Comparative Chloroplast Genomics, Phylogenomics, and Divergence Times of Sassafras (Lauraceae)
Abstract
1. Introduction
2. Results
2.1. Characteristics of Sassafras Chloroplast Genomes
2.2. Enrichment of Chloroplast DNA Genetic Resources of Sassafras
2.3. Phylogenetic Relationships
2.4. Divergence Time of Sassafras
3. Discussion
3.1. Chloroplast Evolution and Development of Genetic Resources for Sassafras
3.2. Phylogenetic Insights
3.3. Divergence Time and Biogeographic History of Sassafras
3.3.1. Divergence Between S. albidum and Its East Asian Species
3.3.2. Divergence Between S. tzumu and S. randaiense
4. Materials and Methods
4.1. Plant Sampling and DNA Extraction of Sassafras
4.2. Illumina Sequencing, Chloroplast Genome De Novo Assembly and Annotation of Sassafras
4.3. Comparative Chloroplast Genome Analysis of Sassafras
4.4. Identification of cp Microsatellites, Repeats, and DNA Barcodes of Sassafras
4.5. Phylogenetic Analysis
4.6. Divergence Time Estimation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Presl, J.S. Sassafras. In O Prirozenosti Rostlin 2. k.w.; Berchtold, F., Presl, J.S., Eds.; Endersa: Praha, Czech Republic, 1825; p. 30. [Google Scholar]
- Hemsley, W.B. Sassafras in China. (Sassafras tzumu, Hemsl.). Bull. Misc. Inf. (R. Gard. Kew) 1907, 1907, 55–56. [Google Scholar] [CrossRef][Green Version]
- Rehder, A. The American and Asiatic Species of Sassafras. J. Arnold Arbor. 1920, 1, 242–245. [Google Scholar] [CrossRef]
- Wen, J. Evolution of Eastern Asian and Eastern North American Disjunct Distributions in Flowering Plants. Annu. Rev. Ecol. Syst. 1999, 30, 421–455. [Google Scholar] [CrossRef]
- Wen, J. Evolution of Eastern Asian–North American Biogeographic Disjunctions: A Few Additional Issues. Int. J. Plant Sci. 2001, 162, S117–S122. [Google Scholar] [CrossRef]
- Berry, E.W. A Revision of the Flora of the Latah Formation; US Government Printing Office: Washington, DC, USA, 1929. [Google Scholar]
- Poole, I.; Richter, H.G.; Francis, J.E. Evidence for Gondwanan Origins for Sassafras (Lauraceae)? Late Cretaceous Fossil Wood of Antarctica. IAWA J. 2000, 21, 463–475. [Google Scholar] [CrossRef]
- Kostermans, A.J.G.H. Lauraceae. Commun. (Pengumuman) For. Res. Inst. 1957, 57, 1–64. [Google Scholar]
- Van der Werff, H.; Richter, H.G. Toward an Improved Classification of Lauraceae. Ann. Mo. Bot. Gard. 1996, 83, 409–418. [Google Scholar] [CrossRef]
- Rohwer, J.G. Toward a Phylogenetic Classification of the Lauraceae: Evidence from matK Sequences. Syst. Bot. 2000, 25, 60–71. [Google Scholar] [CrossRef]
- Chanderbali, A.S.; van der Werff, H.; Renner, S.S. Phylogeny and Historical Biogeography of Lauraceae: Evidence from the Chloroplast and Nuclear Genomes. Ann. Mo. Bot. Gard. 2001, 88, 104–134. [Google Scholar] [CrossRef]
- Rohwer, J.G.; Rudolph, B. Jumping Genera: The Phylogenetic Positions of Cassytha, Hypodaphnis, and Neocinnamomum (Lauraceae) Based on Different Analyses of trnK Intron Sequences. Ann. Mo. Bot. Gard. 2005, 92, 153–178. [Google Scholar]
- Li, J.; Christophel, D.C.; Conran, J.G.; Li, H.W. Phylogenetic Relationships within the ‘Core’ Laureae (Litsea complex, Lauraceae) Inferred from Sequences of the Chloroplast Gene matK and Nuclear Ribosomal DNA ITS Regions. Plant Syst. Evol. 2004, 246, 193–214. [Google Scholar] [CrossRef]
- Nie, Z.L.; Wen, J.; Sun, H. Phylogeny and Biogeography of Sassafras (Lauraceae) Disjunct between Eastern Asia and Eastern North America. Plant Syst. Evol. 2007, 267, 191–203. [Google Scholar] [CrossRef]
- Rohde, R.; Rudolph, B.; Ruthe, K.; Lorea-Hernández, F.G.; de Moraes, P.L.R.; Li, J.; Rohwer, J.G. Neither Phoebe nor Cinnamomum—The Tetrasporangiate Species of Aiouea (Lauraceae). Taxon 2017, 66, 1085–1111. [Google Scholar] [CrossRef]
- Song, Y.; Yu, W.B.; Tan, Y.H.; Liu, B.; Yao, X.; Jin, J.J.; Padmanaba, M.; Yang, J.B.; Corlett, R.T. Evolutionary Comparisons of the Chloroplast Genome in Lauraceae and Insights into Loss Events in the Magnoliids. Genome Biol. Evol. 2017, 9, 2354–2364. [Google Scholar] [CrossRef]
- Zhao, M.L.; Song, Y.; Ni, J.; Yao, X.; Tan, Y.H.; Xu, Z.F. Comparative Chloroplast Genomics and Phylogenetics of Nine Lindera Species (Lauraceae). Sci. Rep. 2018, 8, 8844. [Google Scholar] [CrossRef] [PubMed]
- Jo, S.; Kim, Y.K.; Cheon, S.H.; Fan, Q.; Kim, K.J. Characterization of 20 Complete Plastomes from the Tribe Laureae (Lauraceae) and Distribution of Small Inversions. PLoS ONE 2019, 14, e0224622. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.-F.; Ma, H.; Ci, X.-Q.; Li, L.; Song, Y.; Liu, B.; Li, H.-W.; Wang, S.-L.; Qu, X.-J.; Hu, J.-L.; et al. Can Plastid Genome Sequencing be Used for Species Identification in Lauraceae? Bot. J. Linn Soc. 2021, 197, 1–14. [Google Scholar] [CrossRef]
- Song, Y.; Yu, W.B.; Tan, Y.H.; Jin, J.J.; Wang, B.; Yang, J.B.; Liu, B.; Corlett, R.T. Plastid Phylogenomics Improve Phylogenetic Resolution in the Lauraceae. J. Syst. Evol. 2020, 58, 423–439. [Google Scholar] [CrossRef]
- Yang, Z.; Ferguson, D.K.; Yang, Y. New Insights into the Plastome Evolution of Lauraceae Using Herbariomics. BMC Plant Biol. 2023, 23, 387. [Google Scholar] [CrossRef]
- Song, Y.; Yu, Q.-F.; Zhang, D.; Chen, L.-G.; Tan, Y.-H.; Zhu, W.; Su, H.-L.; Yao, X.; Liu, C.; Corlett, R.T. New Insights into the Phylogenetic Relationships within the Lauraceae from Mitogenomes. BMC Biol. 2024, 22, 241. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Liu, B.; Song, Y.; Meng, H.-H.; Ci, X.-Q.; Conran, J.G.; de Kok, R.P.J.; de Moraes, P.L.R.; Ye, J.-W.; Tan, Y.-H.; et al. Global Advances in Phylogeny, Taxonomy and Biogeography of Lauraceae. Plant Divers. 2025, 47, 341–364. [Google Scholar] [CrossRef] [PubMed]
- Luo, A.; Zhang, C.; Zhou, Q.-S.; Ho, S.Y.W.; Zhu, C.-D.; Ree, R. Impacts of Taxon-Sampling Schemes on Bayesian Tip Dating Under the Fossilized Birth-Death Process. Syst. Biol. 2023, 72, 781–801. [Google Scholar] [CrossRef] [PubMed]
- Xiao, T.W.; Ge, X.J. Plastome Structure, Phylogenomics, and Divergence Times of Tribe Cinnamomeae (Lauraceae). BMC Genom. 2022, 23, 642. [Google Scholar] [CrossRef]
- Qin, Z.; Zheng, Y.J.; Gui, L.J.; Xie, G.A.; Wu, Y.F. Codon Usage Bias Analysis of Chloroplast Genome of Camphora Tree (Cinnamomum camphora). Guihaia 2018, 38, 1346–1355. [Google Scholar] [CrossRef]
- Li, H.W.; Liu, B.; Davis, C.C.; Yang, Y. Plastome Phylogenomics, Systematics, and Divergence Time Estimation of the Beilschmiedia Group (Lauraceae). Mol. Phylogenet. Evol. 2020, 151, 106901. [Google Scholar] [CrossRef]
- Li, H.-T.; Luo, Y.; Gan, L.; Ma, P.-F.; Gao, L.-M.; Yang, J.-B.; Cai, J.; Gitzendanner, M.A.; Fritsch, P.W.; Zhang, T.; et al. Plastid Phylogenomic Insights into Relationships of All Flowering Plant Families. BMC Biol. 2021, 19, 232. [Google Scholar] [CrossRef]
- Yang, Z.; Liu, B.; Yang, Y.; Ferguson, D.K. Phylogeny and Taxonomy of Cinnamomum (Lauraceae). Ecol. Evol. 2022, 12, e9378. [Google Scholar] [CrossRef]
- Nie, Z.-L.; Wen, J.; Azuma, H.; Qiu, Y.-L.; Sun, H.; Meng, Y.; Sun, W.-B.; Zimmer, E.A. Phylogenetic and Biogeographic Complexity of Magnoliaceae in the Northern Hemisphere Inferred from Three Nuclear Data Sets. Mol. Phylogenet. Evol. 2008, 48, 1027–1040. [Google Scholar] [CrossRef] [PubMed]
- Nie, Z.-L.; Sun, H.; Beardsley, P.M.; Olmstead, R.G.; Wen, J. Evolution of Biogeographic Disjunction between Eastern Asia and Eastern North America in Phryma (Phrymaceae). Am. J. Bot. 2006, 93, 1343–1356. [Google Scholar] [CrossRef]
- McElwain, J.C.; Yiotis, C.; Lawson, T. Using Modern Plant Trait Relationships between Observed and Theoretical Maximum Stomatal Conductance and Vein Density to Examine Patterns of Plant Macroevolution. New Phytol. 2016, 209, 94–103. [Google Scholar] [CrossRef]
- Chou, Y.-W.; Thomas, P.I.; Ge, X.-J.; LePage, B.A.; Wang, C.-N. Refugia and Phylogeography of Taiwania in East Asia. J. Biogeogr. 2011, 38, 1992–2005. [Google Scholar] [CrossRef]
- Qiu, Y.-X.; Fu, C.-X.; Comes, H.P. Plant Molecular Phylogeography in China and Adjacent Regions: Tracing the Genetic Imprints of Quaternary Climate and Environmental Change in the World’s Most Diverse Temperate Flora. Mol. Phylogenet. Evol. 2011, 59, 225–244. [Google Scholar] [CrossRef]
- Zhisheng, A.; Kutzbach, J.E.; Prell, W.L.; Porter, S.C. Evolution of Asian Monsoons and Phased Uplift of the Himalaya–Tibetan Plateau since Late Miocene Times. Nature 2001, 411, 62–66. [Google Scholar] [CrossRef]
- Bodare, S.; Stocks, M.; Yang, J.-C.; Lascoux, M. Origin and Demographic History of the Endemic Taiwan Spruce (Picea morrisonicola). Ecol. Evol. 2013, 3, 3320–3333. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Yu, T.; Li, J.Q. Phylogenetic and Biogeographic Study of Acer L. section Palmata Pax (Sapindaceae) Based on Three Chloroplast DNA Fragment Sequences. Acta Ecol. Sin. 2020, 40, 5992–6000. [Google Scholar] [CrossRef]
- Chung, K.F.; van der Werff, H.; Peng, C.I. Observations on the Floral Morphology of Sassafras randaiense (Lauraceae). Ann. Mo. Bot. Gard. 2010, 97, 1–10. [Google Scholar] [CrossRef]
- Guan, B.; Liu, Q.; Liu, X.; Gong, X. Environment Influences the Genetic Structure and Genetic Differentiation of Sassafras tzumu (Lauraceae). BMC Ecol. Evol. 2024, 24, 80. [Google Scholar] [CrossRef] [PubMed]
- Doyle, J.J.; Doyle, J.L. A Rapid DNA Isolation Procedure for Small Quantities of Fresh Leaf Tissue. Phytochem. Bull. 1987, 19, 11–15. [Google Scholar]
- Jin, J.-J.; Yu, W.-B.; Yang, J.-B.; Song, Y.; Depamphilis, C.W.; Yi, T.-S.; Li, D.-Z. GetOrganelle: A Fast and Versatile Toolkit for Accurate De Novo Assembly of Organelle Genomes. Genome Biol. 2020, 21, 241. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.; Chen, H.; Jiang, M.; Wang, L.; Wu, X.; Huang, L.; Liu, C. CPGAVAS2, an Integrated Plastome Sequence Annotator and Analyzer. Nucleic Acids Res. 2019, 47, W65–W73. [Google Scholar] [CrossRef]
- Tillich, M.; Lehwark, P.; Pellizzer, T.; Ulbricht-Jones, E.S.; Fischer, A.; Bock, R.; Greiner, S. GeSeq–Versatile and Accurate Annotation of Organelle Genomes. Nucleic Acids Res. 2017, 45, W6–W11. [Google Scholar] [CrossRef] [PubMed]
- Lohse, M.; Drechsel, O.; Bock, R. OrganellarGenomeDRAW (OGDRAW): A Tool for the Easy Generation of High-Quality Custom Graphical Maps of Plastid and Mitochondrial Genomes. Curr. Genet. 2007, 52, 267–274. [Google Scholar] [CrossRef]
- Frazer, K.A.; Pachter, L.; Poliakov, A.; Rubin, E.M.; Dubchak, I. VISTA: Computational Tools for Comparative Genomics. Nucleic Acids Res. 2004, 32 (Suppl. S2), W273–W279. [Google Scholar] [CrossRef] [PubMed]
- Darling, A.C.E.; Mau, B.; Blattner, F.R.; Perna, N.T. Mauve: Multiple Alignment of Conserved Genomic Sequence with Rearrangements. Genome Res. 2004, 14, 1394–1403. [Google Scholar] [CrossRef]
- Amiryousefi, A.; Hyvönen, J.; Poczai, P. IRscope: An Online Program to Visualize the Junction Sites of Chloroplast Genomes. Bioinformatics 2018, 34, 3030–3031. [Google Scholar] [CrossRef]
- Wright, F. The ‘Effective Number of Codons’ Used in a Gene. Gene 1990, 87, 23–29. [Google Scholar] [CrossRef]
- Sharp, P.M.; Li, W.H. The Codon Adaptation Index–A Measure of Directional Synonymous Codon Usage Bias, and Its Potential Applications. Nucleic Acids Res. 1987, 15, 1281–1295. [Google Scholar] [CrossRef]
- Beier, S.; Thiel, T.; Münch, T.; Scholz, U.; Mascher, M. MISA-web: A Web Server for Microsatellite Prediction. Bioinformatics 2017, 33, 2583–2585. [Google Scholar] [CrossRef] [PubMed]
- Kurtz, S.; Choudhuri, J.V.; Ohlebusch, E.; Schleiermacher, C.; Stoye, J.; Giegerich, R. REPuter: The Manifold Applications of Repeat Analysis on a Genomic Scale. Nucleic Acids Res. 2001, 29, 4633–4642. [Google Scholar] [CrossRef]
- Rozas, J.; Ferrer-Mata, A.; Sánchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sánchez-Gracia, A. DnaSP 6: DNA Sequence Polymorphism Analysis of Large Datasets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Nakamura, T.; Yamada, K.D.; Tomii, K.; Katoh, K. Parallelization of MAFFT for Large-Scale Multiple Sequence Alignments. Bioinformatics 2018, 34, 2490–2492. [Google Scholar] [CrossRef]
- Zhang, D.; Gao, F.; Jakovlić, I.; Zhou, H.; Zhang, J.; Li, W.X.; Wang, G.T. PhyloSuite: An Integrated and Scalable Desktop Platform for Streamlined Molecular Sequence Data Management and Evolutionary Phylogenetics Studies. Mol. Ecol. Resour. 2020, 20, 348–355. [Google Scholar] [CrossRef] [PubMed]
- Xiang, C.; Gao, F.; Jakovlić, I.; Lei, H.; Hu, Y.; Zhang, H.; Zou, H.; Wang, G.; Zhang, D. Using PhyloSuite for Molecular Phylogeny and Tree-Based Analyses. iMeta 2023, 2, e87. [Google Scholar] [CrossRef]
- Guindon, S.; Gascuel, O. A Simple, Fast, and Accurate Algorithm to Estimate Large Phylogenies by Maximum Likelihood. Syst. Biol. 2003, 52, 696–704. [Google Scholar] [CrossRef]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. jModelTest2: More Models, New Heuristics and Parallel Computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef] [PubMed]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; von Haeseler, A.; Lanfear, R. IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; Van Der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian Phylogenetic Inference and Model Choice across a Large Model Space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v4: Recent Updates and New Developments. Nucleic Acids Res. 2019, 47, 256–259. [Google Scholar] [CrossRef] [PubMed]
- Bouckaert, R.; Heled, J.; Kühnert, D.; Vaughan, T.; Wu, C.-H.; Xie, D.; Suchard, M.A.; Rambaut, A.; Drummond, A.J.; Prlic, A. BEAST 2: A Software Platform for Bayesian Evolutionary Analysis. PLoS Comp. Biol. 2014, 10, e1003537. [Google Scholar] [CrossRef]
- Rambaut, A.; Drummond, A.J.; Xie, D.; Baele, G.; Suchard, M.A. Posterior Summarization in Bayesian Phylogenetics Using Tracer 1.7. Syst. Biol. 2018, 67, 901–904. [Google Scholar] [CrossRef] [PubMed]
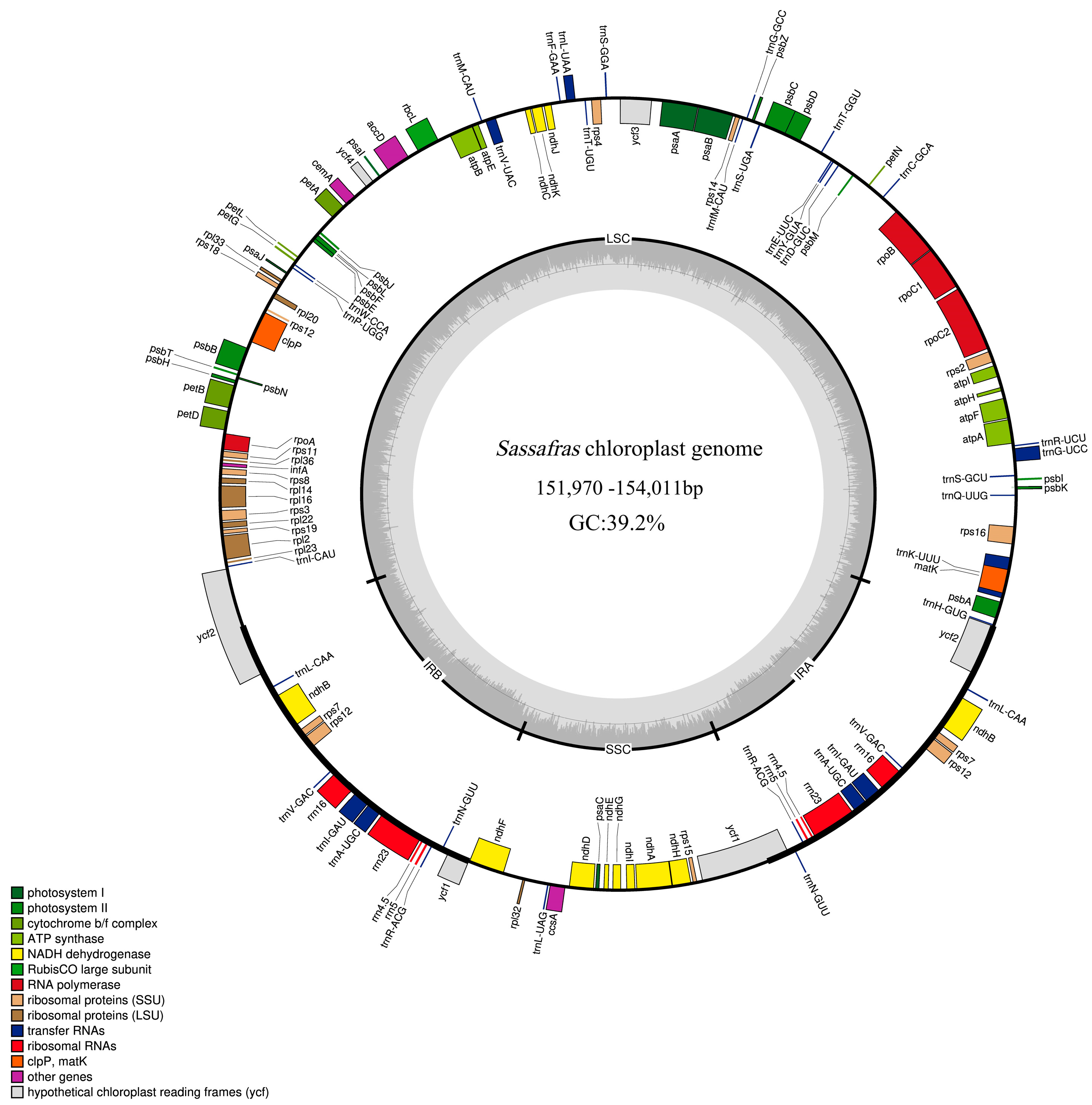
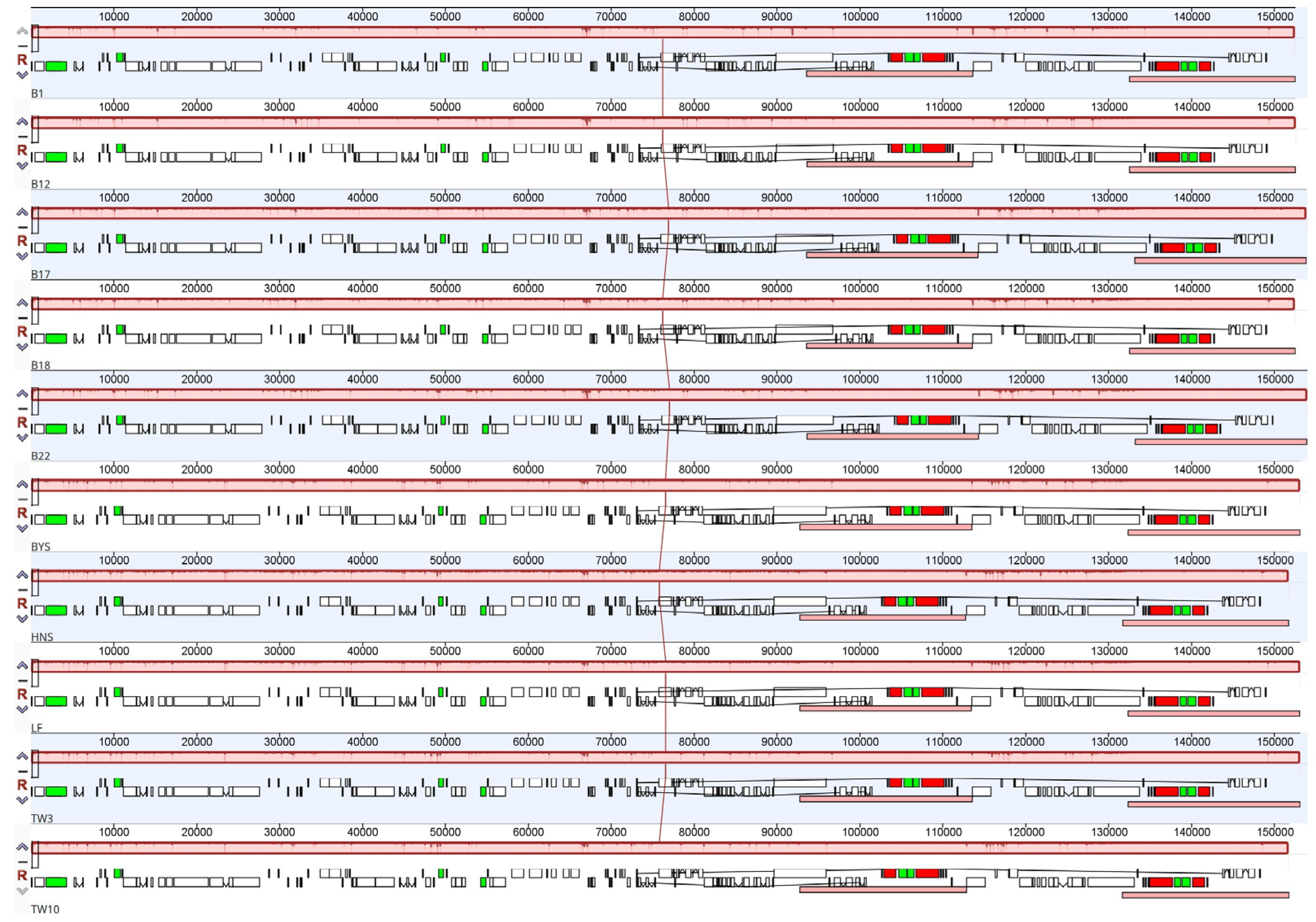

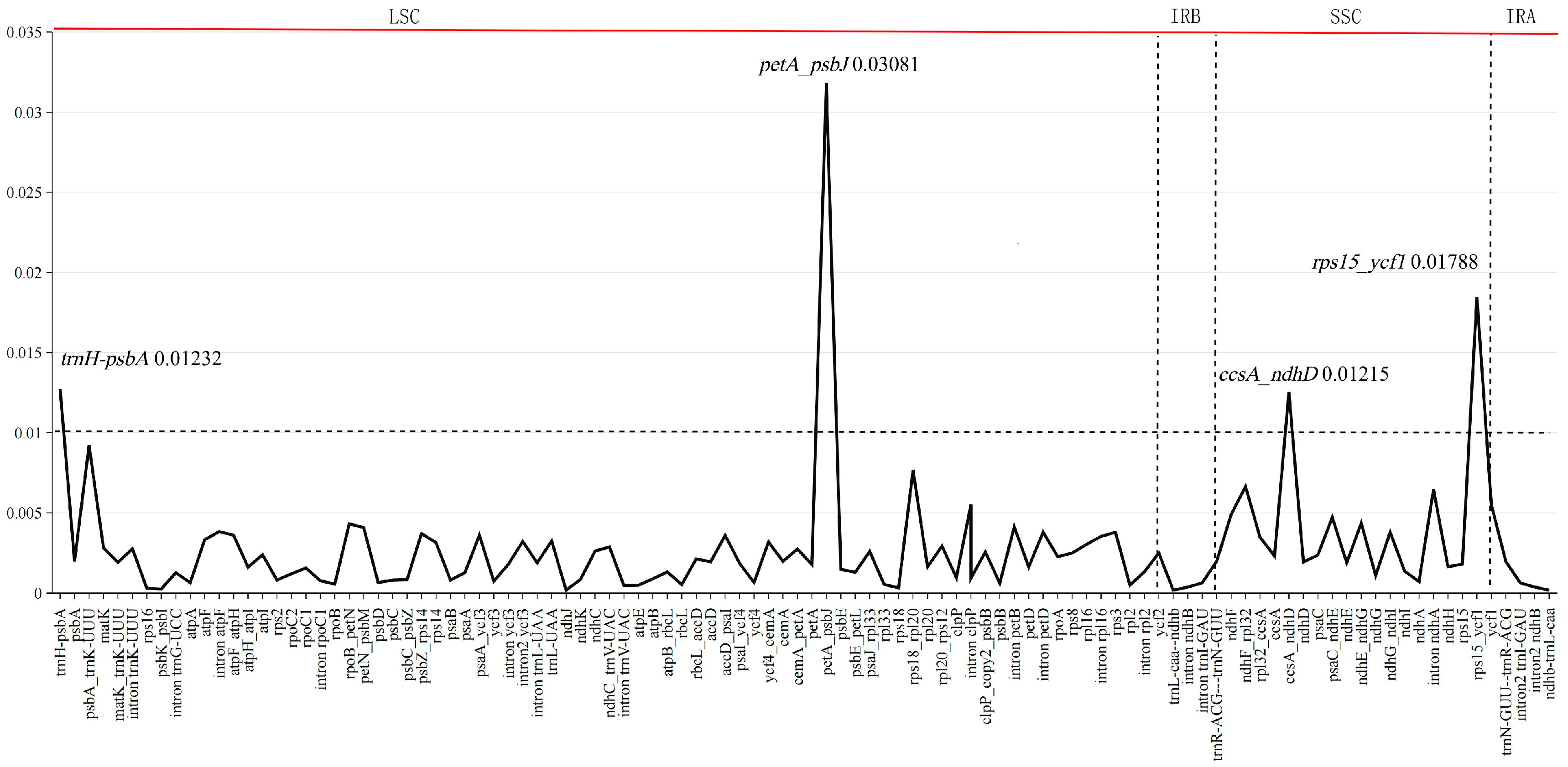
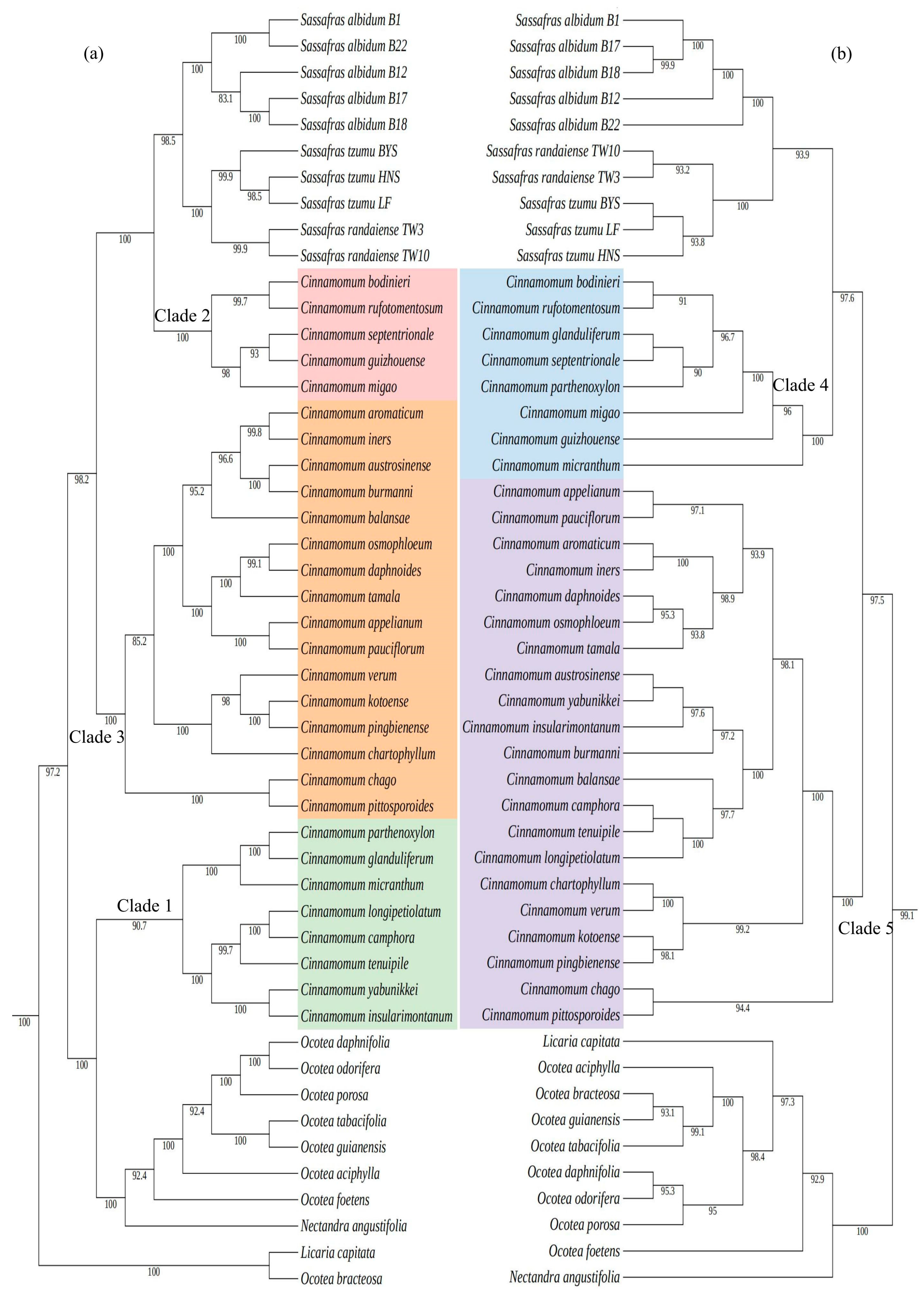

| Species | Collection Number | Genbank ID | Whole Length (bp) | Length of LSC (bp) | Length of IR (bp) | Length of SSC (bp) | Total GC Content (%) | Number of CDS | Number of tRNA | Number of rRNA |
|---|---|---|---|---|---|---|---|---|---|---|
| S. albidum | B1 | MW683126 | 152,562 | 93,587 | 20,054 | 18,867 | 39.2 | 82 | 36 | 8 |
| S. albidum | B12 | MW696794 | 152,621 | 93,600 | 20,077 | 18,867 | 39.2 | 82 | 36 | 8 |
| S. albidum | B17 | MW696797 | 153,910 | 93,562 | 20,732 | 18,884 | 39.2 | 82 | 36 | 8 |
| S. albidum | B18 | MW696798 | 152,567 | 93,564 | 20,059 | 18,885 | 39.2 | 82 | 36 | 8 |
| S. albidum | B22 | MW696799 | 154,001 | 93,634 | 20,750 | 18,867 | 39.2 | 82 | 36 | 8 |
| S. tzumu | BYS | MW696800 | 153,137 | 92,740 | 20,792 | 18,813 | 39.2 | 82 | 36 | 8 |
| S. tzumu | HNS | MW696801 | 151,797 | 92,751 | 20,096 | 18,854 | 39.2 | 82 | 36 | 8 |
| S. tzumu | LF | MW696802 | 153,154 | 92,752 | 20,774 | 18,854 | 39.2 | 82 | 36 | 8 |
| S. randaiense | TW3 | MW696808 | 153,146 | 92,772 | 20,809 | 18,756 | 39.2 | 82 | 36 | 8 |
| S. randaiense | TW10 | MW696807 | 151,790 | 92,772 | 20,131 | 18,756 | 39.2 | 82 | 36 | 8 |
| Groups of Genes | Names of Genes |
|---|---|
| Ribosomal RNAs | rrn4.5 (×2), rrn5 (×2), rrn16 (×2), rrn23 (×2) |
| Transfer RNAs | * trnA-UGC (×2), trnC-GCA, trnD-GUC, trnE-UUC, trnF-GAA, trnG-GCC, * trnG-UCC, trnH-GUG, trnI-CAU, * trnI-GAU (×2), * trnK-UUU, trnL-CAA (×2), * trnL-UAA, trnL-UAG, trnfM-CAU, trnM-CAU, trnN-GUU (×2), trnP-UGG, trnQ-UUG, trnR-ACG (×2), trnR-UCU, trnS-GCU, trnS-GGA, trnS-UGA, trnT-GGU, trnT-UGU, trnV-GAC (×2), * trnV-UAC, trnW-CCA, trnY-GUA |
| Photosystem I | psaA, psaB, psaC, psaI, psaJ |
| Photosystem II | psbA, psbB, psbC, psbD, psbE, psbF, psbH, psbI, psbJ, psbK, psbL, psbM, psbN, psbT, psbZ |
| Cytochrome | petA, * petB, * petD, petG, petL, petN |
| ATP synthase | atpA, atpB, atpE, * atpF, atpH, atpI |
| Rubisco | rbcL |
| NADH dehydrogenease | * ndhA, * ndhB (×2), ndhC, ndhD, ndhE, ndhF, ndhG, ndhH, ndhI, ndhJ, ndhK |
| ATP-dependent protease subunit P | ** clpP |
| Chloroplast envelop membrane protein | cemA |
| Large units | * rpl2, rpl14, * rpl16, rpl20, rpl22, rpl23, rpl32, rpl33, rpl36 |
| Small units | rps2, rps3, rps4, rps7 (×2), rps8, rps11, * rps12 (×2), rps14, rps15, * rps16, rps18, rps19 |
| RNA polymerase | rpoA, rpoB, * rpoC1, rpoC2 |
| Translational initiation factor | infA |
| Miscellaneous proteins | matK, accD, ccsA |
| Hypothetical proteins and conserved reading frame | ** ycf3, ycf4, ycf1, ycf2 |
| Pseudogene | ψ ycf1, ψ ycf2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Z.; Zhang, Y.; Tng, D.Y.P.; Chen, Q.; Wang, Y.; Tian, Y.; Zhou, J.; Wang, Z. Comparative Chloroplast Genomics, Phylogenomics, and Divergence Times of Sassafras (Lauraceae). Int. J. Mol. Sci. 2025, 26, 7357. https://doi.org/10.3390/ijms26157357
Li Z, Zhang Y, Tng DYP, Chen Q, Wang Y, Tian Y, Zhou J, Wang Z. Comparative Chloroplast Genomics, Phylogenomics, and Divergence Times of Sassafras (Lauraceae). International Journal of Molecular Sciences. 2025; 26(15):7357. https://doi.org/10.3390/ijms26157357
Chicago/Turabian StyleLi, Zhiyuan, Yunyan Zhang, David Y. P. Tng, Qixun Chen, Yahong Wang, Yongjing Tian, Jingbo Zhou, and Zhongsheng Wang. 2025. "Comparative Chloroplast Genomics, Phylogenomics, and Divergence Times of Sassafras (Lauraceae)" International Journal of Molecular Sciences 26, no. 15: 7357. https://doi.org/10.3390/ijms26157357
APA StyleLi, Z., Zhang, Y., Tng, D. Y. P., Chen, Q., Wang, Y., Tian, Y., Zhou, J., & Wang, Z. (2025). Comparative Chloroplast Genomics, Phylogenomics, and Divergence Times of Sassafras (Lauraceae). International Journal of Molecular Sciences, 26(15), 7357. https://doi.org/10.3390/ijms26157357






