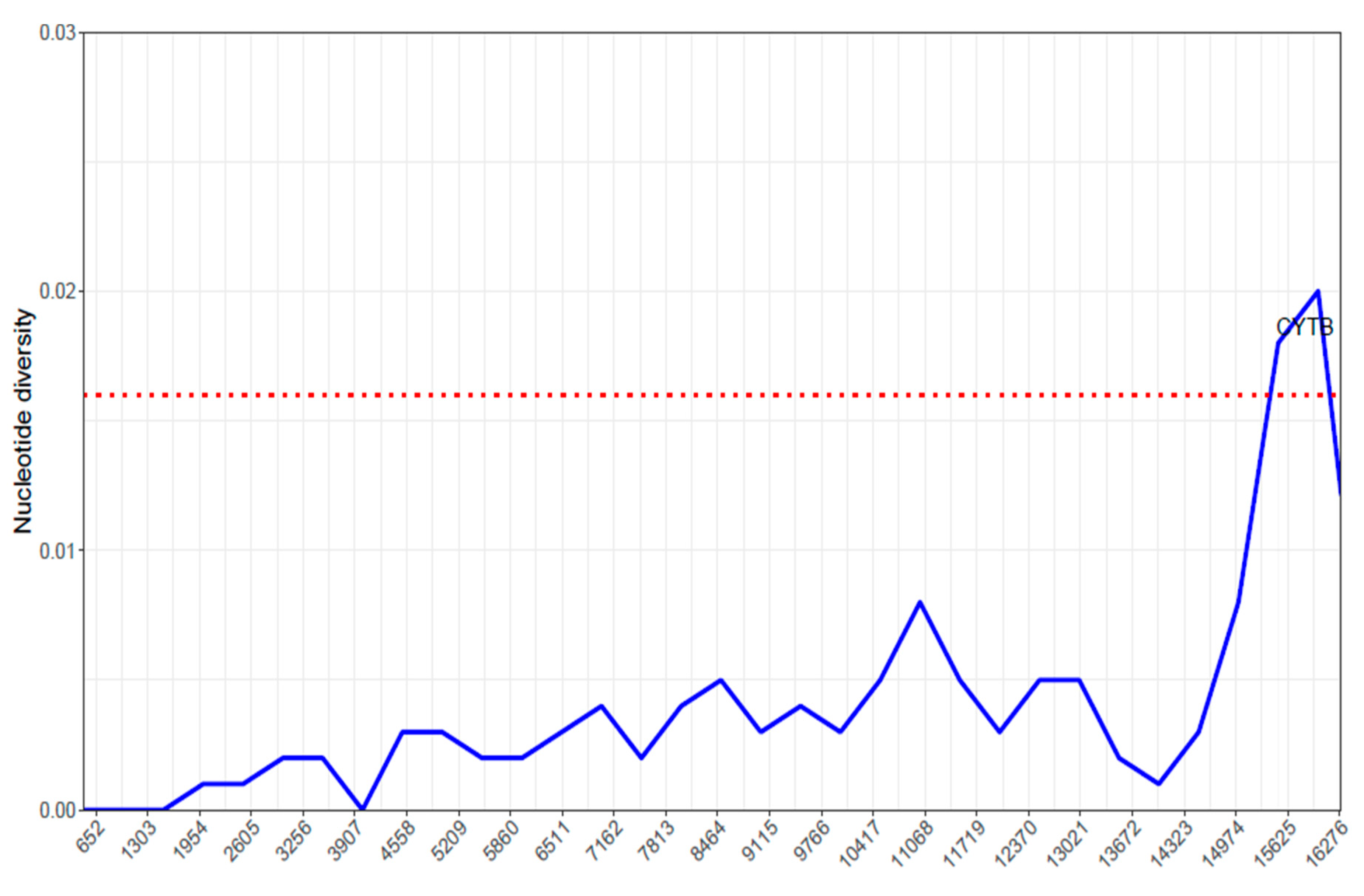
2.5. Nucleotide Diversity
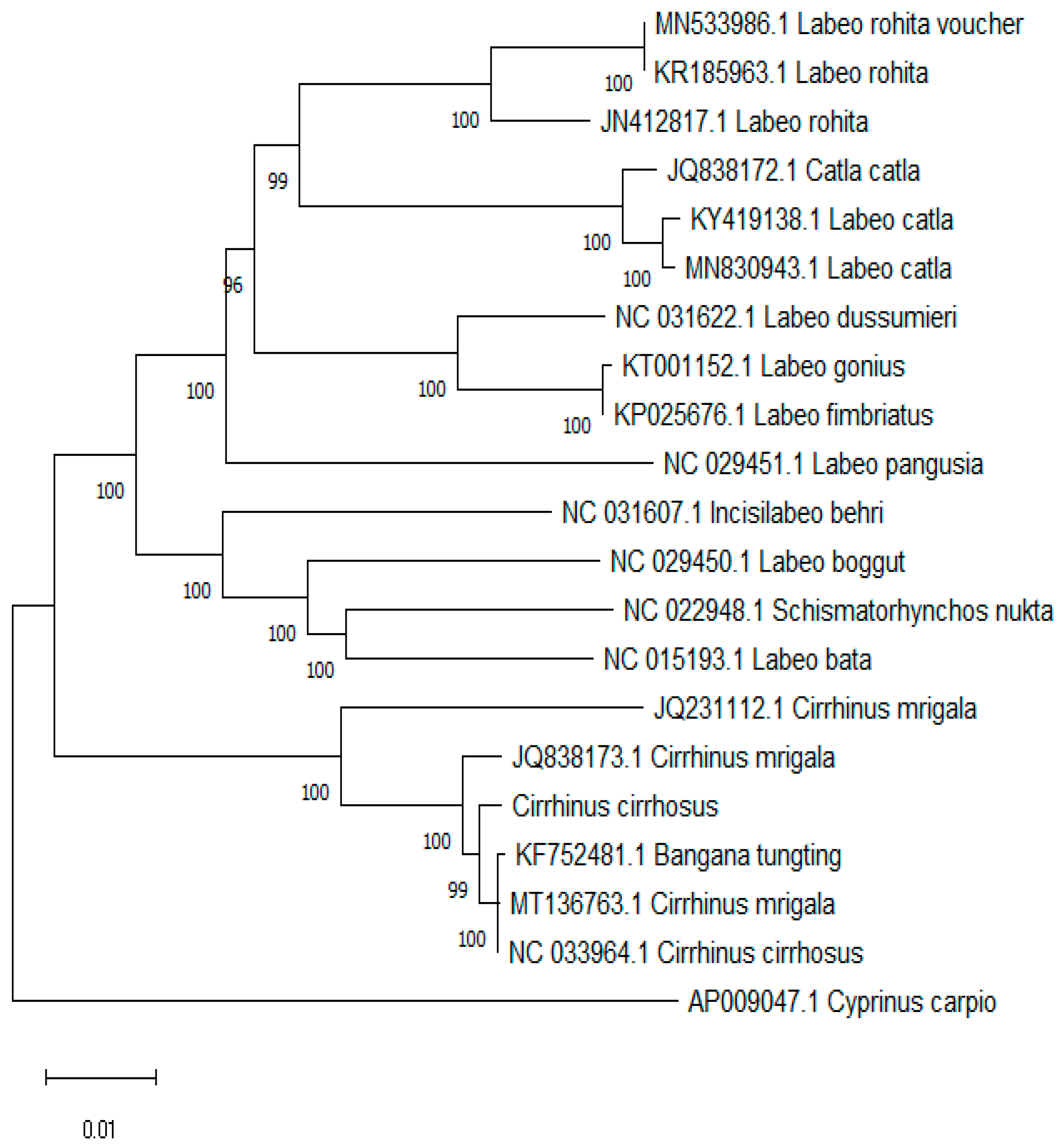
2.6. Phylogenetic Analysis
3. Discussion
4. Materials and Methods
4.1. Ethics Statement
4.2. Sample Collection, DNA Extraction, and Quality Assessment
4.3. Next-Generation Sequencing
4.4. Mitogenome Assembly and Annotation
4.5. Phylogenetic Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- DoF. Yearbook of Fisheries Statistics of Bangladesh, 2021–2022; Fisheries Resources Survey System (FRSS), Department of Fisheries, Ministry of Fisheries and Livestock: Dhaka, Bangladesh, 2022. [Google Scholar]
- Biswas, G.; Jena, J.K.; Singh, S.K.; Patmajhi, P.; Muduli, H.K. Effect of feeding frequency on growth, survival and feed utilization in mrigal, Cirrhinus mrigala, and rohu, Labeo rohita, during nursery rearing. Aquaculture 2006, 254, 211–218. [Google Scholar] [CrossRef]
- Chauhan, T.; Lal, K.K.; Mohindra, V.; Singh, R.K.; Punia, P.; Gopalakrishnan, A.; Sharma, P.C.; Lakra, W.S. Evaluating genetic differentiation in wild populations of the Indian major carp, Cirrhinus mrigala (Hamilton-Buchanan, 1882): Evidence from allozyme and microsatellite markers. Aquaculture 2007, 269, 135–149. [Google Scholar] [CrossRef]
- UCN Bangladesh Red List of Bangladesh Volume 1: Summary; IUCN, International Union for Conservation of Nature, Bangladesh Country Office: Dhaka, Bangladesh, 2015; p. xvi+122.
- Aktera, R.; Sharifb, S.H.; Khanc, M.R. An overview of fish biodiversity and socioeconomic situation of the Padma river’s fisher community in Bangladesh. Big Data Agric. (BDA) 2022, 4, 22–27. [Google Scholar] [CrossRef]
- Jahan, H.; Chakraborty, M.; Alam, M.S.; Begum, R.A. Characterization of complete mitochondrial genome of Labeo rohita from Bangladesh. Bioresearch Commun. (BRC) 2024, 10, 1539–1544. [Google Scholar] [CrossRef]
- Singh, M.; Saini, V.P.; Mohindra, V.; Ojha, M.L.; Lal, K.K.; Singh, R.K. Complete mitochondrial genome of golden variant of freshwater fish Labeo rajasthanicus (Cypriniformes: Cyprinidae): Endemic to India. Mitochondrial DNA Part B 2023, 8, 1364–1367. [Google Scholar] [CrossRef]
- Schroeter, J.C.; Maloy, A.P.; Rees, C.B.; Bartron, M.L. Fish mitochondrial genome sequencing: Expanding genetic resources to support species detection and biodiversity monitoring using environmental DNA. Conserv. Genet. Resour. 2020, 12, 433–446. [Google Scholar] [CrossRef]
- Zhang, R.; Zhu, T.; Luo, Q. The Complete Mitochondrial Genome of the Freshwater Fish Onychostoma ovale (Cypriniformes, Cyprinidae): Genome Characterization and Phylogenetic Analysis. Genes 2023, 14, 1227. [Google Scholar] [CrossRef] [PubMed]
- Ye, X.; Lv, Y.; Wei, L.; Huang, J.; Wen, Y.; Zhang, G.; Zhang, S.; Yang, Z.; Liu, K. The complete mitochondrial genome of Jinbian carp Cyprinus carpio (Cypriniformes: Cyprinidae). Mitochondrial DNA Part B 2018, 3, 1096–1097. [Google Scholar] [CrossRef]
- Zhou, C.; Li, B.; Ma, L.; Zhao, Y.; Kong, X. The complete mitogenome of natural triploid Carassius auratus in Qihe River. Mitochondrial DNA Part A 2014, 27, 605–606. [Google Scholar] [CrossRef]
- Luo, L.; Xu, Y.; Wang, S.; Zhang, R.; Guo, K.; Xu, W.; Zhao, Z. Complete Mitochondrial Genome Sequence and Phylogenetic Analysis of Procambarus clarkii and Cambaroides dauricus from China. Int. J. Mol. Sci. 2023, 24, 11282. [Google Scholar] [CrossRef]
- Satoh, T.P.; Miya, M.; Mabuchi, K.; Nishida, M. Structure and variation of the mitochondrial genome of fishes. BMC Genom. 2016, 17, 719. [Google Scholar] [CrossRef] [PubMed]
- Joseph, J.; Sreeedharan, S.; George, S.; Antony, M.M. The complete mitochondrial genome of an endemic cichlid Etroplus canarensis from Western Ghats, India (Perciformes: Cichlidae) and molecular phylogenetic analysis. Mol. Biol. Rep. 2022, 49, 3033–3044. [Google Scholar] [CrossRef]
- Reyes, A.; Gissi, C.; Pesole, G.; Saccone, C. Asymmetrical directional mutation pressure in the mitochondrial genome of mammals. Mol. Biol. Evol. 1998, 15, 957–966. [Google Scholar] [CrossRef]
- Desjardins, P.; Morais, R. Sequence and gene organization of the chicken mitochondrial genome: A novel gene order in higher vertebrates. J. Mol. Biol. 1990, 212, 599–634. [Google Scholar] [CrossRef]
- Chang, Y.-S.; Huang, F.-L.; Lo, T.-B. The complete nucleotide sequence and gene organization of carp (Cyprinus carpio) mitochondrial genome. J. Mol. Evol. 1994, 38, 138–155. [Google Scholar] [CrossRef]
- Montaña-Lozano, P.; Balaguera-Reina, S.A.; Prada-Quiroga, C.F. Comparative analysis of codon usage of mitochondrial genomes provides evolutionary insights into reptiles. Gene 2023, 851, 146999. [Google Scholar] [CrossRef] [PubMed]
- Temperley, R.J.; Wydro, M.; Lightowlers, R.N.; Chrzanowska-Lightowlers, Z.M. Human mitochondrial mRNAs—Like members of all families, similar but different. Biochim. Et Biophys. Acta (BBA)-Bioenerg. 2010, 1797, 1081–1085. [Google Scholar] [CrossRef] [PubMed]
- Sharma, M.R.; Koc, E.C.; Datta, P.P.; Booth, T.M.; Spremulli, L.L.; Agrawal, R.K. Structure of the mammalian mitochondrial ribosome reveals an expanded functional role for its component proteins. Cell 2003, 115, 97–108. [Google Scholar] [CrossRef]
- Fernández-Silva, P.; Enriquez, J.A.; Montoya, J. Replication and transcription of mammalian mitochondrial DNA. Exp. Physiol. 2003, 88, 41–56. [Google Scholar] [CrossRef]
- Kryazhimskiy, S.; Plotkin, J.B. The population genetics of dN/dS. PLoS Genet. 2008, 4, e1000304. [Google Scholar] [CrossRef]
- Kimura, M. Preponderance of synonymous changes as evidence for the neutral theory of molecular evolution. Nature 1977, 267, 275–276. [Google Scholar] [CrossRef] [PubMed]
- Nei, M.; Gojobori, T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 1986, 3, 418–426. [Google Scholar] [CrossRef] [PubMed]
- Nei, M.; Kumar, S. Molecular Evolution and Phylogenetics; Oxford University Press: Oxford, UK, 2000. [Google Scholar]
- Nabholz, B.; Glémin, S.; Galtier, N. The erratic mitochondrial clock: Variations of mutation rate, not population size, affect mtDNA diversity across birds and mammals. BMC Evol. Biol. 2013, 13, 54. [Google Scholar] [CrossRef]
- Nabholz, B.; Glémin, S.; Galtier, N. Strong variations of mitochondrial mutation rate across mammals—The longevity hypothesis. Mol. Biol. Evol. 2008, 25, 120–130. [Google Scholar] [CrossRef] [PubMed]

| Gene | Gene Type | Location | Intergenic Nucleotides | Size |
|---|---|---|---|---|
| trnF(gaa) | tRNA | 1–69 | 0 | 69 |
| s-rRNA | rRNA | 70–1026 | 0 | 957 |
| trnV(uac) | tRNA | 1027–1098 | 0 | 72 |
| l-rRNA | rRNA | 1099–2786 | 0 | 1688 |
| trnL(uaa) | tRNA | 2787–2862 | 0 | 76 |
| ND1 | CDS | 2864–3838 | 1 | 975 |
| trnI(gau) | tRNA | 3843–3914 | 4 | 72 |
| trnQ(uug) | tRNA | 3913–3983 | −2 | 71 |
| trnM(cau) | tRNA | 3985–4053 | 1 | 69 |
| ND2 | CDS | 4054–5098 | 0 | 1045 |
| trnW(uca) | tRNA | 5099–5169 | 0 | 71 |
| trnA(ugc) | tRNA | 5172–5240 | 2 | 69 |
| trnN(guu) | tRNA | 5242–5314 | 1 | 73 |
| trnC(gca) | tRNA | 5348–5414 | 33 | 67 |
| trnY(gua) | tRNA | 5415–5484 | 0 | 70 |
| COX1 | CDS | 5486–7036 | 1 | 1551 |
| trnS(uga) | tRNA | 7037–7107 | 0 | 71 |
| trnD(guc) | tRNA | 7111–7182 | 3 | 72 |
| COX2 | CDS | 7196–7886 | 13 | 691 |
| trnK(uuu) | tRNA | 7887–7962 | 0 | 76 |
| ATP8 | CDS | 7964–8128 | 1 | 165 |
| ATP6 | CDS | 8122–8804 | −7 | 683 |
| COX3 | CDS | 8805–9590 | 0 | 786 |
| trnG(ucc) | tRNA | 9591–9662 | 0 | 72 |
| ND3 | CDS | 9663–10,011 | 0 | 349 |
| trnR(ucg) | tRNA | 10,012–10,081 | 0 | 70 |
| ND4L | CDS | 10,082–10,378 | 0 | 297 |
| ND4 | CDS | 10,372–11,752 | −7 | 1381 |
| trnH(gug) | tRNA | 11,753–11,821 | 0 | 69 |
| trnS(gcu) | tRNA | 11,822–11,890 | 0 | 69 |
| trnL(uag) | tRNA | 11,892–11,964 | 1 | 73 |
| ND5 | CDS | 11,968–13,791 | 3 | 1824 |
| ND6 | CDS | 13,788–14,309 | −4 | 522 |
| trnE(uuc) | tRNA | 14,310–14,378 | 0 | 69 |
| CYTB | CDS | 14,384–15,524 | 5 | 1141 |
| trnT(ugu) | tRNA | 15,525–15,596 | 0 | 72 |
| tRNA-Pro | tRNA | 15,596–15,665 | −1 | 70 |
| D-loop | misc_feature | 15,666–16,593 | 0 | 928 |
| Gene | Total Bases | Number of Individual Bases | Percentage of Bases (%) | Percentage Content | AT Skew | GC Skew | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | T | G | C | A | T | G | C | AT% | GC% | ||||
| trnF(gaa) | 69 | 26 | 14 | 14 | 15 | 37 | 22 | 20 | 21 | 59 | 41 | 0.3 | −0.03 |
| s-rRNA | 957 | 305 | 184 | 210 | 258 | 31 | 22 | 21 | 26 | 53 | 47 | 0.24 | −0.10 |
| trnV(uac) | 72 | 20 | 15 | 18 | 19 | 27 | 22 | 25 | 26 | 49 | 51 | 0.14 | −0.02 |
| l-rRNA | 1688 | 624 | 334 | 328 | 402 | 36 | 22 | 19 | 23 | 58 | 42 | 0.30 | −0.10 |
| trnL(uaa) | 76 | 19 | 18 | 20 | 19 | 25 | 24 | 26 | 25 | 49 | 51 | 0.02 | 0.02 |
| ND1 | 975 | 292 | 235 | 141 | 307 | 29 | 26 | 14 | 31 | 55 | 45 | 0.10 | −0.37 |
| trnI(gau) | 72 | 19 | 18 | 17 | 18 | 26 | 26 | 23 | 25 | 52 | 48 | 0.02 | −0.02 |
| trnQ(uug) | 71 | 17 | 24 | 19 | 11 | 23 | 36 | 26 | 15 | 59 | 41 | −0.17 | 0.26 |
| trnM(cau) | 69 | 15 | 13 | 17 | 24 | 21 | 21 | 24 | 34 | 42 | 58 | 0.07 | −0.17 |
| ND2 | 1045 | 353 | 218 | 121 | 353 | 33 | 23 | 11 | 33 | 56 | 44 | 0.23 | −0.48 |
| trnW(uca) | 71 | 26 | 18 | 14 | 13 | 36 | 27 | 19 | 18 | 63 | 37 | 0.18 | 0.03 |
| trnA(ugc) | 69 | 23 | 26 | 13 | 7 | 33 | 39 | 18 | 10 | 72 | 28 | −0.06 | 0.3 |
| trnN(guu) | 73 | 15 | 23 | 21 | 14 | 20 | 33 | 28 | 19 | 53 | 47 | −0.21 | 0.2 |
| trnC(gca) | 67 | 20 | 16 | 18 | 13 | 29 | 26 | 26 | 19 | 55 | 45 | 0.11 | 0.16 |
| trnY(gua) | 70 | 12 | 19 | 25 | 14 | 17 | 28 | 35 | 20 | 45 | 55 | −0.22 | 0.28 |
| COX1 | 1551 | 416 | 446 | 277 | 412 | 26 | 31 | 17 | 26 | 57 | 43 | −0.03 | −0.19 |
| trnS(uga) | 71 | 16 | 20 | 21 | 14 | 22 | 30 | 29 | 19 | 52 | 48 | −0.11 | 0.2 |
| trnD(guc) | 72 | 25 | 21 | 12 | 14 | 34 | 31 | 16 | 19 | 65 | 35 | 0.08 | −0.07 |
| COX2 | 691 | 215 | 175 | 111 | 190 | 31 | 26 | 16 | 27 | 57 | 43 | 0.10 | −0.26 |
| trnK(uuu) | 76 | 23 | 18 | 17 | 18 | 30 | 25 | 22 | 23 | 55 | 45 | 0.12 | −0.02 |
| ATP8 | 165 | 59 | 41 | 19 | 46 | 35 | 27 | 11 | 27 | 62 | 38 | 0.18 | −0.41 |
| ATP6 | 683 | 208 | 184 | 90 | 201 | 30 | 28 | 13 | 29 | 58 | 42 | 0.06 | −0.38 |
| COX3 | 786 | 224 | 199 | 125 | 238 | 28 | 27 | 15 | 30 | 55 | 45 | 0.05 | −0.31 |
| trnG(ucc) | 72 | 25 | 21 | 12 | 14 | 34 | 31 | 16 | 19 | 65 | 35 | 0.08 | −0.07 |
| ND3 | 349 | 96 | 98 | 53 | 102 | 27 | 29 | 15 | 29 | 56 | 44 | −0.01 | −0.31 |
| trnR(ucg) | 70 | 20 | 18 | 14 | 18 | 28 | 27 | 20 | 25 | 55 | 45 | 0.05 | −0.12 |
| ND4L | 297 | 78 | 75 | 48 | 96 | 26 | 26 | 16 | 32 | 52 | 48 | 0.01 | −0.33 |
| ND4 | 1381 | 449 | 345 | 181 | 406 | 32 | 26 | 13 | 29 | 58 | 42 | 0.13 | −0.38 |
| trnH(gug) | 69 | 26 | 19 | 10 | 14 | 37 | 29 | 14 | 20 | 66 | 34 | 0.15 | −0.16 |
| trnS(gcu) | 69 | 23 | 18 | 13 | 15 | 33 | 28 | 18 | 21 | 61 | 39 | 0.12 | −0.07 |
| trnL(uag) | 73 | 23 | 19 | 16 | 15 | 31 | 28 | 21 | 20 | 59 | 41 | 0.09 | 0.03 |
| ND5 | 1824 | 612 | 463 | 220 | 529 | 33 | 26 | 12 | 29 | 59 | 41 | 0.13 | −0.41 |
| ND6 | 522 | 59 | 227 | 176 | 60 | 11 | 45 | 33 | 11 | 56 | 44 | −0.58 | 0.49 |
| trnE(uuc) | 69 | 18 | 24 | 17 | 10 | 26 | 36 | 24 | 14 | 62 | 38 | −0.14 | 0.25 |
| CYTB | 1141 | 339 | 305 | 161 | 336 | 29 | 28 | 14 | 29 | 57 | 43 | 0.05 | −0.35 |
| trnT(ugu) | 72 | 18 | 18 | 16 | 20 | 25 | 26 | 22 | 27 | 51 | 49 | 0 | −0.11 |
| tRNA-Pro | 70 | 17 | 24 | 20 | 9 | 24 | 36 | 28 | 12 | 60 | 40 | −0.17 | 0.37 |
| D-loop | 928 | 320 | 309 | 117 | 182 | 34 | 35 | 12 | 19 | 69 | 31 | 0.017 | −0.21 |
| Sequence | Ka | Ks | Ka/Ks | p-Value (Fisher) | Substitutions | Syn-Subs | Non-syn-Subs |
|---|---|---|---|---|---|---|---|
| ND1 | 0.00155 | 0.004498 | 0.344636 | 0.580742 | 2 | 0.882608 | 1.11739 |
| ND2 | 1.75 × 10−5 | 0.017535 | 0.001 | 0.001957 | 3 | 2.98596 | 0.014036 |
| COX1 | 1.39 × 10−5 | 0.013891 | 0.001 | 0.000623 | 4 | 3.98442 | 0.01558 |
| COX2 | 1.25 × 10−5 | 0.012476 | 0.001 | 0.023376 | 2 | 1.9941 | 0.005899 |
| ATP8 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ATP6 | 2.26 × 10−5 | 0.022563 | 0.001 | 0.002104 | 4 | 3.98957 | 0.010428 |
| COX3 | 0.001867 | 0.006831 | 0.273377 | 0.54937 | 2 | 0.922892 | 1.07711 |
| ND3 | 3.42 × 10−5 | 0.034179 | 0.001 | 0.00688 | 3 | 2.99182 | 0.008185 |
| ND4L | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ND4 | 0.002198 | 0.018689 | 0.117584 | 0.005217 | 7 | 4.81737 | 2.18263 |
| ND5 | 2.09 × 10−5 | 0.020923 | 0.001 | 3.87 × 10−6 | 7 | 6.97138 | 0.028618 |
| ND6 | 1.37 × 10−5 | 0.013684 | 0.001 | 0.053704 | 1 | 0.994184 | 0.005816 |
| CYTB | 3.50 × 10−5 | 0.035013 | 0.001 | 1.40 × 10−6 | 8 | 7.9705 | 0.029501 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huda, T.; Kabir, M.A.; Rabbane, M.G. Complete Mitochondrial (mtDNA) Genome Analysis of Economically Significant Fish Cirrhinus cirrhosus in Bangladesh. Int. J. Mol. Sci. 2025, 26, 7473. https://doi.org/10.3390/ijms26157473
Huda T, Kabir MA, Rabbane MG. Complete Mitochondrial (mtDNA) Genome Analysis of Economically Significant Fish Cirrhinus cirrhosus in Bangladesh. International Journal of Molecular Sciences. 2025; 26(15):7473. https://doi.org/10.3390/ijms26157473
Chicago/Turabian StyleHuda, Tajmirul, Md. Alamgir Kabir, and Md. Golam Rabbane. 2025. "Complete Mitochondrial (mtDNA) Genome Analysis of Economically Significant Fish Cirrhinus cirrhosus in Bangladesh" International Journal of Molecular Sciences 26, no. 15: 7473. https://doi.org/10.3390/ijms26157473
APA StyleHuda, T., Kabir, M. A., & Rabbane, M. G. (2025). Complete Mitochondrial (mtDNA) Genome Analysis of Economically Significant Fish Cirrhinus cirrhosus in Bangladesh. International Journal of Molecular Sciences, 26(15), 7473. https://doi.org/10.3390/ijms26157473






