Characterization and Complete Genomic Analysis of a Novel Bacteriophage BUCT775 for Acinetobacter baumannii and Its Elimination Efficiency in the Environment
Abstract
1. Introduction
2. Results and Discussion
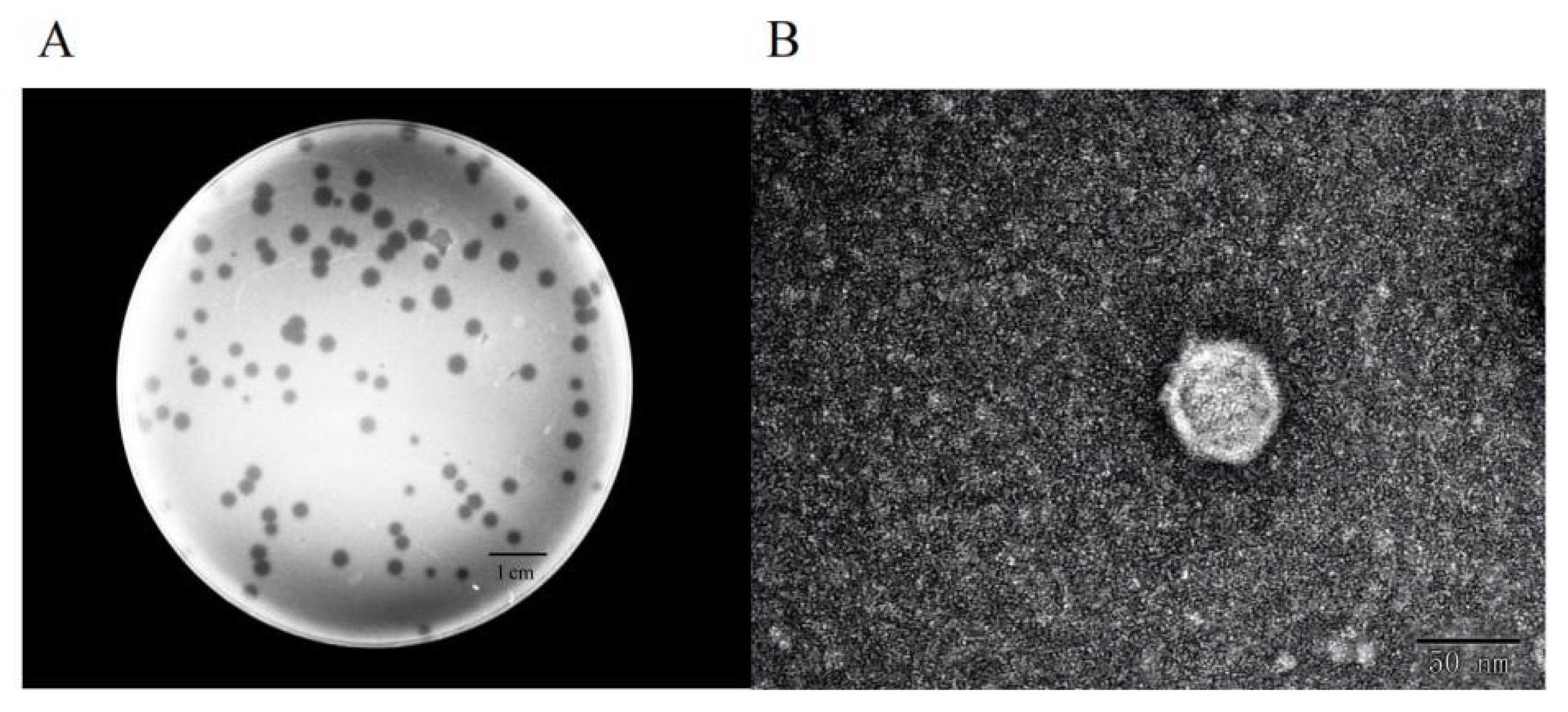
2.1. Morphology
2.2. Host Range
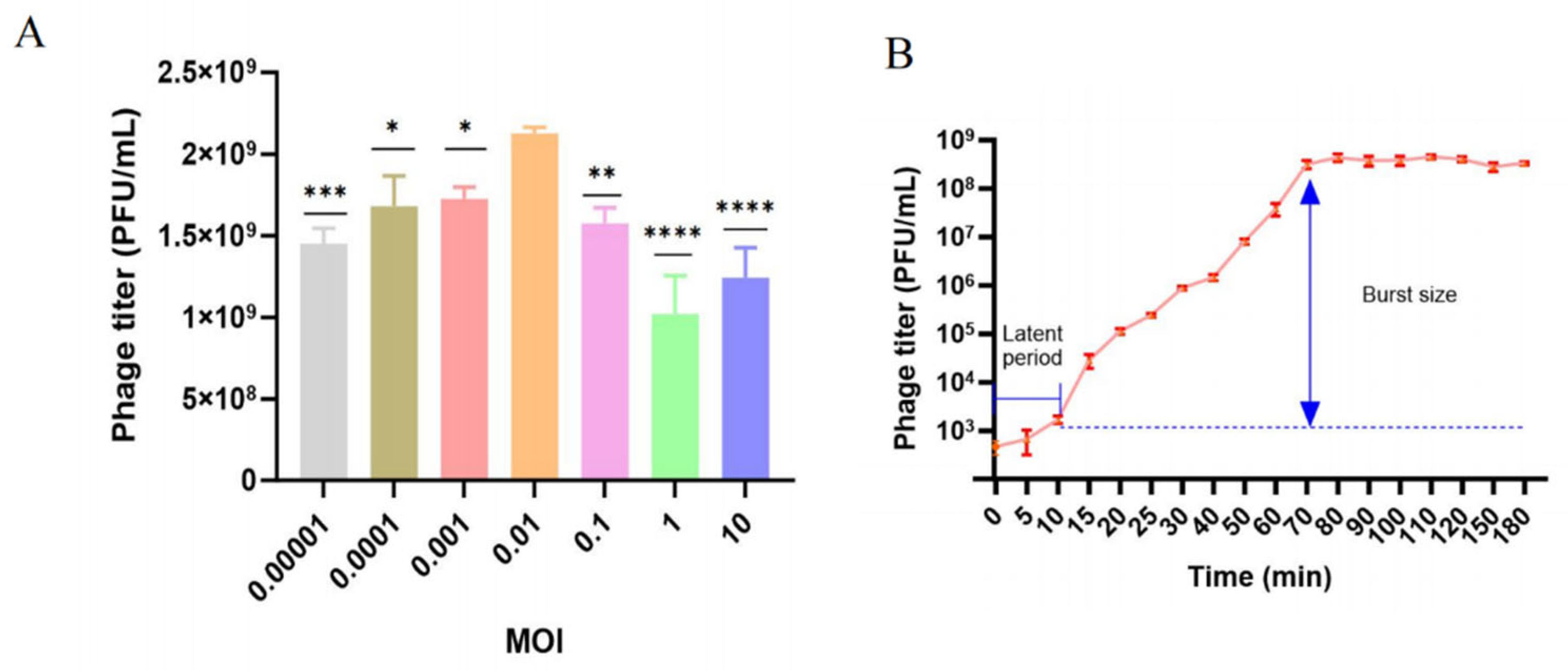
2.3. Optimal Multiplicity of Infection and One-Step Growth Curve
2.4. Thermal and pH Stability of Phage BUCT775
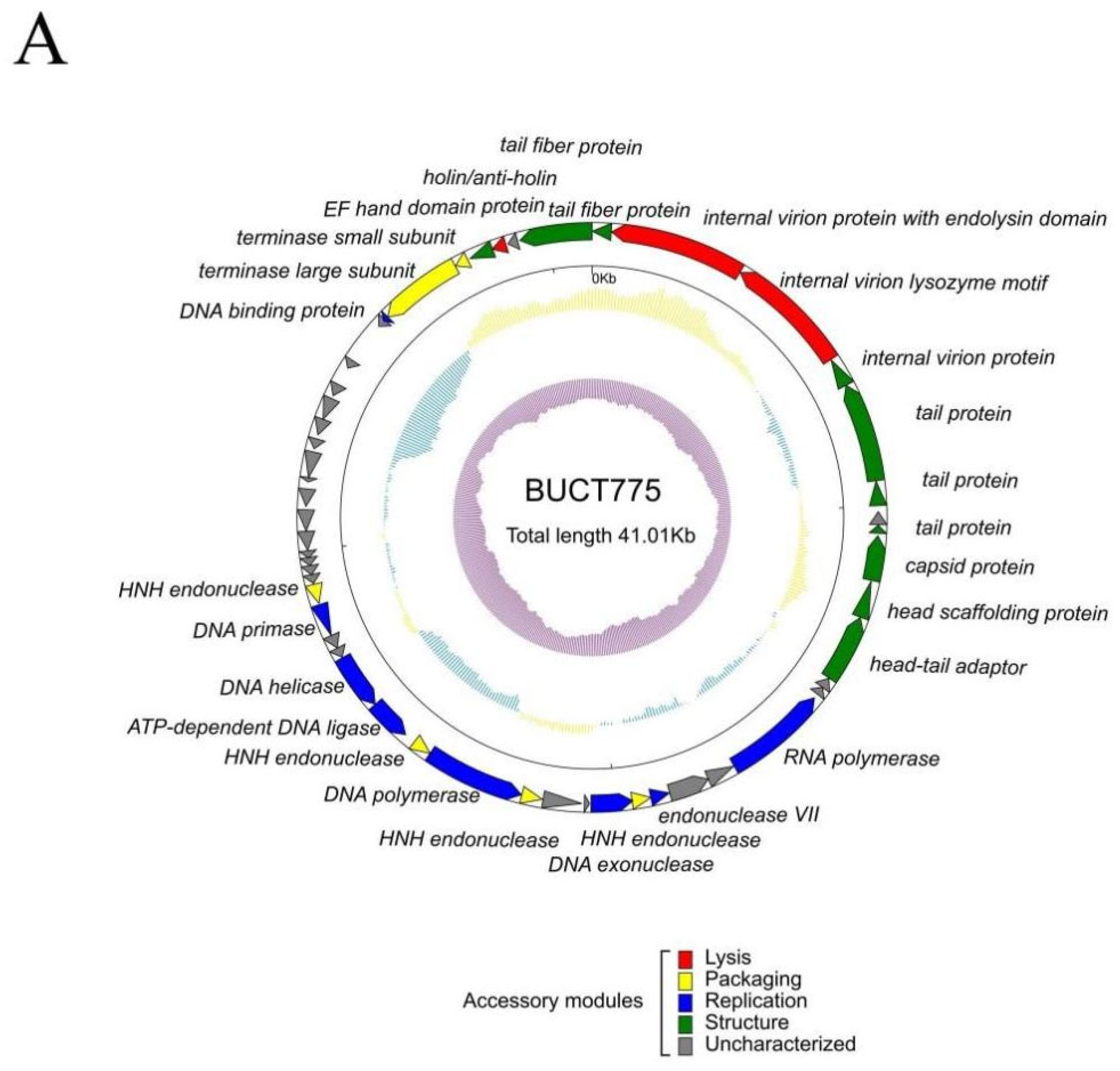
2.5. Genomic and Phylogenetic Analysis of Phage BUCT775
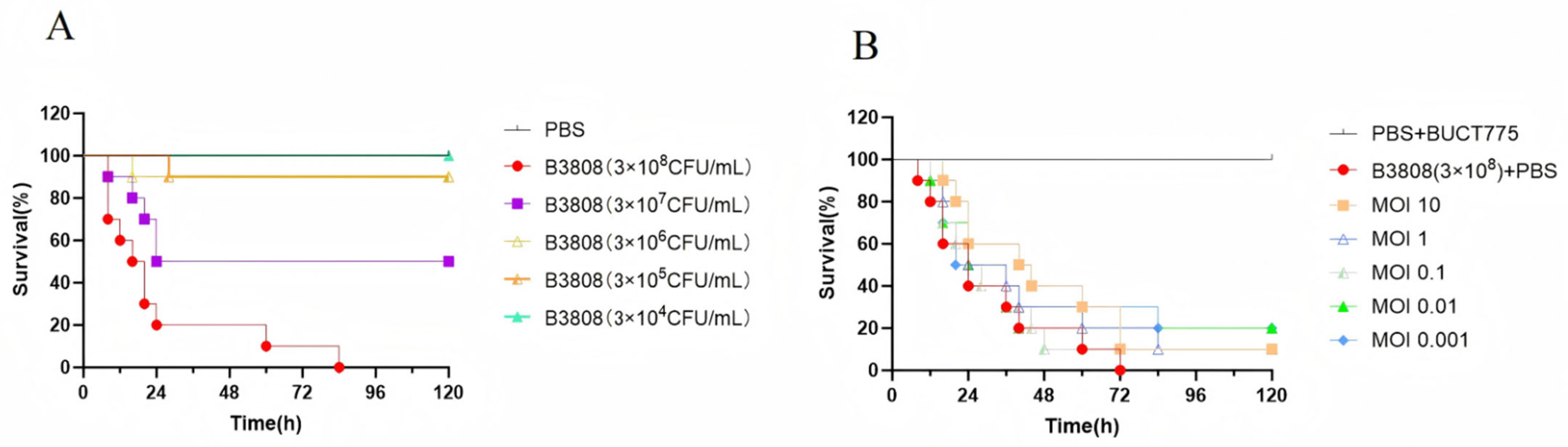
2.6. Antibacterial Effects of Phage BUCT775 In Vivo
2.7. Efficacy of Phage BUCT775 in Eliminating Environmental Bacteria
3. Materials and Methods
3.1. Bacterial Strains and Culture Conditions
3.2. Phage Isolation and Purification
3.3. Transmission Electron Microscopy
3.4. Determination of Phage Host Range
3.5. Optimal Multiplicity of Infection (MOI)
3.6. One-Step Growth Curve
3.7. Thermal and pH Sensitivity
3.8. Genomic Sequencing and Bioinformatic Analysis of the Phage
3.9. Galleria mellonella Model for In Vivo Phage Therapy
3.10. Disinfection Assay Using Phage BUCT775
3.11. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Giamarellou, H.; Antoniadou, A.; Kanellakopoulou, K. Acinetobacter baumannii: A Universal Threat to Public Health? Int. J. Antimicrob. Agents 2008, 32, 106–119. [Google Scholar] [CrossRef]
- Vazquez-Lopez, R.; Solano-Galvez, S.G.; Vignon-Whaley, J.J.J.; Vaamonde, J.A.A.; Alonzo, L.A.P.; Resendiz, A.R.; Alvarez, M.M.; Lopez, E.N.V.; Franyuti-Kelly, G.; Alvarez-Hernandez, D.A.; et al. Acinetobacter baumannii Resistance: A Real Challenge for Clinicians. Antibiotics 2020, 9, 205. [Google Scholar] [CrossRef]
- Tu, Q.; Pu, M.; Li, Y.; Wang, Y.; Li, M.; Song, L.; Li, M.; An, X.; Fan, H.; Tong, Y. Acinetobacter baumannii Phages: Past, Present and Future. Viruses 2023, 15, 673. [Google Scholar] [CrossRef] [PubMed]
- Wong, D.; Nielsen, T.B.; Bonomo, R.A.; Pantapalangkoor, P.; Luna, B.; Spellberg, B. Clinical and Pathophysiological Overview of Acinetobacter Infections: A Century of Challenges. Clin. Microbiol. Rev. 2017, 30, 409–447. [Google Scholar] [CrossRef]
- Antunes, L.C.; Visca, P.; Towner, K.J. Acinetobacter baumannii: Evolution of a Global Pathogen. Pathog. Dis. 2014, 71, 292–301. [Google Scholar] [CrossRef]
- Fournier, P.E.; Richet, H.; Weinstein, R.A. The Epidemiology and Control of Acinetobacter baumannii in Health Care Facilities. Clin. Infect. Dis. 2006, 42, 692–699. [Google Scholar] [CrossRef] [PubMed]
- Longo, B.; Annalisa, P.; Ida, L.; Agapito, T.; Fiorella, D.S.; Stella, G.; Paola, P.; Monica, M.; Anna Maria, D.; Italo, V.; et al. Molecular Findings and Antibiotic-Resistance in an Outbreak of Acinetobacter baumannii in an Intensive Care Unit. Ann. Dell’istituto Super. Di Sanita 2007, 43, 83–88. [Google Scholar]
- Ababneh, Q.; Aldaken, N.; Jaradat, Z.; Al-Rousan, E.; Inaya, Z.; Alsaleh, D.; Alawneh, D.; Al Sbei, S.; Saadoun, I. Predominance of Extensively-Drug Resistant Acinetobacter baumannii Carrying Bla Oxa-23 in Jordanian Patients Admitted to the Intensive Care Units. PLoS ONE 2025, 20, e0317798. [Google Scholar] [CrossRef]
- Lucidi, M.; Capecchi, G.; Spagnoli, C.; Basile, A.; Artuso, I.; Persichetti, L.; Fardelli, E.; Capellini, G.; Visaggio, D.; Imperi, F.; et al. The Response to Desiccation in Acinetobacter baumannii. Virulence 2025, 16, 2490209. [Google Scholar] [CrossRef]
- Shi, J.; Cheng, J.; Liu, S.; Zhu, Y.; Zhu, M. Acinetobacter baumannii: An Evolving and Cunning Opponent. Front. Microbiol. 2024, 15, 1332108. [Google Scholar] [CrossRef]
- Geisinger, E.; Huo, W.; Hernandez-Bird, J.; Isberg, R.R. Acinetobacter baumannii: Envelope Determinants That Control Drug Resistance, Virulence, and Surface Variability. Annu. Rev. Microbiol. 2019, 73, 481–506. [Google Scholar] [CrossRef]
- Sarshar, M.; Behzadi, P.; Scribano, D.; Palamara, A.T.; Ambrosi, C. Acinetobacter baumannii: An Ancient Commensal with Weapons of a Pathogen. Pathogens 2021, 10, 387. [Google Scholar] [CrossRef]
- Harding, C.M.; Hennon, S.W.; Feldman, M.F. Uncovering the Mechanisms of Acinetobacter baumannii Virulence. Nat. Rev. Microbiol. 2018, 16, 91–102. [Google Scholar] [CrossRef]
- Li, S.K.; Jiang, G.L.; Wang, S.K.; Wang, M.; Wu, Y.L.; Zhang, J.Z.; Liu, X.; Zhong, L.; Zhou, M.; Xie, S.C.; et al. Emergence and Global Spread of a Dominant Multidrug-Resistant Clade within Acinetobacter baumannii. Nat. Commun. 2025, 16, 2787. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.; McClean, S. Mapping Global Prevalence of Acinetobacter Baumannii and Recent Vaccine Development to Tackle It. Vaccines 2021, 9, 570. [Google Scholar] [CrossRef] [PubMed]
- Mohd Sazlly Lim, S.; Abidin, A.Z.; Liew, S.M.; Roberts, J.A.; Sime, F.B. The Global Prevalence of Multidrug-Resistance among Acinetobacter baumannii Causing Hospital-Acquired and Ventilator-Associated Pneumonia and Its Associated Mortality: A Systematic Review and Meta-Analysis. J. Infect. 2019, 79, 593–600. [Google Scholar] [CrossRef] [PubMed]
- WHO. Who Bacterial Priority Pathogens List, 2024: Bacterial Pathogens of Public Health Importance to Guide Research, Development and Strategies to Prevent and Control Antimicrobial Resistance; World Health Organization: Geneva, Switzerland, 2024. [Google Scholar]
- Zhen, X.; Lundborg, C.S.; Sun, X.; Gu, S.; Dong, H. Clinical and Economic Burden of Carbapenem-Resistant Infection or Colonization Caused by Klebsiella Pneumoniae, Pseudomonas Aeruginosa, Acinetobacter baumannii: A Multicenter Study in China. Antibiotics 2020, 9, 514. [Google Scholar] [CrossRef]
- Hendrix, R.W. Bacteriophages: Evolution of the Majority. Theor. Popul. Biol. 2002, 61, 471–480. [Google Scholar] [CrossRef]
- Strathdee, S.A.; Hatfull, G.F.; Mutalik, V.K.; Schooley, R.T. Phage Therapy: From Biological Mechanisms to Future Directions. Cell 2023, 186, 17–31. [Google Scholar] [CrossRef]
- Gordillo Altamirano, F.L.; Barr, J.J. Phage Therapy in the Postantibiotic Era. Clin. Microbiol. Rev. 2019, 32, e00066-18. [Google Scholar] [CrossRef]
- Chen, L.K.; Chang, J.C.; Chu, H.T.; Chen, Y.T.; Jiang, H.L.; Wang, L.S.; Teh, S.H.; Yang, H.H.; Chen, D.S.; Li, Y.Z.; et al. Preoptimized Phage Cocktail for Use in Aerosols against Nosocomial Transmission of Carbapenem-Resistant Acinetobacter Baumannii: A 3-Year Prospective Intervention Study. Ecotoxicol. Environ. Saf. 2022, 236, 113476. [Google Scholar] [CrossRef]
- Chen, Z.; Yang, Y.; Li, G.; Huang, Y.; Luo, Y.; Le, S. Effective Elimination of Bacteria on Hard Surfaces by the Combined Use of Bacteriophages and Chemical Disinfectants. Microbiol. Spectr. 2024, 12, e03797-23. [Google Scholar] [CrossRef]
- Sisakhtpour, B.; Mirzaei, A.; Karbasizadeh, V.; Hosseini, N.; Shabani, M.; Moghim, S. The Characteristic and Potential Therapeutic Effect of Isolated Multidrug-Resistant Acinetobacter baumannii Lytic Phage. Ann. Clin. Microbiol. Antimicrob. 2022, 21, 1. [Google Scholar] [CrossRef]
- Kitti, T.; Thummeepak, R.; Thanwisai, A.; Boonyodying, K.; Kunthalert, D.; Ritvirool, P.; Sitthisak, S. Characterization and Detection of Endolysin Gene from Three Acinetobacter Baumannii Bacteriophages Isolated from Sewage Water. Indian J. Microbiol. 2014, 54, 383–388. [Google Scholar] [CrossRef] [PubMed]
- Mardiana, M.; Teh, S.H.; Lin, L.C.; Lin, N.T. Isolation and Characterization of a Novel Siphoviridae Phage, Vb_Abas_Tcup2199, Infecting Multidrug-Resistant Acinetobacter baumannii. Viruses 2022, 14, 1240. [Google Scholar] [CrossRef]
- Kering, K.K.; Zhang, X.; Nyaruaba, R.; Yu, J.; Wei, H. Application of Adaptive Evolution to Improve the Stability of Bacteriophages During Storage. Viruses 2020, 12, 423. [Google Scholar] [CrossRef]
- Tan, Y.; Su, J.; Luo, D.; Liang, B.; Liu, S.; Zeng, H. Isolation and Genome-Wide Analysis of the Novel Acinetobacter Baumannii Bacteriophage Vb_Abam_Ab3p2. Arch. Virol. 2024, 169, 66. [Google Scholar] [CrossRef] [PubMed]
- Cahill, J.; Young, R. Chapter Two—Phage Lysis: Multiple Genes for Multiple Barriers. In Advances in Virus Research; Margaret, K., Thomas, C.M., Marilyn, J.R., Eds.; Academic Press: Cambridge, MA, USA, 2019; pp. 33–70. [Google Scholar]
- Han, P.; Pu, M.; Li, Y.; Fan, H.; Tong, Y. Characterization of Bacteriophage Buct631 Lytic for K1 Klebsiella Pneumoniae and Its Therapeutic Efficacy in Galleria Mellonella Larvae. Virol. Sin. 2023, 38, 801–812. [Google Scholar] [CrossRef]
- Jeon, J.; Park, J.H.; Yong, D. Efficacy of Bacteriophage Treatment against Carbapenem-Resistant Acinetobacter Baumannii in Galleria Mellonella Larvae and a Mouse Model of Acute Pneumonia. BMC Microbiol. 2019, 19, 70. [Google Scholar] [CrossRef] [PubMed]
- Anderson, D.J.; Chen, L.F.; Weber, D.J.; Moehring, R.W.; Lewis, S.S.; Triplett, P.F.; Blocker, M.; Becherer, P.; Schwab, J.C.; Knelson, L.P.; et al. Enhanced Terminal Room Disinfection and Acquisition and Infection Caused by Multidrug-Resistant Organisms and Clostridium difficile (the Benefits of Enhanced Terminal Room Disinfection Study): A Cluster-Randomised, Multicentre, Crossover Study. Lancet 2017, 389, 805–814. [Google Scholar] [CrossRef]
- Schwarz, C.; Mathieu, J.; Laverde Gomez, J.A.; Yu, P.; Alvarez, P.J. Renaissance for Phage-Based Bacterial Control. Environ. Sci. Technol. 2022, 56, 4691–4701. [Google Scholar] [CrossRef]
- Chen, L.K.; Liu, Y.L.; Hu, A.; Chang, K.C.; Lin, N.T.; Lai, M.J.; Tseng, C.C. Potential of Bacteriophage Φab2 as an Environmental Biocontrol Agent for the Control of Multidrug-Resistant Acinetobacter baumannii. BMC Microbiol. 2013, 13, 154. [Google Scholar] [CrossRef]
- Hegarty, B. Making Waves: Intelligent Phage Cocktail Design, a Pathway to Precise Microbial Control in Water Systems. Water Res. 2025, 268, 122594. [Google Scholar] [CrossRef]
- Kasman, L.M.; Kasman, A.; Westwater, C.; Dolan, J.; Schmidt, M.G.; Norris, J.S. Overcoming the Phage Replication Threshold: A Mathematical Model with Implications for Phage Therapy. J. Virol. 2002, 76, 5557–5564. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. Spades: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The Rast Server: Rapid Annotations Using Subsystems Technology. BMC Genom. 2008, 9, 75. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, Y.; Yoshida, T.; Kuronishi, M.; Uehara, H.; Ogata, H.; Goto, S. Viptree: The Viral Proteomic Tree Server. Bioinformatics 2017, 33, 2379–2380. [Google Scholar] [CrossRef] [PubMed]
- Tsai, C.J.Y.; Loh, J.M.S.; Proft, T. Galleria mellonella infection Models for the Study of Bacterial Diseases and for Antimicrobial Drug Testing. Virulence 2016, 7, 214–229. [Google Scholar] [CrossRef] [PubMed]
- Qian, X.; Li, Y.; Zeng, H.; Yin, H.; Wang, Y.; Huang, H.; Gong, Q.; Li, D.; Xue, F.; Tang, F.; et al. Evaluation of the Bactericidal Efficacy of EAEC Phages PNJ1809-11 and PNJ1809-13 as Environmental Disinfectants. Acta Microbiol. Sin. 2021, 61, 2018–2034. (In Chinese) [Google Scholar]



| Species | Strains | ST Type (Pasteur) | Susceptibility | Efficiency of Plating (EOP) | Origin |
|---|---|---|---|---|---|
| Acinetobacter baumannii | 3808 | ST2 | + | Host strain | The Affiliated Hospital of Qingdao University |
| T1319 | ST2 | − | 0 | Peking University Third Hospital | |
| T2297 | ST40 | − | 0 | Peking University Third Hospital | |
| T2321 | ST2 | − | 0 | Peking University Third Hospital | |
| T2347 | ST2 | − | 0 | Peking University Third Hospital | |
| T2404 | ST2 | − | 0 | Peking University Third Hospital | |
| T2634 | ST2 | + | 0.1 | Peking University Third Hospital | |
| T2673 | ST2 | − | 0 | Peking University Third Hospital | |
| T2747 | ST2 | − | 0 | Peking University Third Hospital | |
| T2784 | ST40 | − | 0 | Peking University Third Hospital | |
| T3153 | ST40 | − | 0 | Peking University Third Hospital | |
| T3311 | ST113 | − | 0 | Peking University Third Hospital | |
| T3640 | ST2 | − | 0 | Peking University Third Hospital | |
| T3666 | ST2 | + | 1 | Peking University Third Hospital | |
| T3789 | ST2 | + | 1 | Peking University Third Hospital |
| ORF | Strand | Start | Stop | Length (AA) | Predicted Protein Function | Best-Match BLASTp Result | Query Cover | E-Values | Identity | Accession |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | - | 436 | 2 | 144 | tail fiber protein | Acinetobacter phage vB_AbaP_B4 | 100% | 4 × 10−94 | 99.31% | WNO29457.1 |
| 2 | - | 3541 | 443 | 1032 | internal virion protein with endolysin domain | Acinetobacter phage Abp1 | 100% | 0.0 | 99.81% | YP_008058238.1 |
| 3 | - | 6436 | 3551 | 961 | internal virion lysozyme motif | Acinetobacter phage Abp1 | 100% | 0.0 | 99.79% | YP_008058237.1 |
| 4 | - | 7120 | 6449 | 223 | internal virion protein | Acinetobacter phage Abp1 | 100% | 8 × 10−159 | 100.00% | YP_008058236.1 |
| 5 | - | 9411 | 7120 | 763 | tail protein | Acinetobacter phage SWH-Ab-1 | 100% | 0.0 | 99.21% | YP_009949054.1 |
| 6 | - | 9980 | 9420 | 186 | tail protein | Acinetobacter phage Abp1 | 100% | 5 × 10−133 | 100% | YP_008058234.1 |
| 8 | - | 10,611 | 10,426 | 61 | tail protein | Acinetobacter phage Abp1 | 100% | 1 × 10−32 | 100.00% | YP_008058232.1 |
| 9 | - | 11,698 | 10,667 | 343 | capsid protein | Acinetobacter phage APK127v | 100.00% | 0.0 | 99.42% | URQ05181.1 |
| 10 | - | 12,574 | 11,714 | 286 | head scaffolding protein | Acinetobacter phage Abp1 | 100.00% | 0.0 | 100.00% | YP_008058230.1 |
| 11 | - | 14,139 | 12,583 | 518 | head–tail adaptor | Acinetobacter phage Abp1 | 100% | 0.0 | 100% | YP_008058229.1 |
| 14 | - | 17,116 | 14,699 | 805 | RNA polymerase | Acinetobacter phage Abp1 | 100% | 0.0 | 99.88% | YP_008058226.1 |
| 17 | - | 19,154 | 18,714 | 146 | endonuclease VII | Acinetobacter phage Abp1 | 100% | 5 × 10−105 | 100% | YP_008058223.1 |
| 18 | - | 19,588 | 19,151 | 145 | HNH endonuclease | Acinetobacter phage Abp1 | 100% | 3 × 10−104 | 100% | YP_008058222.1 |
| 19 | - | 20,525 | 19,569 | 318 | DNA exonuclease | Acinetobacter phage SWH-Ab-3 | 100% | 0.0 | 100.00% | YP_009949090.1 |
| 22 | - | 22,121 | 21,642 | 159 | HNH endonuclease | Acinetobacter phage SWH-Ab-1 | 100% | 8 × 10−115 | 98.74% | YP_009949038.1 |
| 23 | - | 24,430 | 22,130 | 766 | DNA polymerase | Acinetobacter phage SWH-Ab-1 | 100% | 0.0 | 99.87% | YP_009949037.1 |
| 24 | - | 24,873 | 24,427 | 148 | HNH endonuclease | Acinetobacter phage phiAB1 | 100% | 4 × 10−104 | 100% | YP_009189356.1 |
| 25 | - | 26,111 | 25,131 | 326 | ATP-dependent DNA ligase | Acinetobacter phage SWH-Ab-1 | 100% | 0.0 | 99.39% | YP_009949035.1 |
| 26 | - | 27,412 | 26,114 | 432 | DNA helicase | Acinetobacter phage SWH-Ab-1 | 100% | 0.0 | 100% | YP_009949034.1 |
| 29 | - | 28,733 | 27,978 | 251 | DNA primase | Acinetobacter phage Abgy2021-6-2 | 100% | 0.0 | 100% | WPF70317.1 |
| 30 | - | 29,212 | 28,763 | 149 | HNH endonuclease | Acinetobacter phage SWH-Ab-3 | 100% | 5 × 10−107 | 100% | YP_009949077.1 |
| 46 | - | 35,819 | 35,685 | 44 | DNA binding protein | Acinetobacter phage IME-200 | 100% | 2 × 10−21 | 100.00% | YP_009216494.1 |
| 47 | - | 37,753 | 35,816 | 645 | terminase large subunit | Acinetobacter phage Abp1 | 100% | 0.0 | 100.00% | YP_008058244.1 |
| 48 | - | 38,071 | 37,763 | 102 | terminase small subunit | Acinetobacter phage Abp1 | 100% | 2 × 10−66 | 100.00% | YP_008058243.1 |
| 49 | - | 38,688 | 38,131 | 185 | EF hand domain protein | Acinetobacter phage Abp1 | 100% | 8 × 10−131 | 100.00% | YP_008058242.1 |
| 50 | - | 39,010 | 38,675 | 111 | holin/anti-holin | Acinetobacter phage Abp1 | 100% | 3 × 10−73 | 100.00% | YP_008058241.1 |
| 52 | - | 41,003 | 39,342 | 553 | tail fiber protein | Acinetobacter phage SWH-Ab-3 | 100% | 0.0 | 99.83% | YP_009949108.1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Y.; Huang, Y.; Zhu, D.; Zhang, L.; Zhang, J.; Tong, Y.; Li, M. Characterization and Complete Genomic Analysis of a Novel Bacteriophage BUCT775 for Acinetobacter baumannii and Its Elimination Efficiency in the Environment. Int. J. Mol. Sci. 2025, 26, 7279. https://doi.org/10.3390/ijms26157279
Liu Y, Huang Y, Zhu D, Zhang L, Zhang J, Tong Y, Li M. Characterization and Complete Genomic Analysis of a Novel Bacteriophage BUCT775 for Acinetobacter baumannii and Its Elimination Efficiency in the Environment. International Journal of Molecular Sciences. 2025; 26(15):7279. https://doi.org/10.3390/ijms26157279
Chicago/Turabian StyleLiu, Yuxuan, Yunfei Huang, Dongxiang Zhu, Lefei Zhang, Jianwei Zhang, Yigang Tong, and Mengzhe Li. 2025. "Characterization and Complete Genomic Analysis of a Novel Bacteriophage BUCT775 for Acinetobacter baumannii and Its Elimination Efficiency in the Environment" International Journal of Molecular Sciences 26, no. 15: 7279. https://doi.org/10.3390/ijms26157279
APA StyleLiu, Y., Huang, Y., Zhu, D., Zhang, L., Zhang, J., Tong, Y., & Li, M. (2025). Characterization and Complete Genomic Analysis of a Novel Bacteriophage BUCT775 for Acinetobacter baumannii and Its Elimination Efficiency in the Environment. International Journal of Molecular Sciences, 26(15), 7279. https://doi.org/10.3390/ijms26157279






